Abstract
Aim: SARS-associated coronavirus (SARS-CoV) has been confirmed as the pathogen for severe acute respiratory syndrome (SARS). The aim of our study was to construct a sensitive and specific real-time quantitative fluorescent (QF) reverse transcriptase (RT)-PCR method for the detection of SARS-CoV RNA.
Methods: Stored blood specimens from 44 patients with confirmed SARS were used along with blood samples from two sets of controls, 30 healthy volunteers who had no contact with SARS patients, and 30 healthy doctors and nurses who had contact with SARS patients but were without symptoms of SARS. Two pairs of primers were synthesized by the Shanghai Sangon Company according to SARS-CoV BJ01 strain sequence (AY278488), and then a pair of primers were designed and compared with a pair of primers published by WHO.
Results: Using serial dilutions of SARS-CoV, the 44 blood samples from SARS patients specimens were tested. Using a 0.01% dilution of SARS-CoV, all 44 clinical samples tested positive in our assay. In comparison, using a 0.1% dilution of SARS-CoV, 26 of the 44 samples tested positive using the WHO primers. In the QF-RT-PCR assay, there was a linear amplification from 100 copies to 108 copies of the control RNA per RT-PCR and at least 10 copies, and sometimes even 1 copy, of target RNA tested positive in our assay.
Conclusion: The primer we developed is sufficiently sensitive and specific to diagnose symptomatic SARS-CoV infections and for monitoring virus load.
Keywords: Severe Acute Respiratory Syndrome, Severe Acute Respiratory Syndrome, Wide Linear Range, Atypical Pneumonia, Severe Acute Respiratory Syndrome Patient
By the beginning of November 2003, the epidemic of severe atypical pneumonia, designated severe acute respiratory syndrome (SARS) by the World Health Organization (WHO) and first observed in the Chinese province of Guangdong in November 2002, had infected 8439 people and caused 812 deaths in 33 countries.[1,2] The SARS-associated coronavirus (CoV) was confirmed as the pathogen.[3–8] In the absence of effective drugs or a vaccine against SARS, controlling this disease relied on the rapid identification of cases and the appropriate management of individuals who had come into close contact with them. However, this disease is characterized by fever, nonproductive cough, dyspnea, chest pain, lung infiltrates and fibrosis, and a decreased lymphocyte count,[9] and it has similar symptoms to other acute febrile illnesses, such as influenza and others atypical pneumonia.
It has been difficult to differentiate between SARS and other acute febrile illnesses in clinical diagnosis; therefore, laboratory methods for SARS diagnosis are very important. Until now, laboratory tests for SARS have primarily: (i) been PCR-based and virus isolation to detect viral RNA in patient secretions/excretions; and (ii) antibody-based to detect their immune responses, such as the titers of antibodies in the blood. However, these methods are hampered by the need for virus isolation to take place under biosafety level 3 or 4 condition; serology tests may not become positive for one or more weeks after the onset of symptoms.
Developing an rapid early diagnostic method has been a high priority for the monitoring and control of this disease. Researchers in several countries have been working towards developing fast and accurate laboratory diagnostic tests for SARS-CoV.[10] In this report, we describe a sensitive and specific quantitative fluorescent (QF)-RT-PCR assay for the detection of SARS-CoV RNA.
Materials and Methods
Serial Dilutions of Acute Respiratory Syndrome (SARS)-Associated Coronavirus (CoV)
A confluent layer of Vero E6 cell was infected with the BJ01 strain of SARS-CoV. After 72 hours, the cells were harvested and inactivated at 65°C for 2 hours. Inactivated cell cultures were diluted in 10-fold dilutions with healthy blood (10%, 1%, 0.1%, 0.01%, 0.001%, and 0.0001%).
Clinical Specimens
We used stored blood specimens from 44 patients (taken 1–3 weeks after the onset of disease) who fulfilled the clinical WHO case definition of SARS, the diagnosis was subsequently confirmed by seroconversion and positive SARS-CoV isolation.[11] Control blood samples were collected from two additional groups: 30 healthy volunteers who had no contact with SARS patients, and 30 healthy doctors and nurses, who had contact history in a hospital but were without symptoms of SARS 14 days after sampling. Whole blood samples were collected in a vacuum container with EDTA and transferred to the laboratory at 4°C, RNA was then extracted immediately.
Standard SARS-CoV Panel
SARS-CoV virions of known concentration (SARS-CoV Quality Assurance Laboratory, National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China) were serially diluted in SARS-CoV-seronegative human plasma to predicted final RNA concentrations of 3.6 × 103, 4.8 × 103, 6 × 106, 6 × 105, 6 × 104, 6 × 103, 1.2 × 103, 1.2 × 102, 6 × 101, 3 × 101 copies/mL. The dilutions were performed once, and sufficient numbers of aliquots from each dilution were stored at -80°C.
Primers
Two pairs of primers were synthesized by the Shanghai Sangon Company (Shanghai, China) according to SARS-CoV BJ01 strain sequence (AY278488) [table I].
Table I.

Two pairs of primers used for acute respiratory syndrome (SARS)-associated coronavirus (CoV) RNA detection
Preparation of Standards for SARS-CoV RNA Quantification
Plasmid pGEM-T (Promega, Madison, WI, USA), which contains a 450 bp fragment between 26092 and 26541 of SARS-CoV (BJ01 strain) downstream of the T7 promoter, was linearized with Pst I, purified with a PCR purification kit (Shanghai Biotech, Shanghai, China), and transcribed with T7 RNA polymerase using the RiboMax™ Express Large Scale RNA production system (Promega, Madison, WI, USA). The template DNA was degraded with 5U of RNase-free Dnase I, and the RNA transcripts were purified twice with an RNeasy kit (QIAGEN GmbH, Hilden, Germany). The RNA was quantified spectrophotometrically at 260nm, diluted to 107 copies/µL, divided into aliquots, and stored at -80°C. Diluted SARS-CoV transcripts (107 to 0.01 copies/μL) were used to generate a standard curve for each single-copy assay.
RNA Extraction and Reverse Transcription
Total RNA was extracted using the QIAamp RNA Blood Mini Kit (52304; Qiagen, GmbH, Hilden, Germany) according to the manufacturer’s instructions. For blood samples, 1mL of sample was mixed with 5mL buffer EL to lyse red cell and then centrifuged at 400×g for 10 minutes at 4°C. The supernatant was removed and the deposits were collected for RNA extraction.
For the other samples in this study, 100μL volume was directly used for RNA extraction. The RNA was re-dissolved in 30μL of diethyl pirocarbonate (DEPC)-treated water containing 1U DNase I. After incubation at 37°C for 15 minutes followed by inactivation at 95°C for 10 minutes, RNA was used immediately or stored at −80°C. All steps were performed in a biosafety level 2 laboratory.
Synthesis of cDNA was undertaken by reverse transcription from 9μL of RNA extracted at 45°C for 45 minutes followed by incubation at 95°C for 3 minutes, in 20μL solution containing 50 mmol/L Tris-HCl (pH 8.3), 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L DTT, 100ng of hexamer random primers (Shanghai Sangong Company, Shanghai, China), 200U of Superscript II (18064-014; Invitrogen, Carlsbad, CA, USA), 25U of RNasin (N2511; Promega, Madison, WI, USA), and 0.5 mmol/L dNTPs.
PCR
PCR was carried out in a 25µL mixture containing 4µL complementary DNA (cDNA), 10 mmol/L Tris-HCl (pH 8.4), 50 mmol/L KCl, 2.5 mmol/L MgCl2, 100 µmol/L dNTPs, 1U Taq Pol (M1661, Promega, Madison, WI, USA), 0.25 µmol/L forward primer, and 0.25 µmol/L of reverse primer. 35 cycles of amplification (94°C for 30 seconds, 54°C for 30 seconds, and then 72°C for 30 seconds) were undertaken after denaturing at 95°C for 4 minutes. Amplified products were detected by agarose gel electrophoresis and sequence identification was undertaken for those RT-PCR products which were unmatched in amplification by two pairs of primers. Both forward and reverse sequencing were undertaken on an ABI PRISM® 377 DNA Sequencer (Applied Biosystems, Foster City, CA, USA) in both directions.
Quantitative Fluorescent Reverse Transcriptase-PCR (QF-RT-PCR)
Serial dilutions of standard SARS-CoV RNA containing the target sequence, RNAs of standard SARS-CoV panel, RNAs of blood samples of 44 SARS patients, and RNAs of blood samples of 60 healthy individuals were tested using QF-RT-PCR. QF-RT-PCR was carried out in a 30µL mixture containing 10µL of RNA, 15µL 2U TaqMan® Universal PCR Master Mix (4304437; Applied Biosystems, Foster City, CA, USA), 0.25 µmol/L forward primer Af, 0.25µmol/L reverse primer Ar, and 0.25 µmol/L of probe (5′-FAM-AGAAGATCAGGAACTCCTTCAGA-TAMRA-3′) in a fluorometric PCR instrument (7900HT; Applied Biosystems, Foster City, CA, USA). 45 cycles of amplification (95°C for 15 seconds, 58°C for 1 minute; fluorescence was recorded at 58°C) were performed after denaturing at 95°C for 10 minutes and 42°C for 45 minutes.
Results
Comparison of Sensitivity and Specificity of Two Pairs of Primers
To compare the sensitivity of two pairs of primers, serial dilutions of SARS-CoV and the 44 positive clinical specimens in cell cultures were tested. 0.01% dilution of SARS-CoV and all clinical samples tested positive in our primers, but only 0.1% dilution of SARS-CoV and 26 of the 44 clinical samples tested positive in this WHO primers assay (figure 1). Those samples that only tested positive in our primers were sequenced and the sequence was shown to be identical to the published SARS-CoV sequence.[8] To determine the specificity, the 60 control samples were tested and all tests gave a negative result.
Fig. 1.
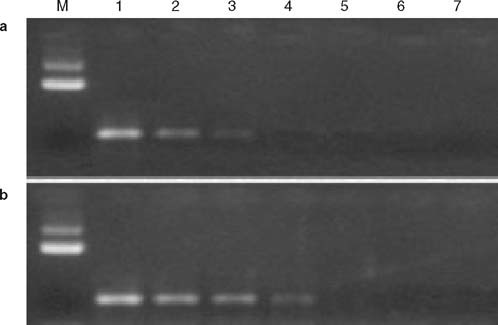
Electrophoretic analysis of reverse transcriptase (RT)-PCR products with 1.0% agarose gel. M is the DNA molecular markers, (a) PCR products amplified by WHO primers; and (b) the PCR products amplified our primers. 1–6 are the serial 10-fold dilutions of severe acute respiratory syndrome (SARS)-associated coronavirus (CoV) RNA (10%, 1%, 0.1%, 0.01%, 0.001%, and 0.0001%), 7 is diethyl pirocarbonate water.
Establishment of a Real-Time RT-PCR Assay for Rapid Detection of SARS-CoV
Sensitivity and Specificity Analyzes
To determine the sensitivity, a standard SARS-CoV panel and the 44 clinical samples were tested. All 44 clinical samples tested positive. In the standard SARS-CoV panel test, the sample of 60 copies/mL sometimes tested positive and the sample of 30 copies/mL always tested negative. To determine the specificity, 60 control samples were tested and all 60 samples tested negative.
Dynamic Range of QF-RT-PCR
To determine the dynamic range, serial dilutions of the standard SARS-CoV RNA containing the target sequence was tested. A wide linear range (beginning at 100 copies and extending through 108 copies of the control plasmid) was established and at least 10 copies, and sometimes even 1 copy, of target RNA tested positive in our assay (figure 2).
Fig. 2.
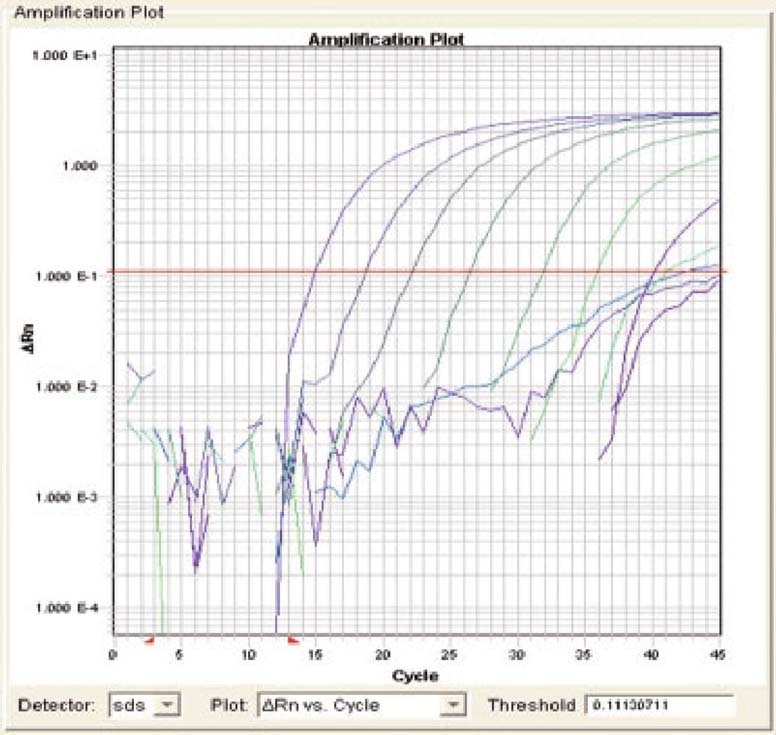
Sensitivity and dynamic range of quantitative fluorescent (QF) reverse transcriptase (RT)-PCR in detection severe acute respiratory syndrome (SARS)-associated coronavirus (CoV) RNA. Serial dilutions of standard SARS-CoV RNA containing the target sequence (107–10−2 copies/μL) were tested. A wide linear range (beginning at 100 copies and extending through 108 copies of the control plasmid per QF-RT-PCR) was established and at least 10 copies, and sometimes even 1 copy, of target RNA tested positive in this assay. The intercept of the magnitude of the fluorescence signal (PCR baseline subtracted curve fit relative fluorescence units [CF RFU]) with the horizontal threshold line represents the threshold cycle value for a given sample.
Accuracy of the QF-RT-PCR Assay
To demonstrate the accuracy of the QF-RT-PCR assay, QF-RT-PCR products of serial dilutions of standard SARS-CoV RNA containing the target sequence were also detected by PCR-ethidium bromide (EB) staining method. QF-RT-PCR with 10 copies has a very faint band (figure 3).
Fig. 3.
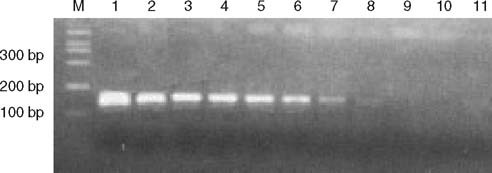
Electrophoretic analysis of reverse transcriptase (RT)-PCR products with 2.5% agarose gel. M is the DNA molecular markers, 1–10 are the serial 10-fold dilutions of standard severe acute respiratory syndrome (SARS)-associated coronavirus (CoV) RNA containing the target sequence with (107–10-2 copies/μL), 11 is diethyl pirocarbonate water.
Discussion
In this study we analyzed blood samples from 44 patients who had been confirmed to have SARS using seroconversion and positive SARS-CoV isolation by infecting Vero E6 cell. Many data showed the blood-borne SARS-CoV loads may be quite low even when viremia occurs. Alternatively, some of the patients may not even develop viremia as most of the viruses are propagating in epithelial tissues, such as the lung. Therefore, for this study we selected those samples that were positive by virus isolation.
The RT-PCR-based test using primer pairs published by WHO is a reliable identification method that was available before the publication of other more sensitive and specific methods.[10] However, the primers published by WHO for SARS-CoV are located in the open reading frame 1 (AY278488, 246-21466). Because SARS-CoV is a sense RNA virus, more target fragments should be near the 3′ terminal of virus genome.
We designed a pair of primers which was expected to amplify a 131 bp length covering the 26241-26371 region of the virus (AY278488) on the basis of published sequences of SARS-CoV. A pair of primers published by WHO was compared with our primers. To confirm the sensitivity of our primers, serial dilutions of SARS-CoV and 44 clinical samples were used. Compared to the WHO primers, our primers appear to have a greater sensitivity. PCR products of 18 samples tested negative in WHO primer assay but positive in our primers assay were sequenced, all matched correctly with SARS-CoV. To determine the specificity, 60 samples from 30 close contacts and 30 healthy medical personnel were tested and all tested negative. These results suggest that our primers have a better sensitivity and specificity.
To determine the sensitivity, specificity, and dynamic range of QF-RT-PCR, a standard SARS-CoV panel, 44 clinical samples, and 60 control samples and serial dilutions of standard SARS-CoV RNA containing the target sequence were subjected to the QF-RT-PCR assay. The results showed that this assay was highly sensitive, specific, and had a wide linear range.
Our primer was able to distinguish 10-fold differences in concentration over a range from 102 to 108, and at least 10 copies, and sometimes even 1 copy, of target RNA tested positive in this assay. In a standard SARS-CoV panel assay, not considering the efficiency of RNA extraction, the deadline of our QF-RT-PCR was between 2 copies/PCR and 4 copies/PCR.
To demonstrate the accuracy of the QF-RT-PCR assay, we compared it with the PCR-EB staining method. Compared with the PCR-EB method, QF-RT-PCR was more sensitive and less time-consuming. These results showed that this QF-RT-PCR assay was a sufficiently sensitive and specific method for diagnosing symptomatic SARS-CoV infections and for monitoring virus load.
Acknowledgements
The authors should like to thank Dr Juan Yu for his feedback on this study.
The authors received research funding from the National High Technology Research and Development Program of China (863 Program) [Grant number 2003AA208216], a program sponsored by the Ministry of Science and Technology, the People’s Republic of China.
Contributor Information
Weijun Chen, Email: chenweijun72@sohu.com.
Jian Wang, Email: wangjian@genomics.org.cn.
References
- 1.World Health Organization. Cumulative number of reported cases of severe acute respiratory syndrome (SARS) [online]. Available from URL: http://www.who.int/csr/sars/country/2003_07_04/en/ [Accessed 2003 Jul 4]
- 2.World Health Organization Severe acute respiratory syndrome (SARS) Wkly Epidemiol Rec. 2003;78:86. [Google Scholar]
- 3.Peiris JSM, Lai ST, Poon LLM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–66. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 6.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–9. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 7.Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 8.Qin E, Zhu Q, Wang J, et al. A complete sequence and comparative analysis of a SARS-associated virus (isolate BJ01) Chin Sci Bull. 2003;48:941–8. doi: 10.1007/BF03184203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization. Preliminary clinical description of severe acute respiratory syndrome [online]. Available from URL: http://www.who.int/csr/sars/clinical/en/ [Accessed 2003 Mar 21]
- 10.World Health Organization. PCR primers for SARS developed by WHO Network Laboratories [online]. Available from URL: http://www.who.int/csr/sars/primers/en [Accessed 2003 Apr 17]
- 11.World Health Organization case definitions for surveillance of severe acute respiratory syndrome (SARS) [online]. Available from URL: http://www.who.int/csr/sars/casedefinition/en/ [Accessed 2003 May 1]


