Abstract
The pharmacological activity of Acacia nilotica’s phytochemical constituents was confirmed with evidence-based studies, but the determination of exact targets that they bind and the mechanism of action were not done; consequently, we aim to identify the exact targets that are responsible for the pharmacological activity via the computational methods. Furthermore, we aim to predict the pharmacokinetics (ADME) properties and the safety profile in order to identify the best drug candidates. To achieve those goals, various computational methods were used including the ligand-based virtual screening and molecular docking. Moreover, pkCSM and SwissADME web servers were used for the prediction of pharmacokinetics and safety. The total number of the investigated compounds and targets was 25 and 61, respectively. According to the results, the pharmacological activity was attributed to the interaction with essential targets. Ellagic acid, Kaempferol, and Quercetin were the best A. nilotica’s phytochemical constituents that contribute to the therapeutic activities, were non-toxic as well as non-carcinogen. The administration of Ellagic acid, Kaempferol, and Quercetin as combined drug via the novel drug delivery systems will be a valuable therapeutic choice for the treatment of recent diseases attacking the public health including cancer, multidrug-resistant bacterial infections, diabetes mellitus, and chronic inflammatory systemic disease.
Keywords: A. nilotica, Ellagic acid, Kaempferol, Quercetin, Multiple mechanisms of action, ADMET and computer-aided drug discovery
Introduction
Acacia nilotica is a tropical and sub-tropical medicinal plant belonging to the Fabaceae family [1]. No doubt, medicinal plants play a vital role in drug discovery, since they are affluent with bioactive phytochemical constituents that are valuable in the treatment of various diseases, particularly those causing recent threats attacking the public health including cancer, multidrug-resistant bacterial infections, diabetes mellitus, and chronic inflammatory systemic diseases [2, 3].
The higher incidence of cancer and mortality rate [4], the emergence of bacterial resistance with the declining in the antibacterial research at several pharmaceutical companies [5], the huge prevalence and complications associated with diabetes mellitus [6] as well as the long-term suffering associated with the chronic inflammatory systemic diseases such as rheumatoid arthritis and multiple sclerosis [7] are leading forces that encourage us to participate in fighting against the probable threats. Such an issue is attained via the discovery and development of efficient innovative anticancer, antibacterial, antidiabetic, and anti-inflammatory drugs. Unfortunately, drug discovery is a time-consuming, costive, as well as difficult process [8, 9]; hence, it necessitated to involve sophisticated techniques in the drug discovery process in order to overcome those limitations. Recently, one of the promising sophisticated techniques is the computational tools (computer-aided drug design) that have a valuable impact in the discovery and development of newer drugs with a reduction in time and cost [8]. They include the ligand-based virtual screening that is based on the searching for the compounds having the highest probability in pharmacological activity [10] and molecular docking that relies on the energy-based scoring function to identify ligand-target complex lowest energy [11]. Moreover, they involve the software of pharmacokinetics, toxicity, and the drug-likeness prediction that work by many algorithms [12] including the graph-based signature [13]. Many studies concerning the application of the computational tools in the discovery of natural-derived drugs were conducted [14–17].
A. nilotica is opulent of many phytochemical constituents including tannins, alkaloids, terpenoids, and flavonoids. Many studies were conducted in it resulting in an evidence-based pharmacological data that revealed the potential pharmacological activities of the phytochemical compounds including anticancer, antibacterial, antidiabetic, anti-inflammatory, and other activity making the plant as a promising source for the development of innovative, safe, biodegradable drugs with great activity. The chemical structure of active A. nilotica’s phytochemical constituents was elucidated, the correlation between the responsible phytochemical constituents for treatment and the diseases were conducted [1], but the determination of exact targets that phytochemical constituents bind and the mechanism of action were not performed; consequently, based on established literature and studies, we aim to identify the exact targets that phytochemical constituents bind to exert the pharmacological activity by utilizing the computational methods as a tool for the study so as to understand the mechanism of action. Within the current drug design pipeline, drug target identification is a very important step in the understanding of the probable mechanism of action, increasing the confidence and reducing the attrition in clinical trials [18]. Furthermore, we aim to predict the pharmacokinetics (ADME) properties and safety profile with the intention of identifying the best drug candidates. The QSAR-based virtual screening is characterized by great and fast throughput with respectable hit rank [10]. Molecular docking is valuable to predict the stability of the ligand-target complex that reflects the biological activity [19]. The pharmacokinetics, toxicity, and drug-likeness prediction are helpful to identify the best drug candidates [12, 20]. To our knowledge, such a study was not conducted before.
Materials and Methods
Ligand-Based Virtual Screening
Ligands Preparation
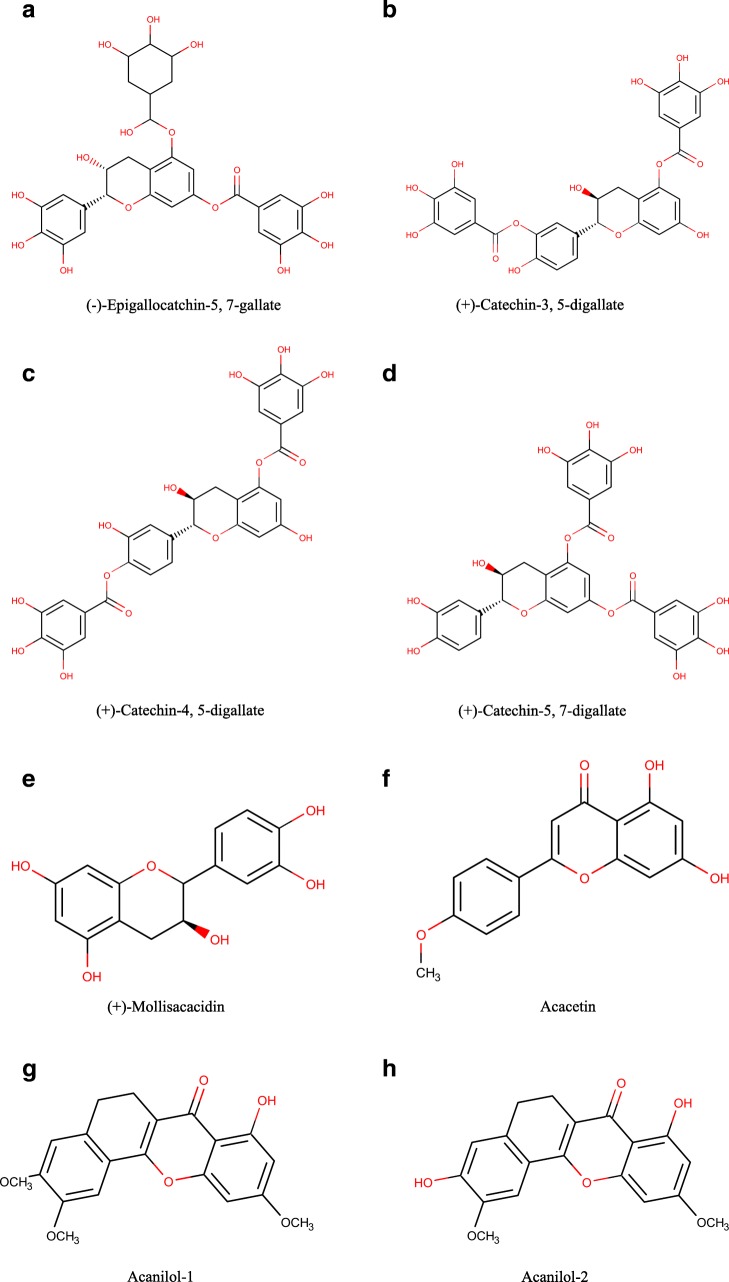
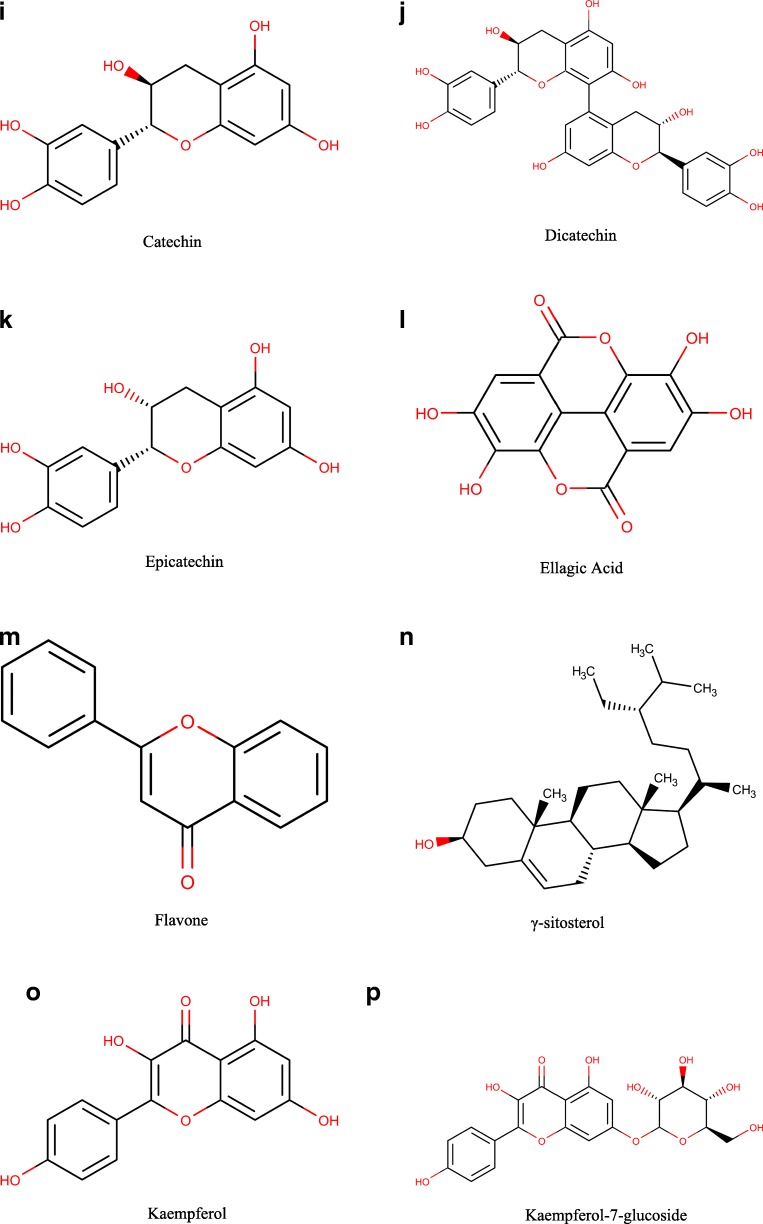
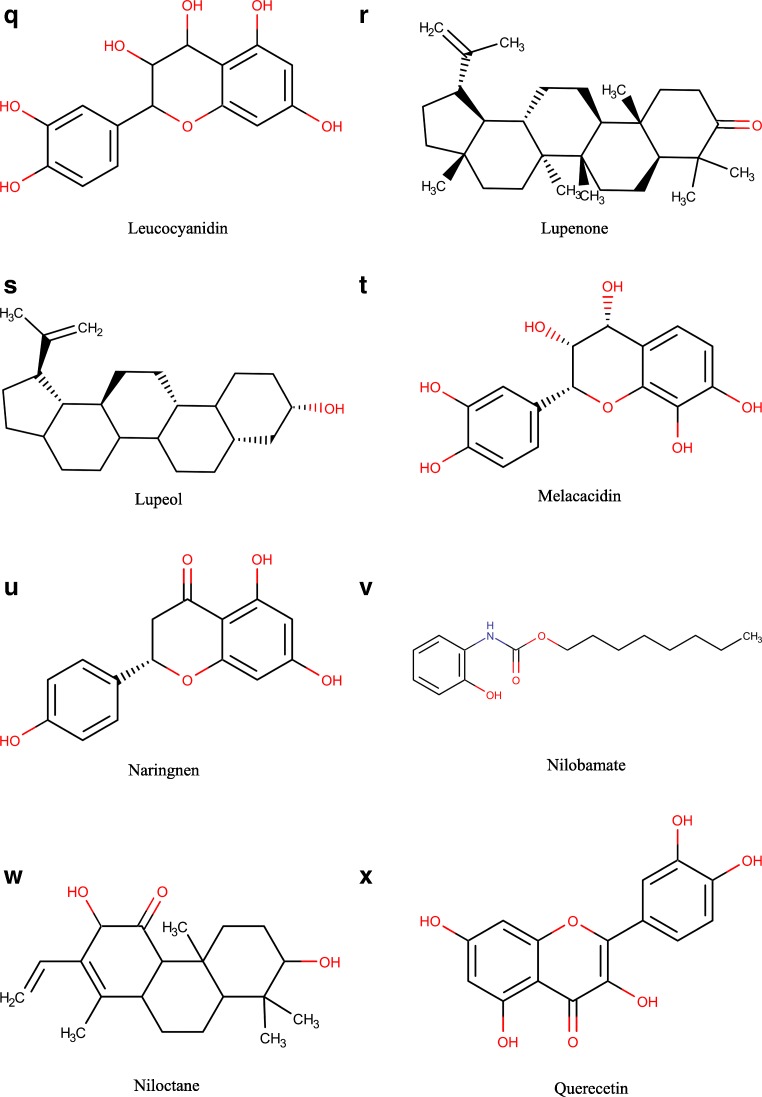
The chemical structure of the reported A. nilotica’s phytochemical constituents (25 compounds) [1] was drawn via Marven Sketch software version 18.5 [21] (Fig. 1). The 3D structure was generated in a mol2 format with Open Babel software [22], minimized and optimized with Cresset Flare software [23] at the accurate type calculation method.
Fig. 1.
a–x The 2D chemical structures of the reported A. nilotica’s phytochemical constituents [1]
Virtual Screening
The screening for the exact target that the phytochemical constituents bind was performed via Similarity Ensemble Search Tool [24] and TargetNet web servers [25]. The compound structures were submitted in smile format. The targets with higher probability score were selected for further validation via molecular docking study (61 targets). The linkage between predicted targets with the diseases was attained via UniProt [26], Pharos [27], and Therapeutic Target Databases [28]. The results are listed in Tables 1, 2, 3, 4, 5, and 6.
Table 1.
Ligand-based virtual screening and molecular docking results regarding the anticancer activity
| Target | Compounds | LBVS | Docking score | |
|---|---|---|---|---|
|
1- Anaplastic lymphoma kinase enzyme “It is pivotal in neural cells proliferation and survival” [26] |
4FNZ* | 2XB7* | ||
| 1- Quercetin | 1.0 | − 8.293 | − 8.31 | |
| a- Staurosporine | − | − 9.306 | 10.95 | |
| • A NZF 1501 | – | − 7.729 | – | |
| • A GUI 1501 | – | – | − 10.6 | |
|
2- Angiopoietin 1 receptor “It regulates the angiogenesis, adhesion, cell spreading and the maintenance of vascular quiescence” [28] |
3L8P* | 4X3J* | ||
| 1- Ellagic acid | 1.0 | − 8.403 | − 7.48 | |
| a- Cabozantinib | – | − 9.041 | − 7.36 | |
| • A 0CE 2207 | – | − 13.37 | – | |
| • A 3WR 1201 | – | – | − 8.56 | |
|
3- Aromatase enzyme “It is involved in the estrogen biosynthesis via conversion of androgens into estrogens” [26] |
3EQM* | 5JKW* | ||
| 1- (+)-Mollisacacidin | 1.0 | − 7.840 | − 8.48 | |
| 2- Acacetin | 0.74 | − 8.317 | − 8.44 | |
| 3- Catechin | 1.0 | − 8.795 | − 8.41 | |
| 4- Ellagic acid | 1.0 | − 8.664 | − 8.65 | |
| 5- Epicatechin | 1.0 | − 8.028 | − 8.45 | |
| 6- Flavone | 1.0 | − 6.450 | − 6.45 | |
| 7- γ-Sitosterol | 1.0 | − 11.51 | − 11.4 | |
| 8- Kaempferol | 1.0 | − 7.406 | − 7.78 | |
| 9- Leucocyanidin | 0.99 | − 11.77 | − 12.1 | |
| 10- Naringenin | 1.0 | − 8.354 | − 8.04 | |
| 11- Quercetin | 1.0 | − 7.579 | − 8.42 | |
| a- Anastrozole | – | − 7.142 | − 8.54 | |
| • A ASD 601 | – | − 9.699 | – | |
| • A TES 601 | – | – | − 9.889 | |
|
4- ATP binding cassette sub-family G member “Has a major role in cancer’s cells multi-drug resistance” [26] |
5NJ3* | 6FEQ* | ||
| 1- (+)-Catechin-3,5,-gallate | 0.95 | − 10.01 | − 11.17 | |
| 2- Acacetin | 1.0 | − 5.343 | − 7.633 | |
| 3- Chalconaringnen-4-O-beta glucopyranoside | 0.98 | − 7.067 | − 10.81 | |
| 4- Ellagic acid | 1.0 | − 5.789 | − 8.654 | |
| 5- Kaempferol | 1.0 | − 5.839 | − 8.052 | |
| 6- Leucocyanidin | 1.0 | − 7.817 | − 10.37 | |
| 7- Naringenin | 1.0 | − 6.023 | − 7.732 | |
| 8- Querecitin | 1.0 | − 5.979 | − 8.520 | |
| • A NAG 702 | – | − 5.220 | – | |
| • A D6T 1002 | – | – | − 14.81 | |
|
5- Aurora A and B kinase enzymes “Aurora A is involved in regulation of cell cycle progression” “Aurora B is a main regulator of mitosis” [16] |
5ORR* | 4B8L* | ||
| 1- Ellagic acid | 1.0 | − 8.327 | − 8.087 | |
| 2- Quercetin | 1.0 | − 6.627 | − 8.572 | |
| a- Axitinib | – | – | − 6.822 | |
| • A ADP 401 | – | − 9.409 | – | |
| • A A0P 1352 | – | – | − 9.203 | |
|
6- Bcl-2-related protein A1 “Pro and anti-apoptotic protein” [29] |
5WHH* | 2VM6* | ||
| 1- (+)-Mollisacacidin | 0.94 | − 7.448 | − 7.925 | |
| 2- Catechin | 0.95 | − 7.922 | − 8.905 | |
| 3- Catechinn-7-O-gallate | 0.95 | − 7.888 | − 8.374 | |
| 4- Ellagic acid | 1.0 | − 8.046 | − 7.130 | |
| 5- Epicatechin | 0.93 | − 7.340 | − 8.150 | |
| 6- Kaempferol | 1.0 | − 7.493 | − 7.061 | |
| 7- Leucocyanidin | 0.976 | − 8.616 | − 9.055 | |
| 8- Naringenin | 0.981 | − 7.774 | − 7.350 | |
| 9- Querecitin | 1.0 | − 8.516 | − 8.115 | |
| a- Venetoclax | – | − 9.339 | − 8.807 | |
|
7- Caspase 9 enzyme “Is an apoptotic initiator, acting as an important therapeutic target” [30] |
1JXQ* | 2AR9* | ||
| 1- (−)-Epigallocatchin-7-gallate | 1.0 | − 11.834 | − 13.18 | |
| 2- Ellagic acid | 0.998 | − 8.206 | − 9.408 | |
| 3- Flavone | 0.90 | − 6.693 | − 7.173 | |
| 4- Gallic acid | 1.0 | − 6.848 | − 7.562 | |
| 5- Kaempferol | 0.846 | − 8.078 | − 8.158 | |
| 6- Quercetin | 1.0 | − 9.214 | − 8.518 | |
| a- Isatin sulfonamide 34 | – | − 10.425 | − 9.046 | |
|
8- Cell division control protein 42 homology (CDC42) “It participates in the oncogenic transformation, invasion, and tumorigenesis [31] |
5UPK* | 4YDH* | ||
| 1- Acacetin | 0.999 | − 8.152 | − 8.270 | |
| 2- Acanilol-1 | – | − 8.348 | − 8.431 | |
| 3- Acanilol-2 | – | − 8.659 | − 8.754 | |
| • C GNP 200 | – | − 13.968 | – | |
| • B GNP 201 | – | – | − 14.66 | |
|
9- Cell division cycle-7-related protein kinase “It is required to initiate the DNA replication” [32] |
4F9B* | 4F99* | ||
| 1- Ellagic acid | 0.993 | − 8.517 | − 8.564 | |
| 2- Flavone | 0.90 | − 6.085 | − 6.069 | |
| • C 0SY 601 | – | − 12.593 | – | |
| • A ADP 601 | – | – | − 8.397 | |
|
10- Cyclin-dependent kinase 1 enzyme “It is a critical regulator of cell cycle progression” [33] |
4YC6* | Swiss | ||
| 1- Ellagic acid | 0.993 | − 7.957 | − 8.845 | |
|
11- Cyclin-dependent kinase CDK 4 and 6 “CDK4 regulates the cell cycle during G (1)/S transition” CDK 6 promotes G (1)/S transition [18] |
2W9Z* | 4AUA* | ||
| 1- Acacetin | 1.0 | − 7.428 | − 8.247 | |
| 2- Acanilol-1 | – | − 7.491 | − 9.171 | |
| 3- Acanilol-2 | – | − 7.413 | − 9.342 | |
| a- Fascaplysin | – | − 6.764 | − 9.109 | |
| • A 4AU 1302 | – | – | − 6.659 | |
|
12- Cyclin-dependent kinase 9 enzyme “It is involved in regulation of Transcription” [18] |
4BCG* | 6GZH* | ||
| 1- Acacetin | 1.0 | − 7.600 | − 7.785 | |
| 2- Acanilol-1 | – | − 8.064 | − 8.435 | |
| 3- Acanilol-2 | – | − 8.852 | − 9.146 | |
| • A T3C 1328 | – | − 8.724 | – | |
| • A LCI 2001 | – | – | − 9.039 | |
|
13- Death-associated protein kinase 1 “It regulates type I Apoptotic, type II autophagic cell deaths” [16] |
5AUV* | 5AUU* | ||
| 1- Quercetin | 1.0 | − 8.275 | − 10.389 | |
| 2- Kaempferol | 1.0 | − 8.106 | − 8.955 | |
| • A AGI 400 | – | − 7.349 | – | |
| • A LU2 400 | – | – | − 10.163 | |
|
14- DNA topoisomerase 1 enzyme “Has an important function in DNA replication” [34] |
1SC7* | 1TL8* | ||
| 1- 1,6-di-O-galloyl-beta | 0.987 | − 14.08 | − 8.781 | |
| 2- Acacetin | 0.67 | − 10.42 | − 6.715 | |
| 3- Kaempferol-7-gluc | 1.0 | − 14.05 | − 9.436 | |
| a- Camptothecin | – | − 12.86 | − 7.811 | |
| b- Edotecarin | – | − 16.28 | − 17.452 | |
| • C M38 990 | – | − 10.74 | – | |
| • D AI3 901 | – | – | − 7.088 | |
|
15- Ephrin type B receptor 4 “It is important in tumor angiogenesis” [16] |
3ZEW* | 6FNI* | ||
| 1- Ellagic acid | 1.0 | − 9.448 | − 9.537 | |
| • A STU 1889 | – | − 11.729 | – | |
| • A DXH 1001 | – | – | − 13.698 | |
|
16- Estrogen receptor beta “has a pivotal role in the development and the progression of tumors via the mitogenic action of estrogens” [35] |
1X7J* | 2NV7* | ||
| 1- (+)-Mollisacacidin | 1.0 | − 9.685 | − 9.507 | |
| 2- Acacetin | 1.0 | − 7.556 | − 7.642 | |
| 3- Catechin | 1.0 | − 9.966 | − 9.551 | |
| 4- Catechin-7-O-gallate | 1.0 | − 9.926 | − 9.978 | |
| 5- Dicatechin | 1.0 | 31.247 | 41.893 | |
| 6- Epicatechin | 1.0 | − 9.975 | − 9.723 | |
| 7- Ellagic acid | 1.0 | − 9.212 | − 8.646 | |
| 8- γ-Sitosterol | 1.0 | − 1.256 | − 2.088 | |
| 9- Kaempferol | 1.0 | − 8.469 | − 9.027 | |
| 10- Leucocyanidin | 1.0 | − 8.452 | − 4.881 | |
| 11- Naringenin | 1.0 | − 9.140 | − 9.363 | |
| 12- Querecitin | 1.0 | − 8.862 | − 9.456 | |
| • A GEN 201 | – | − 9.001 | – | |
| • A 555 501 | – | – | − 9.330 | |
|
17- Focal adhesion kinase enzyme “It is essential in angiogenesis, cell migration and apoptosis” [16] |
4K9Y* | 4D58* | ||
| 1- Ellagic acid | 1.0 | − 8.510 | − 9.052 | |
| 2- Quercetin | 1.0 | − 9.228 | − 7.587 | |
| • A K9Y 701 | – | − 9.549 | – | |
| • B BI9 1690 | – | – | − 8.303 | |
|
18- Glycogen synthase kinase 3 β “It phosphorylates various proteins in the cell cycle and apoptosis. Its inhibitors promote apoptosis” [36] |
4IQ6 | 5K5N | ||
| 1- Ellagic acid | 1.0 | − 9.049 | − 8.588 | |
| • B 6QH 401 | – | – | − 6.632 | |
| • B IQ6 501 | – | − 8.067 | – | |
|
19- Inducible nitric oxide synthase “It produces NO that has a tumoricidal action in macrophage” [16] |
3E7G* | 3NQS* | ||
| 1- (+)-Mollisacacidin | 1.0 | − 10.097 | − 9.977 | |
| 2- Epicatechin | 1.0 | − 10.205 | − 9.723 | |
| 3- Ellagic acid | 1.0 | − 8.337 | − 8.101 | |
| 4- Kaempferol | 0.968 | − 9.407 | − 9.549 | |
| 5- Lupenone | 1.0 | − 10.186 | − 10.537 | |
| 6- Lupeol | 1.0 | − 12.137 | − 10.354 | |
| 7- Naringenin | 0.89 | − 9.896 | − 9.918 | |
| 8- Niloticane | 1.0 | − 10.340 | − 10.484 | |
| 9- γ-Sitosterol | 1.0 | − 11.662 | − 10.763 | |
| 10- Quercetin | 1.0 | − 9.456 | − 9.771 | |
| a- Curcumin | – | − 10.024 | − 9.775 | |
| • A AT2 906 | – | − 8.470 | − 8.788 | |
|
20- Induced myeloid leukemia differentiation protein MCL-1 “It is involved in regulation of apoptosis and cell survival” [16] |
5UUM* | 6B4L* | ||
| 1- Acacetin | 0.998 | − 8.081 | − 8.386 | |
| 2- Ellagic acid | 0.999 | − 8.125 | − 8.204 | |
| 3- Kaempferol | 1.0 | − 8.208 | − 8.629 | |
| 4- Quercetin | 1.0 | − 8.369 | − 8.047 | |
| a- Obatoclax | – | − 8.357 | − 7.452 | |
| • A CJY 401 | – | – | − 9.461 | |
|
21- Serine/threonine-protein kinase pim-1 “It is involved in tumorigenesis, cell survival, and proliferation” [16] |
6AYD* | 6BSK* | ||
| 1- Acacetin | 0.994 | − 7.992 | − 6.878 | |
| 2- Ellagic acid | 0.9 | − 7.155 | − 7.953 | |
| 3- Kaempferol | 0.99 | − 8.282 | − 7.732 | |
| 4- Quercetin | 1.0 | − 8.875 | − 8.342 | |
| a- Leucettine L41 | – | − 8.785 | − 8.134 | |
| • A C2J 401 | – | − 5.852 | – | |
| • A MVG 405 | – | – | − 8.970 | |
|
22- Matrix metalloproteinase 9 “involved in tumor transformation, progression, survival, angiogenesis and metastasis” [37] |
6ESM* | 5CUH* | ||
| 1- Quercetin | 1.0 | − 11.134 | − 9.847 | |
| • A B9Z 306 | – | − 8.173 | – | |
| • A LTQ 306 | – | – | − 11.035 | |
|
23- M phase inducer phosphatase “It is a key cell cycle regulator” [38] |
4WH7* | 4WH9* | ||
| 1- (+)-Mollisacacidin | 0.947 | − 9.204 | − 8.835 | |
| 2- Catechin | 0.947 | − 8.297 | − 8.199 | |
| 3- Digallic acid | 0.982 | − 10.300 | − 8.991 | |
| 4- Epicatechin | 0.947 | − 8.049 | − 9.179 | |
| 5- Kaempferol | 0.994 | − 7.152 | − 7.524 | |
| 6- γ-Sitosterol | 1.0 | − 7.169 | − 10.413 | |
| 7- Niloticane | 1.0 | − 7.099 | − 7.917 | |
| • A 8H8 607 | – | − 4.390 | – | |
| • A 3M8 601 | – | – | − 6.086 | |
|
24- Nuclear receptor ROR-alpha “It is involved in cell growth, differentiation, and control of metastatic behavior of androgen-independent prostate cancer” [39] |
1N83* | 3B0W* | ||
| 1- Acacetin | 1.0 | − 8.342 | − 8.021 | |
| 2- Ellagic acid | 0.969 | − 8.433 | − 8.275 | |
| 3- Quercetin | 0.857 | − 8.974 | − 9.179 | |
| • A CLR 1000 | – | − 11.358 | – | |
| • B DGX 1 | – | – | − 15.907 | |
|
25- Serine/threonine protein kinase Nek2 “It regulates centrosome separation, bipolar spindle formation in cell mitosis” [16] |
2XNN* | 2WQO* | ||
| 1- Quercetin | 1.0 | − 8.417 | − 9.318 | |
| • A 430 1280 | – | − 7.282 | – | |
| • A VGK 1280 | – | – | − 10.033 | |
|
26- P-glycoprotein 1 and 3 “They involved in multi-drug resistance” [40] |
4XWK* | 2CBZ* | ||
| 1- Acacetin | 1.0 | − 8.064 | − 8.580 | |
| 2- Chalconaringnen-4-O | 0.979 | − 11.266 | − 9.360 | |
| 3- Kaempferol | 1.0 | − 7.418 | − 6.493 | |
| 4- Kaempferol-7-glucoside | 0.97 | − 11.300 | − 9.221 | |
| 5- Querecitin | 1.0 | − 8.002 | − 5.888 | |
| • A 4C8 1301 | – | − 8.874 | – | |
| • A ATP 1873 | – | – | − 9.674 | |
|
27- Platelet-derived growth factor 1 receptor “It has a pro-angiogenic action [41] |
5GRN* | 5K5X* | ||
| 1- Ellagic acid | 1.0 | − 8.681 | − 7.922 | |
| a- Sunitinib | – | − 9.363 | − 8.234 | |
|
28- Proto-oncogene tyrosine protein kinase Src “It participates in cancer cells invasion and progression” [42] |
4MXO* | 4MXY* | ||
| 1- Ellagic acid | 1.0 | − 7.488 | − 7.260 | |
| • B DB8 601 | – | − 8.930 | − 7.870 | |
|
29- Protein kinase C epsilon type “It is essential in cell invasion, adhesion, migration, and regulation of apoptosis” [16] |
1GMI* | 2WH0* | ||
| 1- Kaempferol | 0.787 | − 7.836 | − 6.832 | |
| 2- Naringenin | 0.99 | − 7.543 | − 6.772 | |
|
30- Steroid 17 alpha-hydroxylase “It is a key regulatory enzyme, essential in androgens biosynthesis” [43] |
6CIR* | 5UYS* | ||
| 1- (+)-Mollisacacidin | 0.83 | − 8.744 | − 8.462 | |
| 2- Ellagic acid | 1.0 | − 8.526 | − 8.393 | |
| 3- γ-Sitosterol | 1.0 | − 10.868 | − 10.792 | |
| 4- Epicatechin | 0.838 | − 9.273 | − 9.793 | |
| 5- Kaempferol | 0.955 | − 8.360 | − 8.455 | |
| 6- Niloticane | 0.998 | − 8.485 | − 8.727 | |
| 7- Querecitin | 1.0 | − 8.940 | − 8.497 | |
| a- Galeterone | – | − 10.681 | − 11.190 | |
| • A 3NQ 601 | – | − 10.485 | – | |
| • A 8QD 601 | – | – | − 10.777 | |
|
31- Tankayrase enzyme 1 and 2 “Involved in cell cycle progression and telomere homeostasis” [44] |
4U6A* | 4HKI* | ||
| 1- Acacetin | 0.74 | − 10.054 | − 11.596 | |
| 2- Flavone | 1.0 | − 9.475 | − 9.961 | |
| • A 3DN 1402 | – | − 11.568 | – | |
| • A FLN 1204 | – | – | − 10.237 | |
|
32- Telomerase reverse transcriptase enzyme “It is involved in the regulation of transcription and has a major role in the activation of telomerase at cancer” [45] |
5UGW* | 5NPT* | ||
| 1- (+)-Mollisacacidin | 0.991 | − 6.778 | − 7.634 | |
| 2- Acacetin | 1.0 | − 5.023 | − 7.100 | |
| 3- Ellagic acid | 1.0 | − 5.547 | − 8.204 | |
| 4- Catechin | 0.991 | − 6.668 | − 7.590 | |
| 5- Epicatechin | 0.991 | − 6.774 | − 7.570 | |
| 6- Kaempferol | 1.0 | − 5.993 | − 7.128 | |
| 7- Leucocyanidin | 0.912 | − 7.246 | − 10.797 | |
| 8- Naringenin | 1.0 | − 5.848 | − 7.543 | |
| 9- Quercetin | 1.0 | − 5.720 | − 7.434 | |
| a- Berberine | – | − 22.039 | − 9.507 | |
| • A GSH 1201 | – | − 7.916 | – | |
|
33- Transcription factor p65 “It promotes tumor cells proliferation, suppresses the apoptosis, attracts the angiogenesis, metastasis, remodels the local metabolism and energizes the immune system to favor tumor growth” [46] |
2RAM* | 5 U01* | ||
| 1- (+)-Mollisacacidin | 0.987 | − 8.153 | − 8.549 | |
| 2- (+)-Catechin-5,7 digllate | 0.998 | − 11.618 | − 11.996 | |
| 3- Acacetin | 1.0 | − 6.599 | − 7.443 | |
| 4- Catechin-7-O-gallate | 0.883 | − 8.719 | − 9.457 | |
| 5- Ellagic acid | 0.994 | − 7.309 | − 7.547 | |
| 6- Epicatechin | 0.987 | − 8.661 | − 9.370 | |
| 7- Kaempferol | 1.0 | − 7.535 | − 7.202 | |
| 8- Leucocyanidin | 0.986 | − 9.687 | − 9.905 | |
| 9- Quercetin | 1.0 | − 7.730 | − 8.485 | |
| a- Bortezomib | – | − 6.232 | − 6.396 | |
|
34- Tyrosine-protein kinase Lyn “It is involved in the control of proliferation and the inhibition of apoptosis” [47] |
5XY1* | 3A4O* | ||
| 1- Ellagic acid | 1.0 | − 7.446 | − 8.238 | |
| • A 8H0 601 | – | − 8.054 | – | |
| • X STU 902 | – | – | − 10.207 | |
|
35- Vascular endothelial growth factor receptor 3 “It promotes tumor angiogenesis” [48] |
3WZD* | 5EW3* | ||
| 1- Quercetin | 1.0 | − 8.561 | − 8.602 | |
| a- Axitinib | – | − 8.699 | − 11.087 | |
| b- Cabozantinib | – | − 6.576 | − 7.936 | |
| • A LEV 1201 | – | − 8.648 | – | |
| • A 5T2 1201 | – | – | − 9.156 | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicates positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID. Swiss means that the 3D structure of the target was modeled using SWISS-MODEL web server [49]
Table 2.
Ligand-based virtual screening and molecular docking results regarding the antibacterial activity
| Target | Compounds | LBVS | Docking Score | ||
|---|---|---|---|---|---|
|
1- 3-oxyacyl-[acyl-carrier protein] reductase FabG “It catalyzes the first reductive step in the elongation cycle of fatty acid biosynthesis” [26] |
4BNT* | 5OVK* | 5END* | ||
| 1- Acacetin | 0.68 | − 9.017 | − 7.925 | − 7.891 | |
| 2- Catechin | 0.60 | − 10.605 | − 9.857 | − 8.487 | |
| 3- Epicatechin | 0.60 | − 10.538 | − 8.816 | − 8.518 | |
| 4- Kaempferol | 1.0 | − 9.184 | − 8.071 | − 7.497 | |
| 5- Quercetin | 1.0 | − 10.068 | − 8.324 | − 8.186 | |
| 6- C 36E 1248 | – | − 5.612 | – | – | |
| 7- D NDP 301 | – | – | − 14.254 | – | |
|
2- Enoyl-acyl carrier protein reductase “It is a limiting step enzyme in fatty acid biosynthesis, has no homolog in mammals” [50] |
4M87* | 4NR0* | 4O1M* | ||
| 1- (−)-Epigallocatchin-7-gallate | 0.43 | − 11.920 | − 12.991 | − 13.66 | |
| 2- (+)-Catechin-4, 5,digallate | 0.55 | − 13.771 | − 13.034 | − 14.25 | |
| 3- (+)-Catechin-5-gallate | 0.6 | − 12.496 | − 11.710 | − 13.05 | |
| 4- Quercetin | 1.0 | − 10.248 | − 10.823 | − 10.12 | |
| a- Isonazid | – | − 5.757 | − 5.210 | − 5.594 | |
| b- Triclosan | – | − 7.254 | − 7.110 | − 7.585 | |
| • `A NAD 301 | – | − 13.553 | − 13.298 | − 11.81 | |
|
3- D-alanine D-alanine ligase enzyme “It is an essential bacterial enzyme in peptidoglycan biosynthesis” [51] |
6DGI* | 5C1P* | 3R23* | ||
| 1- Quercetin | 1.0 | − 6.267 | − 8.968 | − 7.719 | |
| a- Adenosine-5′-diphosphate | – | − 3.430 | − 8.350 | − 5.561 | |
| • A GOL 401 | – | − 5.112 | – | – | |
| • D ADP 401 | – | – | − 7.924 | – | |
|
4- AmpC Beta-lactamase enzyme “It is responsible for hydrolysis of beta-lactams, with substrate specificity toward cephalosporins, has an important role in cephalosporins resistance” [16] |
2HDQ* | 2PU2* | 2R9W* | ||
| 1- D-pinitol | 0.876 | − 8.334 | − 7.930 | − 7.400 | |
| 2- Niloticane | 0.992 | − 8.691 | − 8.713 | − 7.221 | |
| 3- Quercetin | 1.0 | − 9.481 | − 9.128 | − 8.917 | |
| a- Clavulanic acid | – | − 8.808 | − 7.732 | − 8.108 | |
| • A C21 501 | – | − 6.937 | – | – | |
| • B DK2 701 | – | – | − 9.133 | – | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicate positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID
Table 3.
Ligand-based virtual screening and molecular docking results regarding the antiviral and the antiplasmodial activity
| Target | Compounds | LBVS | Docking score | |
|---|---|---|---|---|
|
1- HIV integrase enzyme “Antiviral target acts on the essential step in viral replication cycle via catalyzes of viral DNA integration into host DNA” [52] |
3ZT4* | 5KGX* | ||
| 1- Acacetin | 1.0 | − 8.771 | − 4.595 | |
| 2- Digallic acid | 1.0 | − 11.163 | − 7.227 | |
| 3- Ellagic acid | 0.75 | − 9.509 | − 4.765 | |
| 4- Kampeferol | 1.0 | − 8.889 | − 5.473 | |
| 5- Naringenin | 0.76 | − 9.118 | − 5.388 | |
| 6- Querecitin | – | − 9.897 | − 5.120 | |
| • A 7SK 301 | – | – | − 4.705 | |
| • A ZT2 1217 | – | – | − 8.030 | |
|
2- Corona virus replicase polyprotein 1 ab “Antiviral target involved in transcription and replication of viral RNAs and interacts with host 40S ribosomal subunit leading to translation inhibition” [26] |
5NH0* | 5N5Ov* | ||
| 1- Quercetin | 1.0 | − 9.436 | − 9.848 | |
| • A 8X8 301 | – | − 7.612 | – | |
| • A 8O5 401 | – | – | − 10.887 | |
|
3- Toll-like receptor 9 “Antiviral and antibacterial target acts as innate immune receptor acts in recognition of microbial DNA” [53] |
5Y3L* | 5ZLN* | ||
| 1- Acacetin | 1.0 | − 7.024 | − 6.001 | |
| 2- Ellagic acid | 0.999 | − 7.956 | − 6.225 | |
| 3- Epicatechin | 0.819 | − 8.138 | − 6.796 | |
| 4- Kaempferol | 1.0 | − 7.726 | − 6.369 | |
| 5- Leucocyanidin | 0.992 | − 9.222 | − 8.689 | |
| 6- Naringenin | 0.889 | − 7.664 | − 7.244 | |
| 7- Niloctne | 1.0 | − 7.967 | − 6.212 | |
| 8- Quercetin | 1.0 | − 7.481 | − 7.048 | |
|
4- β-Hydroxy acyl-ACP dehydratase FabZ “Antiplasmodial target involved in the fatty acid biosynthesis” [54] |
3AZB* | 3AZA* | ||
| 1- (+) Mollisacacidin | 0.6 | − 8.297 | − 10.946 | |
| 2- Acacetin | 0.67 | − 6.787 | − 8.454 | |
| 3- Catechin | 0.60 | − 9.251 | − 11.096 | |
| 4- Epicatechin | 0.60 | − 8.857 | − 11.216 | |
| 5- Kaempferol | 0.78 | − 7.318 | − 9.544 | |
| 6- Quercetin | 1.0 | − 10.906 | − 9.286 | |
| • G KM1 | – | − 7.913 | – | |
| • B KM0 2 | – | – | − 6.603 | |
|
5- Hydroxyacyl-[acyl-carrier-Protein] dehydratase “Antiplasmodial and antibacterial target responsible for fatty acid biosynthesis” [55] |
3ED0* | 3CF9* | ||
| 1- Quercetin | 1.0 | − 10.032 | − 9.607 | |
| • A EMO 163 | – | − 8.454 | – | |
| • A AGI 161 | – | – | − 7.841 | |
|
6- MO15-related protein kinase pfmrk enzyme “Antiplasmodial target that is a cyclin-dependent kinase enzyme plays a central role in the regulation of cell cycle” [56] |
Raptor x | Phyre 2 | ||
| 1- Acacetin | 0.74 | − 8.125 | − 9.482 | |
|
7- M18 aspartyl aminopeptidase enzyme “Antiplasmodial target involved in host erythrocyte invasion and the degradation of host hemoglobin” [57] |
4EME* | Phyre 2 | ||
| 1- (+) Mollisacacidin | 1.0 | − 10.319 | − 8.855 | |
| 2- Epicatechin | 1.0 | − 9.281 | − 9.895 | |
| 3- Querecitin | 1.0 | − 10.223 | − 9.218 | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicate positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID. Phyre2 and Raptor x means the 3D structure of target was modeled by Phyre2 [58] and RaptorX [59] web servers, respectively
Table 4.
Ligand-based virtual screening and molecular docking results regarding the antidiabetic activity
| Target | Compounds | LBVS | Docking Score | |
|---|---|---|---|---|
| 1- Insulin receptor | 2W12* | 4OGA* | ||
| 1- Ellagic acid | 1.0 | − 9.239 | − 8.785 | |
| a- Ceritinib | – | − 8.150 | − 8.093 | |
|
2- Glycogen phosphorylase (muscle) “An important allosteric enzyme in carbohydrate metabolism” [16], “potential target in type 2 diabetes mellitus” [56] |
2ZB2* | 3CEJ* | ||
| 1- Quercetin | 1.0 | − 11.15 | − 9.523 | |
| • A A46 850 | – | − 11.44 | − | |
| • A AVF 833 | – | − 11.52 | ||
|
3- Sodium/glucose co-transporter 2 “The interaction with this enzyme inhibits the renal glucose reabsorption, leading to a reduction in plasma glucose level” [60] |
2XQ2* | 3DH4* | ||
| 1- 1,6-di-O-galloyl-beta-D-glucose | 1.0 | − 12.05 | − 12.05 | |
| 2- Chalconaringnen-4-O-beta. | 1.0 | − 10.53 | − 10.53 | |
| 3- Niloticane | 0.961 | − 7.634 | − 7.822 | |
| 4- Canagliflozin | – | − 11.11 | − 8.030 | |
|
4- Aldose reductase enzyme “It is involved in the development of the secondary diabetic complications” [61] |
3RX4* | 3V36* | ||
| 1- (+)-Mollisacacidin | 1.0 | − 9.074 | − 11.20 | |
| 2- Acacetin | 0.74 | − 9.196 | − 10.17 | |
| 3- Chalconaringnen-4-O-beta-glucose | 0.74 | − 12.16 | − 14.31 | |
| 4- Catechin | 1.0 | − 9.581 | − 11.74 | |
| 5- Dicatechin | 0.959 | − 12.70 | − 10.04 | |
| 6- Ellagic acid | 1.0 | − 8.425 | − 8.886 | |
| 7- Epicatecin | 1.0 | − 9.513 | − 11.73 | |
| 8- Kaempferol | 1.0 | − 8.185 | − 10.86 | |
| 9- Kaempferol-7-glucoside | 0.97 | − 12.35 | − 14.27 | |
| 10- Leucocyanidin | 1.0 | − 11.45 | − 14.21 | |
| 11- Melacacidin | 1.0 | − 9.75 | − 12.90 | |
| 12- Naringenin | 1.0 | − 9.618 | − 9.869 | |
| 13- Quercetin | 1.0 | − 8.824 | − 10.81 | |
| a- Epalrestat | – | − 8.849 | − 10.50 | |
| • A SFI 317 | – | − 8.393 | − 8.61 | |
|
5- Beta-secretase enzyme “It is down-regulator of insulin receptors amounts and signaling in the liver” [62] |
5MXD* | 4BEL* | ||
| 1- Ellagic acid | 1.0 | − 7.493 | − 7.677 | |
| 2- Quercetin | 1.0 | − 6.955 | − 7.408 | |
| a- 5,5-Diphenyliminohydantoin | – | − 4.885 | − 5.654 | |
| • A III 701 | – | − 5.569 | – | |
| • A B3P 1399 | – | – | − 6.797 | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicate positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID
Table 5.
Ligand-based virtual screening and molecular docking results regarding the anti-inflammatory activity
| Target | Compounds | LBVS | Docking score | |
|---|---|---|---|---|
| 1- Arachidonate 15-lipoxygenase. “Has an important role in the immune and the inflammatory responses” [16]. | 4NRE* | 2P0M* | ||
| 1- (−)-Epigallocatchin-7-gallate | 1.0 | − 11.046 | − 12.03 | |
| 2- (+)-Mollisacacidin | 1.0 | − 7.845 | − 9.535 | |
| 3- Acacetin | 1.0 | − 7.693 | − 9.283 | |
| 4- Catechin | 0.984 | − 7.864 | − 10.05 | |
| 5- Dicatechin | 1.0 | − 9.003 | − 11.19 | |
| 6- Ellagic acid | 1.0 | − 6.923 | − 8.525 | |
| 7- Epicatechin | 1.0 | − 8.210 | − 9.662 | |
| 8- Kaempferol | 1.0 | − 7.659 | − 9.058 | |
| 9- Leucocyanidin | 1.0 | − 10.035 | − 12.76 | |
| 10- Naringenin | 1.0 | − 7.857 | − 8.760 | |
| 11- Quercetin | 1.0 | − 7.865 | − 9.732 | |
| a- Diethylcarbamazine | – | − 7.511 | − 7.623 | |
| • A C8E 702 | – | − 5.722 | – | |
| • B RS7 841 | – | – | − 6.830 | |
| 2- Cycloxygenase-2 enzyme (COX-2). “It generates the inflammatory mediator’s Prostaglandins from the arachidonic acid” [62]. | 5IKQ* | 5F1A* | ||
| 1- (+)-Mollisacacidin | 0.801 | − 10.127 | − 8.246 | |
| 2- Acacetin | 1.0 | − 8.944 | − 7.561 | |
| 3- Ellagic acid | 1.0 | − 7.932 | − 7.969 | |
| 4- Kaempferol | 1.0 | − 9.149 | − 7.444 | |
| 5- Leucocyanidin | 0.907 | − 8.121 | − 5.193 | |
| 6- Naringenin | 0.991 | − 9.426 | − 8.345 | |
| 7- Querecitin | 1.0 | − 9.281 | − 7.759 | |
| a- Diclofenac | – | − 8.628 | − 7.131 | |
| b- Indomethacin | – | − 8.771 | − 7.162 | |
| • A JMS 602 | – | − 8.632 | – | |
| • A SAL 601 | – | – | − 5.384 | |
| 3- Macrophage migration inhibitory factor. “It is a pro-inflammatory cytokine counteracts the anti-inflammatory activity of glucocorticoids” [16]. | 5XEJ* | 6CB5* | ||
| 1- (−)-Epigallocatchin-5, 7-gallate | 1.0 | − 8.500 | − 11.51 | |
| 2- (+)-Catechin-5,7-digallate | 1.0 | − 10.457 | − 12.24 | |
| 3- (+)-Mollisacacidin | 1.0 | − 6.527 | − 9.613 | |
| 4- Catechin | 1.0 | − 6.836 | − 9.855 | |
| 5- Dicatechin | 0.966 | − 7.621 | − 8.843 | |
| 6- Epicatechin | 1.0 | − 7.600 | − 9.411 | |
| 7- Kaempferol | 1.0 | − 5.332 | − 8.927 | |
| 8- Leucocyanidin | 1.0 | − 7.623 | − 9.649 | |
| 9- Naringenin | 1.0 | − 6.853 | − 8.881 | |
| 10- Querecitin | 1.0 | − 5.987 | − 9.257 | |
| a- 3,4-Dihydroxycinnamic acid | – | − 7.846 | − 10.35 | |
| • A 6UV 204 | – | − 8.978 | – | |
| • A EV7 201 | – | – | − 11.79 | |
| 4- Phospholipase A2 enzyme. “It is responsible for the release of the arachidonic acid from arachidonyl phospholipids, thereby involved in the initiation of the inflammatory response” [16]. | 2B96* | 5OW8* | ||
| 1- γ-Sitosterol | 0.831 | − 8.764 | − 7.798 | |
| 2- Digallic acid | 0.884 | − 8.626 | − 8.788 | |
| 3- Kaempferol | 0.938 | − 8.563 | − 7.556 | |
| 4- Quercetin | 1.0 | − 8.655 | − 8.101 | |
| a- Prostaglandin A2 | – | − 7.640 | − 5.746 | |
| • A ANN 501 | – | − 5.760 | – | |
| 5- Receptor-interacting serine/ threonine protein kinase 2. “It is involved in the formation of the productive inflammatory Response” [63]. | 6FU5* | 6HMX* | ||
| 1- Acacetin | 0.962 | − 8.185 | − 6.108 | |
| 2- Digallic acid | 0.924 | − 12.137 | − 9.105 | |
| 3- Ellagic acid | 1.0 | − 8.111 | − 7.246 | |
| 4- Kaempferol | 0.919 | − 8.138 | − 6.606 | |
| 5- Niloctne | 1.0 | − 10.128 | − 7.078 | |
| 6- Quercetin | 0.906 | − 8.320 | − 6.902 | |
| • A E7N 400 | – | − 9.371 | – | |
| 6- Xanthine dehydrogenase/ oxidase enzyme. “It contributes to uric acid formation and generation of reactive oxygen species” [16]. | 2E1Q* | 3AM9* | ||
| 1- (+)-Mollisacacidin | 1.0 | − 11.185 | − 11.260 | |
| 2- 1,6-di-O-galloyl-beta-D-glu. | 1.0 | − 14.648 | − 13.042 | |
| 3- Acacetin | 1.0 | − 9.690 | − 9.847 | |
| 4- Catechin | 1.0 | − 11.260 | 91.380 | |
| 5- Digallic acid | 1.0 | − 10.059 | − 10.049 | |
| 6- Ellagic acid | 1.0 | − 9.337 | − 11.830 | |
| 7- Epicatechin | 1.0 | − 11.367 | − 11.669 | |
| 8- Kaempferol | 1.0 | − 9.900 | − 10.184 | |
| 9- Leucocyanidin | 1.0 | − 10.999 | − 11.794 | |
| 10- Quercetin | 1.0 | − 11.177 | − 11.371 | |
| a- Allopurinol | – | − 6.680 | − 6.120 | |
| • A MTE | – | − 13.547 | − 13.141 | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicate positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID
Table 6.
Ligand-based virtual screening and molecular docking results regarding the antidiarrheal, anti-platelets, and anticholinesterase activity
| Target | Compounds | LBVS | Docking Score | |
|---|---|---|---|---|
|
1- Mu-type opioid receptor “Antidiarrheal target, the action on mu and delta opioid receptors leads to inhibition of diarrhea without constipation [64] |
4DKL* | 5C1M* | ||
| 1- (+)-Mollisacacidin | 0.994 | − 7.279 | − 9.051 | |
| 2- Catechin | 0.994 | − 7.822 | − 8.727 | |
| 3- Dicatechin | 0.983 | − 8.880 | − 12.268 | |
| 4- Epicatechin | 0.994 | 7.392 | − 8.836 | |
| a- Eluxadoline | – | − 6.868 | − 10.799 | |
| • A 4VO 401 | – | – | − 11.228 | |
|
2- Delta-type opioid receptor Antidiarrheal target |
4N6H* | 4EJ4* | ||
| 1- (+)-Catechin-3, 5-digallate | 0.998 | − 11.44 | − 10.808 | |
| 2- Dicatechin | 0.999 | − 12.06 | − 11.488 | |
| a- Eluxadoline | – | − 7.834 | − 7.712 | |
| • A EJ4 1219 | – | − 11.01 | – | |
| • A EJ4 500 | – | – | − 10.392 | |
|
3- P2Y12 receptor “Anti-platelets target has a central role in platelet activation” [65] |
4PXZ* | 4NTJ* | ||
| 1- 1, 6-di-O-galloyl-beta-D-glucose | 0.756 | − 11.99 | − 11.397 | |
| 2- Clopidogrel | – | − 5.521 | − 6.873 | |
| • A 6AD 1201 | – | − 20.29 | – | |
| • A AZJ 1201 | – | − 19.78 | – | |
|
4- Acetylcholinesterase enzyme “The reversible inhibition of enzyme acetylcholinesterase increases the concentration of acetylcholine that is helpful in neurodegenerative disorders like Alzheimer’s disease [66] |
1H22* | 1ODC* | ||
| 1- (+)-Catechin-4,5-digallate | 0.73 | − 12.837 | − 13.137 | |
| 2- (+)-Mollisacacidin | 0.984 | − 10.697 | − 11.074 | |
| 3- Acacetin | 0.74 | − 8.728 | − 10.147 | |
| 4- Catechin | 0.984 | − 11.143 | − 11.397 | |
| 5- Digallic acid | 0.94 | − 12.683 | − 13.272 | |
| 6- Epicatechin | 0.984 | − 11.053 | − 10.701 | |
| 7- Flavone | 1.0 | − 7.519 | − 7.521 | |
| 8- Leucocyanidin | 0.99 | − 12.967 | − 13.153 | |
| 9- Lupenone | 1.0 | − 10.976 | − 8.968 | |
| 10- Lupeol | 0.99 | − 11.737 | − 10.071 | |
| 11- Melacacidin | 0.99 | − 11.162 | − 11.291 | |
| 12- Niloticane | 1.0 | − 10.396 | − 10.735 | |
| a- Neostigmine | – | − 7.919 | − 8.196 | |
| • A E10 1536 | – | − 11.084 | − 11.266 | |
| • A A8B 1538 | – | – | − 11.266 | |
In Compounds, the numbers 1, 2, 3, … indicate A. nilotica’s phytochemical constituents, letters a, b, … indicates positive controls, • indicates the co-crystallized ligands, and the italic emphasis indicates compounds with the higher scores. At Ligand-Based Virtual Screening Score (LBVS sco.), en dash (–) means that the compound was not screened. Asterisk (*) indicates the PDB ID
Molecular Docking
Target Preparation
The 3D structure of selected targets from virtual screening was obtained from the RCSB protein data bank [67]. The structure with better resolution and validation scores was selected for the study. In order to validate the docking results, multiple 3D X-ray crystallographic structures for the same target were downloaded in PDB format. For the structures that have no practically determined 3D structure, Phyre2 [58], SWISS-MODEL web server [49], and RaptorX [59] web servers were used for 3D structure modeling, then downloaded in PDB format. The target preparation was carried out in Cresset Flare software [23] according to the default settings. After preparation, the targets 3D structures were minimized via Cresset Flare software [23] at the normal type calculation method. The targets were input to the software in PDB format.
Ligands Preparation
The preparation of reported A. nilotica’s phytochemical constituents for molecular docking study was carried out as described above.
Molecular Docking of Phytochemical Constituents With the Predicted Targets
The docking calculations were carried out in Cresset Flare software [23] in normal mode and default settings. The grid box was defined according to the co-crystallized ligands, but in the absence of co-crystallized ligands, the grid box was defined via picking of active site amino acids. Beside the A. nilotica’s phytochemical constituents, drugs that are well known to bind with the predicted targets (selected randomly from Therapeutic Target [28] and Pharos [27] databases) and the co-crystallized ligands were used as positive controls. The compounds and the targets were input in mol2 and PDB format, respectively. The results are listed in Tables 1, 2, 3, 4, 5, 6, and 7 and Figs. 1, 2, 3, 4, and 5.
Table 7.
The predicted pharmacokinetics properties of phytochemical constituents having higher affinity scores (part A)
| Phytochemical constituent | Intestinal absorption | BBB permeability | Human Vd (L/kg) | Total clearance (mg/kg/day) | Renal OCT2 substrate |
|---|---|---|---|---|---|
| (−)-Epigallocatechin-5,7-gallate | Low (14.341%) | No | 1.29 | 0.23 | No |
| (−)-Epigallocatechin-7-gallate | Low (47.214%) | No | 1.46 | 3.4 | No |
| (+)-Catechin-3,5,-digallate | Low (44.42%) | No | 0.97 | 0.3 | No |
| (+)-Mollisacacidin | High (72.264%) | No | 1.33 | 1.85 | No |
| 1,6-di-O-galloyl-beta-D-glucose | Low (28.679%) | No | 2.65 | 5.4 | No |
| Acacetin | High (94.546%) | No | 0.78 | 5.9 | No |
| Acanilol-1 | High (98.82%) | No | 0.34 | 5.5 | No |
| Acanilol-2 | High (96.44%) | No | 0.3 | 4.6 | No |
| Catechin | High (72.264%) | No | 1.33 | 1.9 | No |
| Catechin-7-O-gallate | (54.376%) | No | 1.40 | 1.0 | No |
| Chalconaringnen-4-O-beta-glucopyranoside | Low (17.445%) | No | 0.51 | 1.79 | No |
| Dicatechin | (69.966%) | No | 0.75 | 1.73 | No |
| Digallic acid | Low (47.548%) | No | 1.26 | 3.5 | No |
| Ellagic acid | High (76.935%) | No | 0.97 | 4.1 | No |
| Epicatechin | High (72.264%) | No | 1.33 | 1.85 | No |
| Flavone | High (94.935%) | Yes | 0.96 | 2.12 | No |
| γ-Sitosterol | High (95.884%) | No | 1.32 | 4.25 | No |
| Kaempferol | High (75.481)% | No | 1.02 | 4.52 | No |
| Kaempferol-7-glucoside | Low (44.274%) | No | 0.67 | 5.17 | No |
| Leucocyanadin | (65.231%) | No | 2.8 | 1.47 | No |
| Lupenone | High (100%) | No | 1.1 | 1.27 | No |
| Lupeol | High (98.249%) | No | 0.7 | 4.19 | No |
| Melacacidin | (67.928%) | No | 4.7 | 1.01 | No |
| Naringnen | High (89.345%) | No | 0.53 | 1.33 | No |
| Naringnen-7-O-beta-glucopyranoside | (56.167%) | No | 1.3 | 1.74 | No |
| Nilobamate | High (88.608%) | Yes | 1.5 | 21.29 | No |
| Niloctane | High (95.567%) | Yes | 1.1 | 7.17 | Yes |
| Quercetin | High (75.36%) | No | 2.2 | 3.83 | No |
The italic emphasis indicates desirable prosperity
BBB blood-brain barrier, Vd volume of distribution, Renal OCT2 human organic cation transporter 2 [68]
Fig. 2.
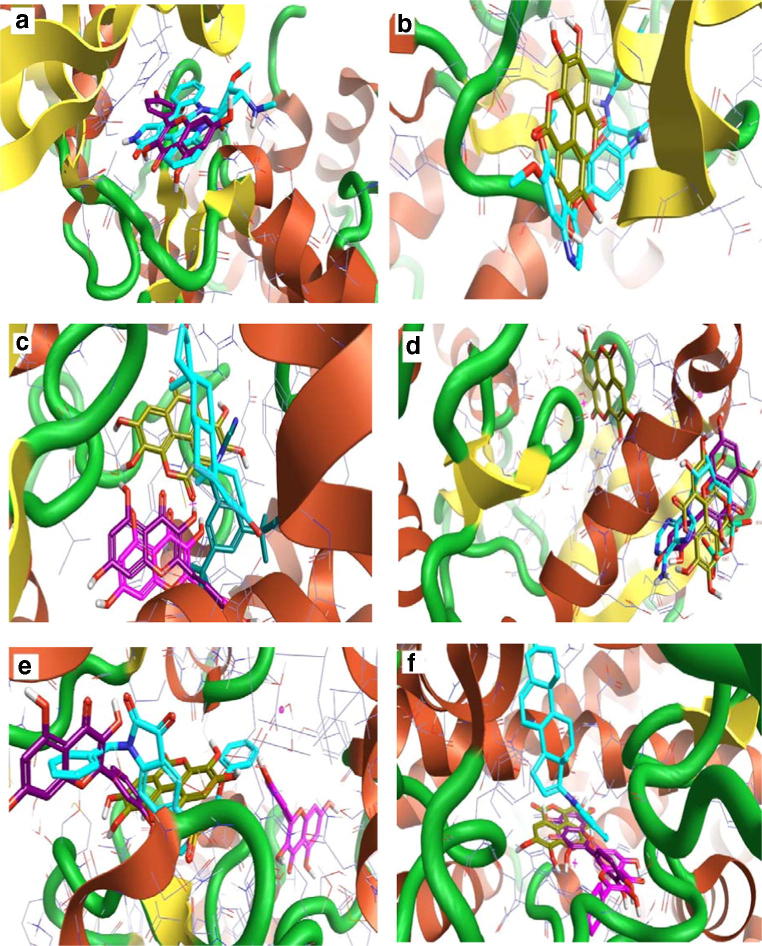
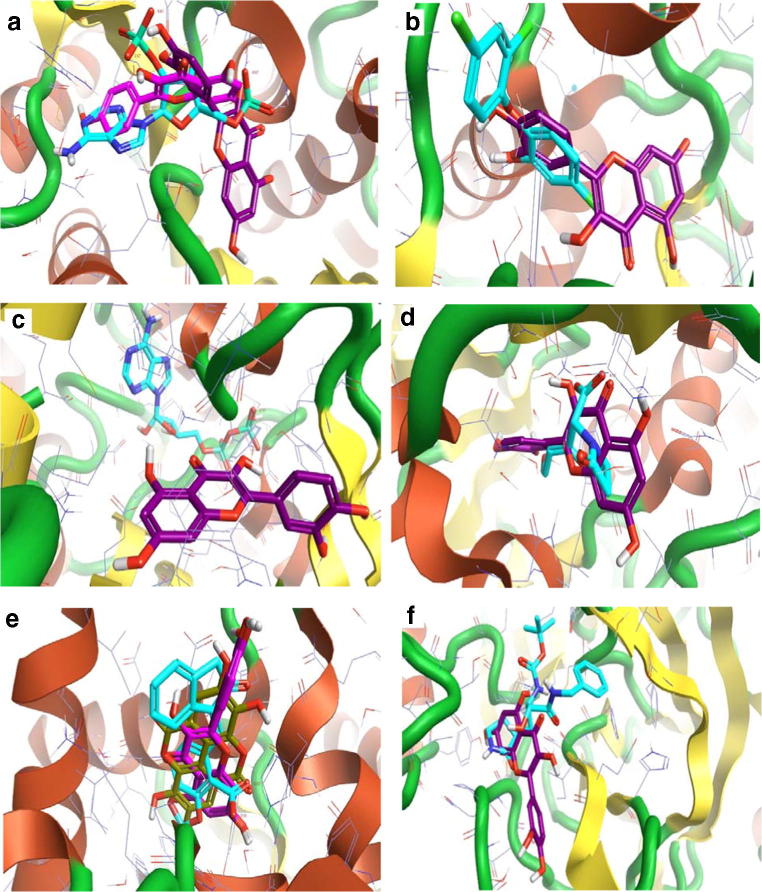
The 3D interaction between the best compounds with some of their predicted anticancer targets. a Quercetin (violet) with anaplastic lymphoma kinase enzyme. Staurosporine (turquoise) as control. b Ellagic acid (dark yellow) with angiopoietin 1 receptor. Cabozantinib (turquoise) as control. c Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with the aromatase enzyme. Anastrozole (teal) and the co-crystallized ligand A ASD 601(turquoise) as a control. d Ellagic acid (dark yellow) and quercetin (violet) with Aurora A kinase enzyme. The co-crystallized ligand A ADP 401(turquoise) as a control. e Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with caspase 9 enzyme. The Isatin sulfonamide 34 (turquoise) as a control. Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with steroid 17 alpha-hydroxylase enzyme. Galeterone (turquoise) as a control
Fig. 3.
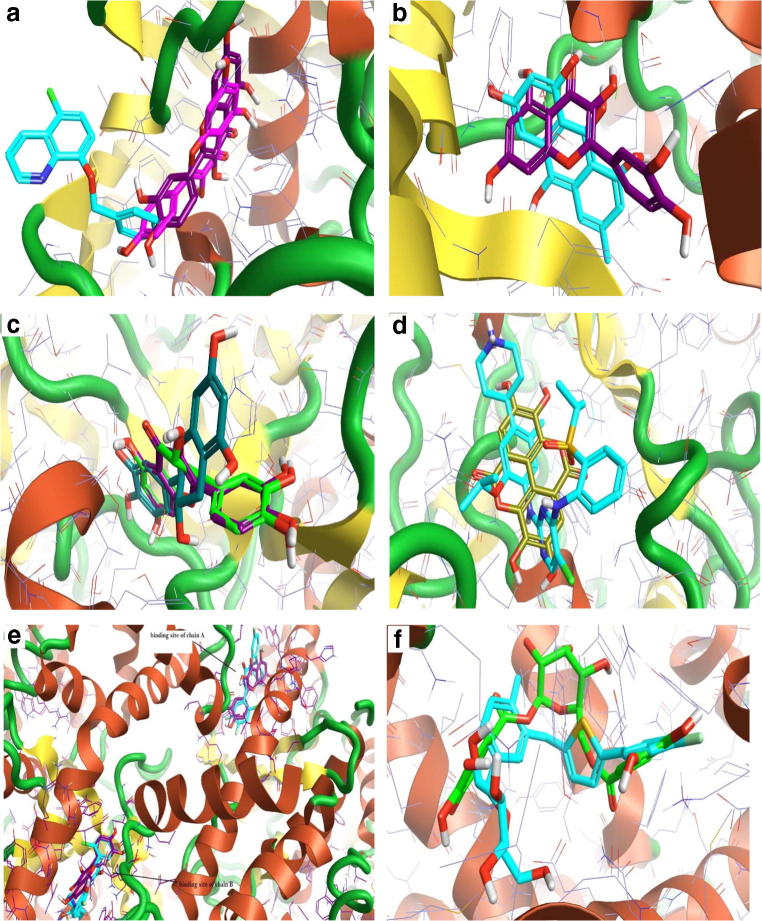
The 3D interaction between the best compounds with their predicted antibacterial and antiviral targets. a Kaempferol (pink), and quercetin (violet) with 3-oxyacyl-[acyl-carrier protein] reductase FabG. The co-crystallized ligand D NDP 301 (turquoise) as a control. b Quercetin (violet) with enoyl-acyl carrier protein reductase. Triclosan (turquoise) as a control. c Quercetin (violet) with D-alanine D-alanine ligase enzyme. The co-crystallized ligand D ADP 401(turquoise) as a control. d Quercetin (violet) with AmpC beta-lactamase enzyme. Clavulanic acid (turquoise) as a control. e Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with HIV integrase enzyme. The co-crystallized ligand A ZT2 1217 (turquoise) as a control. f Quercetin (violet) with corona virus replicase polyprotein 1 ab. The co-crystallized ligand A 8X8 301 (turquoise) as a control
Fig. 4.
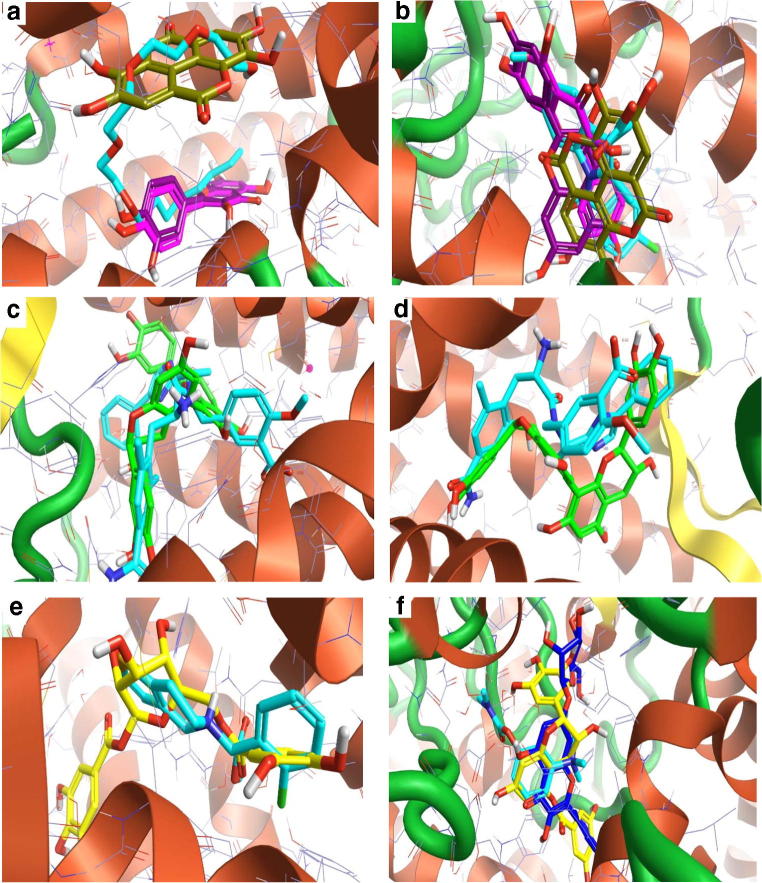
The 3D interaction between the best compounds with some of their predicted antiplasmodial and antidiabetic targets. a Kaempferol (pink), and quercetin (violet) with β-hydroxy acyl-ACP dehydratase FabZ. The co-crystallized ligand B KM0 2 (turquoise) as a control. b Quercetin (violet) with hydroxyacyl-[acyl-carrier-Protein] dehydratase. The co-crystallized ligand A EMO 163 (turquoise) as a control. c (+)-Mollisacacidin (green), epicatechin (teal), and quercetin (violet) with M18 aspartyl aminopeptidase enzyme. d Ellagic acid (dark yellow) with insulin receptor. Ceritinib (turquoise) as a control. e Quercetin (violet) with glycogen phosphorylase (muscle). The co-crystallized ligand AVF 833 (turquoise) as a control. f 1,6-di-O-galloyl-beta-D-glucose (green) with sodium /glucose co-transporter 2. Canagliflozin (turquoise) as a control
Fig. 5.
The 3D interaction between the best compounds with some of their predicted anti-inflammatory, antidiarrheal, and anti-platelets targets and acetyl cholinesterase enzyme. a Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with arachidonate 15-lipoxygenase enzyme. The co-crystallized ligand A C8E 702 (turquoise) as a control. b Ellagic acid (dark yellow), kaempferol (pink), and quercetin (violet) with cyclooxygenase-2 enzyme. Indomethacin (turquoise) as a control. c Dicatechin with Mu-type opioid receptor. Eluxadoline (turquoise) as a control. d Dicatechin with Delta-type opioid receptor. Eluxadoline (turquoise) as a control. e 1,6-di-O-galloyl-beta-D-glucose (yellow) with P2Y12 receptor. Clopidogrel (turquoise) as a control. f Digallic acid (yellow) and leucocyanidin (blue) with acetylcholinesterase enzyme. Neostigmine (turquoise) as a control
The Pharmacokinetics and Toxicity Prediction
The intestinal absorption, volume of distribution, blood-brain barrier, p-glycoprotein and cytochrome-P enzymes inhibition, the renal OCT2 substrate probability, and total clearance were predicted via pkCSM [13] and SwissADME web servers [12]. Moreover, the hepatotoxicity, skin sensitization, the hERG potassium channel inhibition, AMES toxicity, human maximum tolerated dose, carcinogenicity, oral rate acute, and chronic toxicity were predicted via pkCSM web server [13] at the default settings via submitting of the chemical structures in smile format. The results of pharmacokinetics are listed in Tables 7 and 8 and toxicity in Table 9.
Table 8.
The predicted pharmacokinetics properties of phytochemical constituents having higher affinity scores (part B)
| Phytochemical constituent | CYP2D6 and CYP3A4 substrate | CYP enzymes inhibition | P-gp substrate | P-gp I or II inhibition |
|---|---|---|---|---|
| (−)-Epigallocatechin-5, 7-gallate | No | Non inhibitor | Substrate | P-gp II |
| (−)-Epigallocatechin-7-gallate | No | CYP1A2, CYP3A4 | Substrate | Non inhibitor |
| (+)-Catechin-3,5,-digallate | No | CYP2C9 | Substrate | P-gp II |
| (+)-Mollisacacidin | No | Non inhibitor | Substrate | Non inhibitor |
| 1,6-di-O-galloyl-beta-D-glucose | No | Non inhibitor | Substrate | Non inhibitor |
| Acacetin | No | CYP1A2, CYP2C19, CYP2C9, CYP2D6 CYP3A4 inhibitor | Substrate | Non inhibitor |
| Catechin | No | Non inhibitor | Substrate | Non inhibitor |
| Catechin-7-O-gallate | No | CYP1A2 inhibitor | substrate | P-gp II |
| Chalconaringnen-4-O-beta-glucopyranoside | No | Non inhibitor | Substrate | Non inhibitor |
| Dicatechin | No | Non inhibitor | Substrate | P-gp I and II |
| Digallic acid | No | CYP1A2, CYP3A4 inhibitor | Substrate | Non inhibitor |
| Ellagic acid | No | CYP1A2 inhibitor | Substrate | Non inhibitor |
| Epicatechin | No | Non inhibitor | Substrate | Non inhibitor |
| Flavone | CYP3A4 | CYP1A2, CYP2C19, CYP2C9, and CYP2D6 | Substrate | P-gp II |
| γ-Sitosterol | CYP3A4 | Non inhibitor | No | P-gp I and II |
| Kaempferol | No | CYP1A2 inhibitor | Substrate | Non inhibitor |
| Kaempferol-7-glucoside | No | Non inhibitor | Substrate | Non inhibitor |
| Leucocyanadin | No | Non inhibitor | Substrate | Non inhibitor |
| Lupenone | CYP3A4 | Non inhibitor | No | P-gp I and II |
| Lupeol | CYP3A4 | Non inhibitor | No | P-gp II |
| Melacacidin | No | Non inhibitor | Substrate | Non inhibitor |
| Naringenin | No | CYP1A2, CYP2C19, CYP3A4 | Substrate | P-gp II |
| Naringenin-7-O-beta-glucose | No | Non inhibitor | Substrate | P-gp II |
| Nilobamate | No | Non inhibitor | Substrate | Non inhibitor |
| Niloctane | No | Non inhibitor | Substrate | Non inhibitor |
| Quercetin | No | CYP1A2 inhibitor | Substrate | Non inhibitor |
The italic emphasis indicates desirable prosperity, the bold emphasis indicates undesirable prosperity
CYP cytochrome-P enzyme, P-gp P-glycoprotein transporter
Table 9.
The predicted toxicity of phytochemical constituents having higher affinity scores
| Phytochemical constituent | AMES tox. | hERG I or II inhibition | Hepatotoxicity | Skin sensitization | Carcinogenicity | Human maximum tolerated dose (mg/kg/day) | Oral rat acute toxicity (mol/kg) | Oral rat chronic tox. (mg/kg_bw/day) |
|---|---|---|---|---|---|---|---|---|
| (−)-Epigallocatechin-5,7-gallate | No | hERG II | No | No | No | 2.73 | 2.507 | 6.693 |
| (−)-Epigallocatechin-7-gallate | Yes | hERG II | No | No | No | 4.12 | 2.728 | 4.827 |
| (+)-Catechin-3,5,-digallate | No | hERG II | No | No | No | 2.37 | 2.506 | 6.19 |
| (+)-Mollisacacidin | Yes | No | No | No | No | 2.57 | 2.057 | 1.881 |
| 1,6-di-O-galloyl-beta-D-glucose | No | No | No | No | No | 11.83 | 2.736 | 4.203 |
| Acacetin | No | hERG II | No | No | No | 5.74 | 2.558 | 1.467 |
| Acanilol-1 | Yes | hERG II | Yes | No | No | 1.62 | 2.52 | 1.055 |
| Acanilol-2 | Yes | hERG II | No | No | No | 1.65 | 2.256 | 1.865 |
| Catechin | Yes | No | No | No | No | 2.57 | 2.057 | 1.881 |
| Chalconaringnen-4-O-beta-glucopyranoside | Yes | No | No | No | No | 6.76 | 2.633 | 3.95 |
| Dicatechin | No | hERG II | No | No | No | 1.729 | 2.463 | 4.729 |
| Digallic acid | Yes | hERG II | No | No | No | 3.86 | 2.744 | 4.743 |
| Ellagic acid | No | No | No | No | No | 5.77 | 2.401 | 2.013 |
| Epicatechin | Yes | No | No | No | No | 2.57 | 2.057 | 1.881 |
| γ-Sitosterol | No | hERG II | No | No | No | 0.46 | 2.854 | 1.085 |
| Kaempferol | No | No | No | No | No | 8.13 | 2.301 | 2.699 |
| Kaempferol-7-glucoside | Yes | hERG II | No | No | No | 7.64 | 2.468 | 4.273 |
| Leucocyanadin | Yes | No | No | No | No | 9.4 | 2.175 | 2.974 |
| Lupenone | No | hERG II | No | No | No | 3.48 | 2.353 | 1.015 |
| Lupeol | No | No | No | No | No | 0.135 | 2.712 | 1.688 |
| Melacacidin | Yes | No | No | No | No | 10.47 | 2.092 | 3.379 |
| Naringnen | No | hERG II | No | No | No | 2.36 | 2.132 | 1.995 |
| Naringnen-7-O-beta-glucopyranoside | Yes | No | No | No | No | 4.73 | 2.583 | 4.019 |
| Niloctane | No | No | No | No | No | 0.27 | 2.419 | 1.788 |
| Quercetin | No | No | No | No | No | 11.07 | 2.221 | 2.997 |
The italic emphasis indicates desirable prosperity, the bold emphasis indicates undesirable prosperity
AMES tox. AMES toxicity
Drug-Likeness and Medicinal Chemistry Friendliness Prediction
The probability of A. nilotica’s phytochemical constituents to be as drug candidates was carried via applying of Lipinski, Ghose, Veber, Egan, and Muegge filters. In addition, lead likeness and synthetic accessibility were used to predict medicinal chemistry friendliness. The prediction was carried out via SwissADME web server via submitting of the chemical structures in smile format [12]. The results are listed in Table 10.
Table 10.
The predicted lead likeness, drug likeness, and synthetic accessibility score of phytochemical constituents having higher affinity scores
| Phytochemical constituent | Lipinski rule of fivea | Ghose filtersa | Veber filtersa | Egan filtersa | Muegge filtersa | Lead likeness | Synthetic accessibility |
|---|---|---|---|---|---|---|---|
| (−)-Epigallocatchin-5,7-gallate |
3 violations (MW > 500, rotatb. bonds > 10, H-don > 5). |
3 violations (MW > 480, M R > 130, atoms > 70) |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
4 violations (MW > 600, TPSA > 150, H-acc > 10, H-don > 5) |
1 violation (MW > 350) |
5.84 |
| (−)-Epigallocatchin-7-gallate |
2 violations (rotatb. bonds > 10, H-don > 5) |
No violations |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
3 violations (TPSA > 150, H-acc > 10, H-don > 5) |
1 violation (MW > 350) |
4.07 |
| (+)-Catechin-5,7-digallate |
3 violations (MW > 500, rotatb. bonds > 10, H-don >5) |
2 violations (MW > 480, M R > 130) |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
3 violations (TPSA > 150, H-acc > 10, H-don > 5) |
1 violation (MW > 350) |
4.62 |
| (+)-Mollisacacidin | Yes | Yes | Yes | Yes | Yes | Yes | 3.50 |
| 1,6-di-O-galloyl-beta-D-glucose |
1 violation (H-don > 5) |
1 violation (WLOGP < − 0.4) |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
2 violations (TPSA > 150,, H-don > 5) |
Yes | 4.17 |
| Acacetin | Yes | Yes | Yes | Yes | Yes | Yes | 2.98 |
| Catechin | Yes | Yes | Yes | Yes | Yes | Yes | 3.50 |
| Catechin-7-O-gallate |
1 violation (H-don > 5) |
Yes |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
2 violations (TPSA > 150,, H-don > 5) |
1 violation (MW > 350) |
4.03 |
| Dicatechin |
3 violations (MW > 500, H-acc > 10, H-don > 5) |
2 violations (MW > 480, M R > 130) |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
3 violations (TPSA > 150, H-acc > 10, H-don > 5) |
1 violation (MW > 350) |
5.32 |
| Digallic acid | Yes | Yes |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
2 violations (TPSA > 150, H-don > 5) |
Yes | 2.45 |
| Ellagic acid | Yes | Yes |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
Yes | Yes | 3.17 |
| Epicatechin | Yes | Yes | Yes | Yes | Yes | Yes | 3.50 |
| γ-Sitosterol |
1 violation (MLOGP > 4.15) |
3 violations (WLOGP > 5.6, M R > 130, atoms > 70) |
Yes |
1 violation (WLOGP > 5.88) |
2 violations (XLOGP3 > 5, heteroatoms < 2) |
1 violation (MW > 350, XLOGP > 3.5) |
6.30 |
| Kaempferol | Yes | Yes | Yes | Yes | Yes | Yes | 3.14 |
| Kaempferol-7-glucoside |
2 violations (H-acc > 10, H-don > 5) |
Yes |
1 violation (TPSA > 140) |
1 violation (TPSA > 131.6) |
3 violations (TPSA > 150,, H-acc > 10, H-don > 5). |
1 violation (MW > 350) |
5.24 |
| Leucocyanadin | Yes | Yes | Yes | Yes |
1 violation (H-don > 5). |
Yes | 3.76 |
| Melacacidin |
1 violation (H-don > 5). |
Yes | Yes | Yes |
1 violation (H-don > 5). |
Yes | 3.75 |
| Naringnen | Yes | Yes | Yes | Yes | Yes | Yes | 3.01 |
| Niloctane | Yes | Yes | Yes | Yes | Yes | Yes | 5.37 |
| Quercetin | Yes | Yes | Yes | Yes | Yes | Yes | 3.23 |
The synthetic accessibility is from 1 (very easy) to 10 (very difficult). The bold indicates desirable prosperity, the Italic indicates undesirable prosperity as well as the bold-italic indicates the best compounds
MW molecular weight, rotatb. bonds rotatable bonds, M R molar refractivity, H-don hydrogen bond donors, H-acc hydrogen bond acceptors, TPSA topological polar surface area
aLipinski rule of five, Ghose, Veber, Egan, and Muegge describe the relationship between the pharmacokinetic and physiochemical parameters. The parameters including molecular weight, number of rotatable bonds, molar refractivity, number of hydrogen bond donors and acceptors, as well as the topological polar surface area. Each parameter has a specific range that the structure must not be under or above it to be free from violation [12]
Methods Validation
The consistency and the reproducibility of the used tools including the molecular docking were validated by the resubmission of the compounds for many times.
Results
The Anticancer Targets
The anticancer activity of A. nilotica’s was attributed to the suppression of the oncogenic transformations, progression, and development, DNA replication, and transcription. Moreover, the prevention of cancer cells proliferation, invasion, angiogenesis as well as the suppression of drug resistance and the induction of apoptosis.
The anti-breast cancer activity was due to the inhibition of the aromatase enzyme and estrogen receptor beta. In contrast, the anti-prostate cancer activity is due to the control of metastatic behavior of prostate cancer via the interaction with nuclear receptor ROR-alpha and the inhibition of Steroid 17 alpha-hydroxylase (Table 1 and Fig. 2).
.
The Antibacterial Targets
The antibacterial activity of A. nilotica was attributed to the prevention of fatty acids, peptidoglycans biosynthesis as well as the prevention of bacterial resistance to the beta-lactam antibiotics. The fatty acid biosynthesis inhibitory activity was against different types of bacteria including Mycobacterium, Pseudomonas aeruginosa, and Vibrio cholera.
The Antiviral and Antiplasmodial Targets
The antiviral activity was attributed to the action on toll-like receptor 9. The anti-HIV activity is due to the inhibition of HIV integrase enzyme. The anti-coronavirus activity is due to coronavirus replicase polyprotein 1 ab enzyme. The antiplasmodial activity was attributed to the inhibition of enzymes MO15-related protein kinase pfmrk and M18 aspartyl aminopeptidase as well as the prevention of fatty acid biosynthesis via inhibition of the enzymes: β-hydroxy acyl-ACP dehydratase FabZ and hydroxyacyl-[acyl-carrier-Protein] dehydratase (Table 3).
The Antidiabetic Targets
The antidiabetic activity was attributed to the interaction with the insulin receptor, glycogen phosphorylase enzyme, sodium/glucose co-transporter 2 as well as the aldose reductase enzyme (Table 4).
The Anti-Inflammatory Targets
The anti-inflammatory activity was attributed to the inhibition of enzymes: arachidonate 15-lipoxygenase, cyclooxygenase-2 (COX-2), phospholipase A2, receptor-interacting serine/threonine protein kinase 2, and xanthine dehydrogenase/oxidase as well as the interaction with macrophage migration inhibitory factor (Table 5).
The Antidiarrheal, Anti-Platelets, and Anticholinesterase Targets
The antidiarrheal activity was attributed to the interaction with the opioid receptors Mu-type Delta-type. The Anti-platelets activity was attributed to the interaction with the P2Y12 receptor. The inhibition of the enzyme acetylcholinesterase is contributed to the anticholinesterase activity.
The Predicted Pharmacokinetic Properties
According to the results, Acacetin, γ-Sitosterol, Kaempferol, Flavone, Lupenone, Lupeol, Niloctane, and Quercetin had the highest gastrointestinal absorption, tissue distribution (Vd), and respectable total clearance.
Moreover, Flavone, Nilobamate, and Niloctane were permeable to the blood-brain barrier (BBB). Besides, acanilol-1, acanilol-2, γ-sitosterol, flavone, lupenone, and lupeol were found to be subjected to the metabolism via CYP3A4 enzyme (Table 7).
Moreover, (+)-Mollisacacidin, Catechin, Chalconaringnen-4-O-beta-glucopyranoside, Epicatechin, Niloticane, Kaempferol-7-glucoside, Leucocyanidin, and Nilobamate were free from drug-drug interaction via the inhibition of cytochrome-P (CYP) or P-glycoprotein (P-gp) I and II enzymes (Table 8).
The Predicted Toxicity
According to the results, 1,6-di-O-galloyl-beta-D-glucose, ellagic acid, kaempferol, and quercetin were non-toxic as well as non-carcinogen (Table 9).
Drug-Likeness, Lead-Likeness, and Synthetic Accessibility Prediction
According to the results, (+)-Mollisacacidin, Acacetin, Catechin, Epicatechin, Kaempferol, Naringenin, Niloctane, and Quercetin were found to be the best lead and drug candidates with good synthetic accessibility, followed by Digallic acid, Ellagic acid, Leucocyanidin, and Melacacidin (Table 10).
Discussion
Despite the enormous conducted studies on the pharmacology activity of A. nilotica’s [1], the determination of the target that contribute to its activity and the understanding of the mechanism of action as well as to assess the pharmacokinetics, safety, and the drug-likeness probability are important issues that were not conducted yet. Such studies are required to bring the plant in the drug discovery pipeline so as to design a novel drug with broad-spectrum of therapeutic activity and safety.
To identify the targets, TargetNet web servers that utilize a QSAR model based on the chemogenomic data as a predictive algorithm [25] and Similarity Ensemble Search Tool [24] were used. To validate the predicted target from the web servers, a molecular docking study was performed using Cresset Flare software [23] that uses the Lead finder program [69] for docking calculation. Moreover, pkCSM [13] and SwissADME web servers [12] were used to predict the pharmacokinetics (ADME: Absorption, Distribution, Metabolism, and Elimination), toxicity, and the drug-likeness probability.
The total predicted targets form the virtual screening with the highest probability that was validated by the molecular docking were 61 targets.
The interaction of Acacetin with the cell division control protein 42 homolog (CDC42) will prevent the oncogenic transformations. The inhibition of enzymes—anaplastic lymphoma kinase by Quercetin, cyclin-dependent kinases 1, 4, and 6 by Ellagic acid and Acacetin, Aurora A and B by Ellagic acid and Quercetin, serine/threonine protein kinase Nek2 by Quercetin, proto-oncogene tyrosine-protein Kinase Src by Ellagic acid, tankyrase 1 and 2 by Acacetin as well as M phase inducer phosphatase by Digallic acid, Epicatechin, and Kaempferol—will prevent the cancer progression and development.
Moreover, the inhibition of the enzymes—cell division cycle-7-related protein kinase by Ellagic acid, serine/threonine-protein kinase pim-1 by Quercetin, Ellagic acid, and Kaempferol as well as DNA topoisomerase 1 by Kaempferol-7-glucoside and 1,6-di-O-galloyl-beta-D-glucose—will suppress the DNA replication; the inhibition of enzymes—cyclin-dependent kinase 9 by Acacetin and telomerase reverse transcriptase by Leucocyanidin, Quercetin, Ellagic acid, and Kaempferol—will suppress the transcription; the inhibition of tyrosine-protein kinase Lyn enzyme by Ellagic acid will prevent the cancer cells proliferation; as well as the inhibition of angiopoietin-1 receptor, proto-oncogene tyrosine-protein kinase Src by Ellagic acid, and protein kinase C epsilon by kaempferol and Naringnen will suppress the cancer cells invasion.
Furthermore, the inhibition of ephrin type B receptor 4 and platelet-derived growth factor 1 receptor by Ellagic acid, vascular endothelial growth factor receptor 3 by Quercetin, as well as focal adhesion kinase enzyme Ellagic acid and Quercetin will suppress the angiogenesis.
The inhibition of P-glycoprotein 1, 3 transporters by Kaempferol-7-glucoside, Chalconaringnen-4-O-beta-glucopyranoside, Kaempferol, and Quercetin as well as ATP binding cassette sub-family G member 2 by (+)-Catechin-3, 5-digallate, Chalconaringnen-4-O-beta-glucopyranoside, Leucocyanidin, Quercetin, Ellagic acid, and Kaempferol will suppress the cancer cells resistance.
The interaction of Lupeol, Quercetin, Ellagic acid, and Kaempferol with the enzyme inducible nitric oxide synthase on macrophage will promote a tumoricidal action. The inhibition of the enzymes—Bcl-2-related protein A1 by Leucocyanidin, Quercetin, Ellagic acid, and Kaempferol, the induced myeloid leukemia differentiation protein MCL-1 by Acacetin, Quercetin, Ellagic acid, and Kaempferol as well as the interaction with enzymes: caspase 9 by (−)-Epigallocatechin-7-gallate, Quercetin, Ellagic acid, and Kaempferol, death-associated protein kinase 1 by Kaempferol and Quercetin—will induce cancer cell apoptosis. Consequently, those compounds show substantial anticancer activity (Table 1).
The interaction of Kaempferol and Quercetin with the enzymes 3-oxyacyl-[acyl carrier protein] reductase FabG and the interaction of (−)-Epigallocatechin-7-gallate, (+)-Catechin-4, 5-digallate, and Quercetin with enoyl-acyl carrier protein reductase will inhibit the bacterial fatty acids biosynthesis that is essential in the formation of bacterial membrane phospholipids [70] leading to “an impairment in the cellular envelope structure and function, the ability to form biofilms as well as increasing the susceptibility to the environmental stress” [71]. Moreover, the inhibition of the enzyme D-alanine D-alanine ligase by Quercetin will suppress the peptidoglycans biosynthesis that is vital in bacterial cell structure causing loss of bacterial cell integrity [72]. Therefore, those compounds have significant antibacterial activity (Table 2).
The interaction of Leucocyanidin, Ellagic acid, Kaempferol, and Quercetin with Toll-like receptor 9 will activate this innate immune receptor that helps in the recognition of microbial DNA [53]. The interaction of Digallic acid, Acacetin, Ellagic acid, Kaempferol, and Quercetin with HIV integrase enzyme will inhibit the viral DNA integration into host DNA leading to the suppression of replication cycle [52]. The interaction of Quercetin with Coronavirus replicase polyprotein 1 ab will inhibit the transcription and replication of viral RNAs [26]. Thus, those A. nilotica’s phytochemical constituents exhibit considerable antiviral activity (Table 3).
The interaction of Acacetin with the enzyme MO15-related protein kinase pfmrk will disrupt the regulation of plasmodial cell cycle [56], and the interaction of Quercetin and (+)-Mollisacacidin with the enzyme M18 aspartyl aminopeptidase will prevent the invasion in host erythrocyte and the degradation of host hemoglobin [57]. Furthermore, the interaction of Quercetin with the plasmodial enzymes β-hydroxy acyl-ACP dehydratase FabZ and hydroxyacyl-[acyl-carrier-Protein] dehydratase will inhibit the fatty acid biosynthesis [54, 55] that are important for plasmodial membrane [73]. Subsequently, those A. nilotica’s phytochemical constituents show considerable antiplasmodial activity (Table 3).
The interaction of Ellagic acid with the insulin receptor will promote glucose uptake that lowers the blood glucose level [74]. The interaction of Quercetin with the enzyme glycogen phosphorylase will inhibit the glycogenolysis that reduces the hyperglycemia [75]. Moreover, the interaction of 1,6-di-O-galloyl-beta-D-glucose and Chalconaringnen-4-O-beta-glucopyranoside with the sodium/glucose co-transporter 2 will inhibit the renal glucose reabsorption leading to a reduction in plasma glucose level [60], the interaction of Dicatechin, Kaempferol-7-glucoside, Leucocyanidin, Ellagic acid, Kaempferol, and Quercetin with the aldose reductase enzyme will suppress the development of the secondary Diabetic complications [61], as well as the interaction of Ellagic acid and Quercetin with the beta-secretase enzyme will upregulate the insulin receptors in the liver [62]. Successively, those A. nilotica’s phytochemical constituents have significant antidiabetic activity (Table 4).
The interaction of (−)-Epigallocatechin-7-gallate, Ellagic acid, Kaempferol, and Quercetin with the enzyme arachidonate 15-lipoxygenase and the interaction of Digallic acid, Acacetin, Ellagic acid, Kaempferol, and Quercetin with the receptor-interacting serine/threonine protein kinase 2 will interrupt the inflammatory responses [16, 63].
The interaction of (+)-Mollisacacidin, Naringnen, Ellagic acid, Kaempferol, and Quercetin with Cycloxygenase-2 enzyme (COX-2) will prevent the formation of inflammatory mediators prostaglandins [62], and the interaction of Digallic acid, Kaempferol, and Quercetin with Phospholipase A2 enzyme will prevent the initiation of the inflammatory response [16]. Moreover, the interaction of 1, 6-di-O-galloyl-beta-D-glucose, Digallic acid, Acacetin, Ellagic acid, Kaempferol, and Quercetin with the xanthine dehydrogenase/oxidase enzyme will inhibit the formation of uric acid and reactive oxygen species [16]. Thus, those of A. nilotica’s phytochemical constituents exhibit considerable anti-inflammatory activity (Table 5).
The interaction of Dicatechin with the mu and delta opioid receptors will lead to antisecretory and anti-transit action that will inhibit diarrhea [76]. The interaction of 1,6-di-O-galloyl-beta-D-glucose with P2Y12 receptor will inhibit the platelet activation [65]; consequently, they have considerable antidiarrheal and anti-platelets activity, respectively (Table 6).
The chemogenomic-based QSAR models of TargetNet web server were strictly evaluated and validated leading to respected screening results [25]. Furthermore, the lead finder program [69] on Cresset flare software [23] is characterized by the combination between the genetic algorithm and different optimization strategies leading to great efficiency, robustness, accuracy, and speed of calculations [69]. Musab Ibrahim et al. [77] found the results of a molecular docking study about novel synthesized COX enzyme inhibitors conducted in Cresset Flare software were aligned with results of the conducted in vivo study. Depending on that, the obtained results of the predicted targets could be with considerable accuracy.
Since the pharmacological activity does not depend only on the pharmacodynamic properties, but also on the pharmacokinetic properties. Moreover, as the drug safety, the assessment of drug-likeness probability, and the synthetic accessibility are important issues [78], the identification of the best A. nilotica’s phytochemical constituents will be attained by the assessment of those issues collectively.
The pharmacokinetics is concerning the study of the entrance, movement, changing, and leaving of the drug to the body [79]. The higher absorption from the gastrointestinal tract leads to higher drug concentration on the blood, the higher volume of distribution provides higher supply to the body tissues, and the adequate metabolism and elimination prevent the accumulation of the drug in the body, hence reduce the toxicity [79]. Consequently, the consideration of the pharmacokinetics in drug design is an essential task [80].
For instance, Lupenone is highly lipophilic; hence, it has higher absorption percent (100%); in contrast, the hydrophilic groups of Ellagic acid reduce it absorption percent to 76.935%, however, still it as a high absorption percent. The higher absorption will make Lupenone is highly bioavailable. Niloctane is permeable to BBB; therefore, its concentration that reaches the brain targets is more than Ellagic acid that is not permeable to the BBB. The predicted volume of distribution (Vd) of Melacacidin (4.7 L/kg) is the highest one, meaning that it has the highest distribution in body tissues. In contrary, Nilobamate has the highest predicted total clearance, meaning that it is the fastest one that eliminated from the body (Table 7).
Moreover, the inhibition of cytochrome-P enzyme CYP1A2 by Ellagic acid will decrease the biotransformation of drugs that metabolized by it leading to increase in the concentration of them that may increase the side effects; consequently, the drug-drug interaction must be in consideration. The binding of Dicatechin with the P-glycoprotein may decrease the transportation of drugs transported by this transporter and may involve in the drug resistance by the pumping out mechanism (Table 8).
Furthermore, the predicted AMES toxicity of Epicatechin will lead to genotoxicity and mutagenicity [81], the predicted hERG II potassium channel inhibitory effect of Acacetin “prolongs the QT interval in ECG that increases the risk for potentially fatal ventricular arrhythmias” [82]; subsequently, such drugs will not be considered as drug-likeness candidate (Table 9).
Besides, Quercetin has no violation in Lipinski rule of five; hence, it will a good candidate as an orally active drug as well as it has no violation in Ghose, Veber, and Egan filters; therefore, it will be a good lead-likeness candidate [12] (Table 10).
According to the results of pharmacodynamics, pharmacokinetics, safety, and drug-likeness predictions collectively, Ellagic acid, Kaempferol, and Quercetin were the best A. nilotica’s phytochemical constituents that contribute to the therapeutic activities. The 3D interaction with their predicted targets demonstrates marked ligand superimposing with the control compounds (e.g., Figs. 1a, b, 2e, and 5b); however, it may at the same active site without ligand superimposing (e.g., Figs. 1c and 2c). Ellagic acid interacts with Aurora A kinase enzyme with two binding modes (Fig. 1d). Ellagic acid, Kaempferol, and Quercetin interact with Steroid 17 alpha-hydroxylase enzyme at a binding mode that differs from the binding mode of control Galeterone (Fig. 1f).
They were followed by (+)-Mollisacacidin, Epicatechin, and Melacacidin, those of their predicted AMES toxicity decreased their rank. The predicted hERG II potassium channel inhibitory effect of Acacetin decreased its rank; however, it has good pharmacodynamics and pharmacokinetics profile.
Despite the efficient pharmacodynamics and the respectable safety profile of Ellagic acid, Kaempferol, and Quercetin, practically, each compound suffers from the low bioavailability [83–85], albeit the predicted intestinal absorption of them is high (Table 7). The reduced bioavailability of Ellagic acid is attributed to the poor absorption and rapid elimination from the body [86] (the predicted total clearance of Ellagic acid is high). The higher topological polar surface area (TPSA) (Table 10) contributes to the poor absorption. The reduced absorption of Kaempferol is attributed to the larger particle size and poor water solubility [83]. The reduced bioavailability of Quercetin is attributed to “the poor solubility and crystalline form at body temperature” [85].
Moreover, Ellagic acid, Kaempferol, and Quercetin have many polar phenolic hydroxyl groups (structure l, o, and x); consequently, they are subjected to direct glucuronide conjugation with as phase II metabolism. “Kaempferol and Quercetin are rapidly excreted in urine as glucuronides mainly” [87].
The reduced bioavailability affects pharmacological activity. Hence, to maintain the pharmacological activity, the bioavailability must be enhanced. The nano-suspension form of Kaempferol is increased its absorption and bioavailability [83]. The administration of Isoquercetin (Quercetin-3-glucoside) increases the absorption and bioavailability of Quercetin [88]. The bioavailability Ellagic acid, Kaempferol, and Quercetin is increased bypassing the entero-hepatic phase II conjugation (e.g., formation of ester derivatives) and by using novel drug delivery systems as the liposomes. Furthermore, the co-administration of Ginkgo biloba extract with Kaempferol and Quercetin increased the bioavailability of them [89]. Consequently, the combination of Ellagic acid, Kaempferol, and Quercetin will be optimum treatment choice that maximizes the therapeutic activity and the safety profile as well as overwhelms the limits in the bioavailability. As they naturally are available in one plant, the combination of them at the therapeutic doses will be additive and will not induce drug-drug interactions. The design of multi-target drug is an effective promising approach for the treatment of complex disease [90].
The computational methods including the virtual screening are not to substitute the in vitro and in vivo methods, however, to reduce the time, cost, and the difficultness in the drug target identification [91]. Therefore, this study is an attempt to identify the best A. nilotica’s phytochemical constituents that contribute to its pharmacological activity as well as their targets. It is not meaning that this study alone will be sufficient to judge about the result; however, experimental studies are required to validate the results.
Conclusion
According to the results of pharmacodynamics, pharmacokinetics, safety, and drug-likeness predictions collectively, Ellagic acid, Kaempferol, and Quercetin were the best A. nilotica’s phytochemical constituents that contribute to the therapeutic activities; consequently, we recommend the use of Ellagic acid, Kaempferol, and Quercetin as a combined drug via the novel drug delivery systems for the treatment of recent diseases attacking the public health including cancer, multidrug-resistant bacterial infections, diabetes mellitus, and chronic inflammatory systemic diseases. Moreover, we recommend wet lab studies to validate the results.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that having no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subject performed by any of the authors
Footnotes
This article is part of the Topical Collection on Natural Products: From Chemistry to Pharmacology
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Islam SU, Rather LJ, Mohammad F. Acacia nilotica (L.): A review of its traditional uses, phytochemistry, and pharmacology. Sustain Chem Pharm. 2015;8:2352–5541. [Google Scholar]
- 2.Shakya AK. Medicinal plants: future source of new drugs. Int J Herbal Med. 2016;4:59–64. [Google Scholar]
- 3.Ghulam Mustafa RA, Atta A, Sharif S, Jamil A. Bioactive compounds from medicinal plants and their importance in drug discovery in Pakistan. Matrix Sci Pharma. 2017;1:17–26. doi: 10.26480/msp.01.2017.17.26. [DOI] [Google Scholar]
- 4.N. C. Institute. Cancer statistics. www.cancer.gov. Accessed April 2018.
- 5.Barker JJ. Antibacterial drug discovery and structure-based design. Drug Discov Today. 2006;11:391–404. doi: 10.1016/j.drudis.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 6.A. D. Association. Statistics about diabetes. www.diabetes.org, Accessed March 2018.
- 7.Straub RH, Schradin C. Chronic inflammatory systemic diseases an evolutionary trade-off between acutely beneficial but chronically harmful programs. Evol Med Public Health. 2016;2016:73–51. doi: 10.1093/emph/eow001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eweas AF, Maghrabi IA, Namarneh AI. Advances in molecular modeling and docking as a tool for modern drug discovery. Sch Res Lib Der Pharma Chem. 2014;6:211–228. [Google Scholar]
- 9.Saher Afshan Shaikh TJ. Sandhu G, Soni A, Jayaram B. From drug target to leads-sketching a physico-chemical pathway for lead molecule design in silico. Front Med Chem. 2011;6. [DOI] [PubMed]
- 10.Banegas-Luna AJ, Ceron-Carrasco JP, Perez-Sanchez H. A review of ligand-based virtual screening web tools and screening algorithms in large molecular databases in the age of big data. Future Med Chem. 2018;10:2641–2658. doi: 10.4155/fmc-2018-0076. [DOI] [PubMed] [Google Scholar]
- 11.Guo ZYL, Zheng X, Hu L, Yang Y, Wang JA. A comparison of various optimization algorithms of protein-ligand docking programs by fitness accuracy. J Mol Model. 2014;20:2251–2261. doi: 10.1007/s00894-014-2251-3. [DOI] [PubMed] [Google Scholar]
- 12.Daina A. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pires DEV, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh S, Awasthi M, Tiwari S, Pandey VP, Dwivedi UN. Computational approaches for therapeutic application of natural products in Alzheimer’s disease. Neuromethods. 2018;132:483–511. doi: 10.1007/978-1-4939-7404-7_17. [DOI] [Google Scholar]
- 15.Wadhwa B, Mahajan P, Barik MR, Malik F, Nargotra A. Combining ligand- and structure-based in silico methods for the identification of natural product-based inhibitors of Akt1. Mol Divers. 2019. [DOI] [PubMed]
- 16.Pereira F, Aires-de-Sousa J. Computational methodologies in the exploration of the marine natural product leads. Mar Drugs. 2018;16. [DOI] [PMC free article] [PubMed]
- 17.Worachartcheewan A, Prachayasittikul V, Shoombuatong W, Songtawee N, Simeon S, Prachayasittikul V, et al. Computer-aided drug design of bioactive natural products. Curr Top Med Chem. 2015;15:1780–1800. doi: 10.2174/1568026615666150506151303. [DOI] [PubMed] [Google Scholar]
- 18.Jones LH. An industry perspective on drug target validation. Exp Opin Drug Discovery. 2016;11:623–625. doi: 10.1080/17460441.2016.1182484. [DOI] [PubMed] [Google Scholar]
- 19.Xuan-Yu Meng H-XZ, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pires DEV, Kaminskas LM, Ascher DB. Prediction and optimization of pharmacokinetic and toxicity properties of the ligand. Methods Mol Biol. 2018;1762:271–284. doi: 10.1007/978-1-4939-7756-7_14. [DOI] [PubMed] [Google Scholar]
- 21.Marvin Sketch, www.chemaxon.com/products/marvin. Available: www.chemaxon.com/products/marvin.
- 22.Noel MB, Boyle MO’, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3. [DOI] [PMC free article] [PubMed]
- 23.Cheeseright T, Mackey M, Rose S, Vinter A. Molecular field extrema as descriptors of biological activity: definition and validation. J Chem Inf Model. 2006;46:665–676. doi: 10.1021/ci050357s. [DOI] [PubMed] [Google Scholar]
- 24.Keiser MJ RB, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 25.Zhi-Jiang JY, Che Y-J, Zhu MF, Wen M, Lu A-P, Cao D-S. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J Comput Aided Mol Des. 2016;30(5):413–424. doi: 10.1007/s10822-016-9915-2. [DOI] [PubMed] [Google Scholar]
- 26.T. N. Consortium. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2008;36. [DOI] [PMC free article] [PubMed]
- 27.Nguyen D-T, Mathias S, et al. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45:D995–D1002. doi: 10.1093/nar/gkw1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YH, Yu YY, Li XX, Zhang P, Tang J, Yang QX, Yang QX, Fu TT, Cui XJ, Tu G, Zhang Y, Li S, Yang FY, Sun Q, Qin C, Zeng X, Chen Z, Chen YZ, Zhu F. Therapeutic target database update 2018: enrich resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46:D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anna Frenzel FG, Chmielewski W, Villunger A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 2012;14:584–596. doi: 10.1007/s10495-008-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim B, Srivastava SK, Kim SH. Caspase-9 as a therapeutic target for treating cancer. Expert Opin Ther Targets. 2015;19:113–127. doi: 10.1517/14728222.2014.961425. [DOI] [PubMed] [Google Scholar]
- 31.Stengel K, Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011;23:1415–1423. doi: 10.1016/j.cellsig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montagnoli A, Moll J, Colotta F. Targeting cell division cycle 7 kinase: a new approach for cancer therapy. Clin Cancer Res. 2010;15(16):4503–4508. doi: 10.1158/1078-0432.CCR-10-0185. [DOI] [PubMed] [Google Scholar]
- 33.Jian Kang CMS, Sutherland RL, Musgrove EA. Targeting cyclin-dependent kinase 1 (CDK1) but not CDK4/6 or CDK2 is selectively lethal to MYC-dependent human breast cancer cells. BMC Cancer. 2014;14:32. doi: 10.1186/1471-2407-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lock RB, Ross WE. DNA topoisomerases in cancer therapy. Anticancer Drug Des. 1987;2:151–164. [PubMed] [Google Scholar]
- 35.Gallo D, De Stefano I, Grazia Prisco M, Scambia G, Ferrandina G. Estrogen receptor beta in cancer: an attractive target for therapy. Curr Pharm Des. 2012;18:2734–2757. doi: 10.2174/138161212800626139. [DOI] [PubMed] [Google Scholar]
- 36.Yoshino Y, Ishioka C. Inhibition of glycogen synthase kinase-3 beta induces apoptosis and mitotic catastrophe by disrupting centrosome regulation in cancer cells. Sci Rep. 2015;5:13249. doi: 10.1038/srep13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farina AR, Mackay AR. Gelatinase B/MMP-9 in tumour pathogenesis and progression. Cancers (Basel) 2014;6:240–296. doi: 10.3390/cancers6010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George Lund SD, Borkin D, Ni W, Grembecka J, Cierpicki T. Inhibition of CDC25B phosphatase through disruption of protein-protein interactions. ACS Chem Biol. 2015;10:390–394. doi: 10.1021/cb500883h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moretti RM, Montagnani Marelli M, Motta M, Limonata P. Role of the orphan nuclear receptor ROR alpha in the control of the metastatic behavior of androgen-independent prostate cancer. Oncol Rep. 2002;9:1139–1143. [PubMed] [Google Scholar]
- 40.Lehne G. P-glycoprotein as a drug target in the treatment of multidrug-resistant cancer. Curr Drug Targets. 2000;1:85–99. doi: 10.2174/1389450003349443. [DOI] [PubMed] [Google Scholar]
- 41.Roskoski R., Jr The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;129:65–83. doi: 10.1016/j.phrs.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 42.Roskoski Jr R. Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 43.Sivonova MK, Jurecekova J, Tatarkova Z, Kaplan P, Lichardusova L, Hatok J. The role of CYP17A1 in prostate cancer development: structure, function, mechanism of action, genetic variations and its inhibition. Gen Physiol Biophys. 2017;36:487–499. doi: 10.4149/gpb_2017024. [DOI] [PubMed] [Google Scholar]
- 44.Lehtio L, Chi NW, Krauss S. Tankyrases as drug targets. FEBS J. 2013;280:3576–3593. doi: 10.1111/febs.12320. [DOI] [PubMed] [Google Scholar]
- 45.Leao R, Apolonio JD, Lee D, Figueiredo A, Tabori U, Castelo-Branco P. Mechanism oh human telomerase reverse transcriptase (hTERT) regulation: clinical impact in cancer. J Biomed Sci. 2018;25:22. doi: 10.1186/s12929-018-0422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia Y, Shen S, Verma IM. NF-kB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldenberg-Furmanov M, Stein I, Pikarsky E, Rubin H, Kasem S, Wygoda M, Weinstein I, Reuveniand H, Ben-Sasson SA. Lyn is a target gene for prostate cancer sequence-based inhibition induces regression of human tumor xenografts. Cancer Res. 2004. [DOI] [PubMed]
- 48.Neufeld G, Cohen T, Gengrinovitich S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13:9–22. doi: 10.1096/fasebj.13.1.9. [DOI] [PubMed] [Google Scholar]
- 49.Torsten Schwede JK, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zitko J, Dolezal M. Enoyl-acyl carrier protein reductase inhibitors:n an updated patent review. Expert Opin Ther Pat. 2016;26:1079–1094. doi: 10.1080/13543776.2016.1211112. [DOI] [PubMed] [Google Scholar]
- 51.Hrast M, Vehar B, Turk S, Kong J, Gobec S, Janezic D. Function of the D-alanine:D-alanine ligase lid loop: a molecular modeling and bioactivity study. J Med Chem. 2012;55:6849–6856. doi: 10.1021/jm3006965. [DOI] [PubMed] [Google Scholar]
- 52.Craigie R. The molecular biology of HIV integrase. Future Virol. 2012;7:679–686. doi: 10.2217/fvl.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hemmi A, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 54.Kostrewa D, Winkler F, Folkers G, Scapozza L, Perozzo R. The crystal structure of PfFabZ, the unique β-hydroxy acyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci. 2005;14:1570–1580. doi: 10.1110/ps.051373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, Lou C, Han C, Peng S, Yang Y, Yue J, Shen X, Jiang H. A new β-hydroxy acyl-acyl carrier protein dehydratase (fabZ) from Helicobacter pylori: molecular cloning, enzymatic characterization, and structural modeling. Biochem Biophys Res Commun. 2005;333:1078–1086. doi: 10.1016/j.bbrc.2005.05.197. [DOI] [PubMed] [Google Scholar]
- 56.Li J-L, Robson KJH, Chen J-L, Targett GAT, Baker DA. Pfmrk, a MO15-related protein kinase from Plasmodium falciparum. Eur J Biochem. 1996;241:805–813. doi: 10.1111/j.1432-1033.1996.00805.x. [DOI] [PubMed] [Google Scholar]
- 57.Kumari M, Chandra S, Tiwari N, Subbarao N. 3D QSAR, pharmacophore and molecular docking studies of known inhibitors and designing of novel inhibitors for M18 aspartyl aminopeptidase of Plasmodium falciparum. BMC Struct Biol. 2016;16:12. doi: 10.1186/s12900-016-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley LA, et al. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng J, Xu J. RaptorX: exploiting structure information for protein alignment by statistical inference. Proteins. 2011;79(Suppl 10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mudaliar S, Polidori D, Zambrowicz B, Henry RR. Sodium-glucose cotransporter inhibitors: effects on renal and intestinal glucose transport from bench to beside. Diabetes Care. 2015;38:2344–2353. doi: 10.2337/dc15-0642. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava SK, Ramana KV, Bhatnagar A. Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev. 2005;26:380–392. doi: 10.1210/er.2004-0028. [DOI] [PubMed] [Google Scholar]
- 62.Paul AM, Meakin J, Benabou E, Haas ME, Bonardo B, Grino M, Brunel J-M, Desbois-Mouthon C, Biddinger SB, Govers R, Ashford MLJ, Peiretti F. The beta-secretase BACE1 regulates the expression of the insulin receptor in the liver. Nat Commun. 2018;9:1306. doi: 10.1038/s41467-018-03755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tigno-Aranjuez JT, Benderitter P, Rombouts F, Deroose F, Bai X, Mattioli B, Cominelli F, Pizarro TT, Hoflack J, Abbott DW. In vivo inhibition of RIPK2 kinase alleviates inflammatory disease. J Biol Chem. 2014;289:29651–29664. doi: 10.1074/jbc.M114.591388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shook JE, Lemcke PK, Gehrig CA, Hruby VJ, Burks RF. Antidiarrheal properties of supraspinal mu and delta and kappa opioid receptors: inhibition of diarrhea without constipation. J Pharmacol Exp Ther. 1989;249:83–90. [PubMed] [Google Scholar]
- 65.Dorsam RT, Kunapuli SP. Central role of the P2Y12 receptor in platelet activation. J Clin Invest. 2004;113:340–345. doi: 10.1172/JCI20986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colovic MB, Krstic DZ, Lazarevic-Pasti TD, Bondzic AM, Vasic VM. Acetylcholinesterase inhibitors: pharmacology and toxicology. Curr Neuropharmacol. 2013;11:315–335. doi: 10.2174/1570159X11311030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berman JWHM, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hacker K, Maas R, Kornhuber J, Fromm MF, Zolk O. Substrate-dependent inhibition of the human organic cation transporter OCT2: a comparison of metformin with experimental substrates. PLoS One. 2015;10:e0136451. doi: 10.1371/journal.pone.0136451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strognov OV, Novikov FN, Stroylov VS, Kulkov V, Chilov GG. Lead finder: an approach to improve the accuracy of protein-ligand docking, binding energy estimation, and virtual screening. J Chem Inf Model. 2008;48:2371–2385. doi: 10.1021/ci800166p. [DOI] [PubMed] [Google Scholar]
- 70.Cronan JE, Thomas J. Bacterial fatty acids synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowlett VW, VKPS M, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, et al. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol. 2017. [DOI] [PMC free article] [PubMed]
- 72.Heijenoort J. Formation of glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25R. [DOI] [PubMed] [Google Scholar]
- 73.van Schaijk BC, Kumar TR, Vos MW, Richman A, van Gemert GJ, Li T, Eappen AG, Williamson KC, Morahan BJ, Fishbaugher M, Kennedy M, Camargo N, Khan SM, Janse CJ, Sim KL, Hoffman SL, Kappe SH, Sauerwein RW, Fidock DA, Vaughan AM. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot Cell. 2014;13:550–559. doi: 10.1128/EC.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiang G, Yue S, Yang JJ, Du G, Pang X, Li X, Goswami D, Griffin PR, Ortlund EA, Chan CB, Ye K. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes. 2014;63:1394–1409. doi: 10.2337/db13-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baker DJ, Timmons JA, Greenhaff PL. Glycogen phosphorylase inhibition in type 2 diabetes therapy: a systematic evaluation of metabolic and functional effects in rat skeletal muscles. Diabetes. 2005;54:2453–2459. doi: 10.2337/diabetes.54.8.2453. [DOI] [PubMed] [Google Scholar]
- 76.Holzer P. Opioid receptors in the gastrointestinal tract. Regul Pept. 2009;155:11–17. doi: 10.1016/j.regpep.2009.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ibrahim MM, Elsaman T, Al-Nour MY. Synthesis, anti-inflammatory activity, and in silico study of novel diclofenac and isatin conjugates. Int J Med Chem. 2018;2018:9139786. doi: 10.1155/2018/9139786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morimoto BH, Castelloe E, Fox AW. Safety pharmacology in drug discovery and development. Handb Exp Pharmacol. 2015;229:65–80. doi: 10.1007/978-3-662-46943-9_3. [DOI] [PubMed] [Google Scholar]
- 79.Doogue MP, Polasek TM. The ABCD of clinical pharmacokinetics. Ther Adv Drug Saf. 2013;4:5–7. doi: 10.1177/2042098612469335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hassan M, Sallam H, Hassan Z. The role of pharmacokinetics and pharmacodynamics in early drug development with reference to the cyclin-dependent kinase (Cdk) inhibitor-roscovitine. Sultan Qaboos Univ Med J. 2011;11:165–178. [PMC free article] [PubMed] [Google Scholar]
- 81.McCarren P, Springer C, Whitehead L. An investigation into pharmaceutically relevant mutagenicity data and the influence on Ames predictive potential. J Cheminform. 2011;3:51. doi: 10.1186/1758-2946-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Priest BT, Bell IM, Garcia ML. Role of hERG potassium channel assays in drug development. Channels. 2008;2:87–93. doi: 10.4161/chan.2.2.6004. [DOI] [PubMed] [Google Scholar]
- 83.Qian YS, Ramamurthy S, Candasamy M, Shadab M, Kumar RG, Meka VS. Production, characterization, and evaluation of kaemferol nanosuspension for improving oral bioavailability. Curr Pharm Biotechnol. 2016;17:549–555. doi: 10.2174/1389201017666160127110609. [DOI] [PubMed] [Google Scholar]
- 84.Seeram NP, Lee R, Heber D. Bioavailability of ellagic acid in pomegranate (Punica granum L.) juice. Clin Chim Acta. 2004;348:63–68. doi: 10.1016/j.cccn.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 85.Kasikci MB, Bagdatlioglu N. Bioavailability of quercetin. Curr Res in Nutr Food Sci. 2016;4.
- 86.Hamad AWR, al Momani W, Janakat S, Oran SA. Bioavailability of ellagic acid after single dose administration using HPLC. Pak J Nutr. 2009;8:1661–1664. doi: 10.3923/pjn.2009.1661.1664. [DOI] [Google Scholar]
- 87.Wang FM, Yao TW, Zeng S. Disposition of quercetin and kaempferol in a human following an oral administration of Ginkgo biloba. Eur J Drug Metab Pharmacokinet. 2003;28:173–177. doi: 10.1007/BF03190482. [DOI] [PubMed] [Google Scholar]
- 88.Thilakarathna SH, Rupasinghe HPV. Flavonoid bioavailability and attempts for bioavailability enhancement. Nutrients. 2013;5:3367–3387. doi: 10.3390/nu5093367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen ZP, Sun J, Chen HX, Xiao YY, et al. Comparative pharmacokinetics and bioavailability studies of quercetin, kaempferol, and isorhamnetin after oral administration of Ginkgo biloba extracts, Ginkgo biloba extract phospholipid complexes and Ginkgo biloba extract solid dispersions in rats. Fitoterapia. 2010;81:1045–1052. doi: 10.1016/j.fitote.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 90.Wang T, Liu XH, Guan J, Ge S, Wu MB, Lin JP, Yang LR. Advancement of multi-target drug discoveries and promising applications in the field of Alzheimer’s disease. Eur J Med Chem. 2019;169:200–223. doi: 10.1016/j.ejmech.2019.02.076. [DOI] [PubMed] [Google Scholar]
- 91.Neves BJ, Braga RC, Melo-Filho CC, Teofilo J, Moreira-Filho JT, Muratov EN, et al. QSAR-based virtual screening: advances and applications in drug discovery. Front Pharmacol. 2018. [DOI] [PMC free article] [PubMed]