Cryo-electron microscopy (cryo-EM) is rapidly becoming a leading player in structural biology, while X-ray crystallography still prevails as a powerful traditional tool for studying 3D structures of proteins (Fig. 1). The Taiwan-Japan joint symposium on structural biology using X-ray crystallography and cryo-EM, chaired by myself and Prof. Ken Yokoyama from Kyoto Sangyo University, was held as a session in the BSJ meeting in Miyazaki Japan in the afternoon of 25th September 2019. In this symposium, six researchers from Taiwan and Japan presented cutting-edge results using the two methods as described below.
Fig. 1.
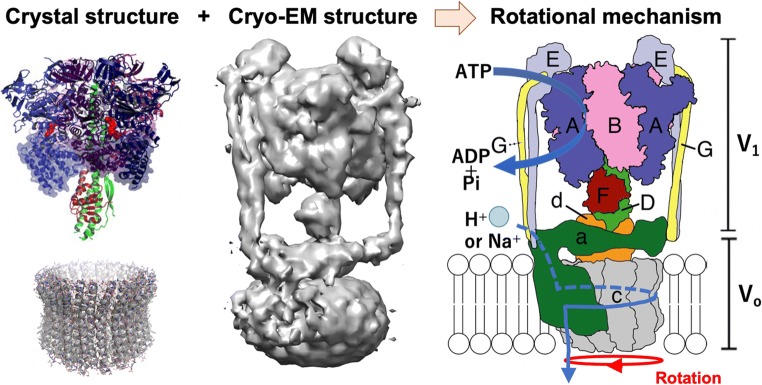
Structure and function of a bacterial V-ATPase. Left: crystal structures of V1 part and Vo-c ring. Center: Cryo-EM structure of a whole V-ATPase complex. Right: Schematic model of the V-ATPase. The V1 part is composed of a hexameric arrangement of alternating A and B subunits responsible for ATP binding and hydrolysis. The Vo part, in which rotational energy is converted to drive H+ (or Na+) translocation, is composed of oligomers of the c subunits and an a subunit. The V1 and Vo parts are connected by a central stalk, which is composed of D, F, and d subunits, and two peripheral stalks, which are composed of E and G subunits of V1. ATP hydrolysis induces the rotation of the central stalk (DFd complex) and an attached c ring, which causes ion pumping at the interface between the c ring and a subunit
The first speaker was Prof. Masahide Kikkawa from the University of Tokyo, who talked about cryo-EM analysis of cilia and microtubule-based motor proteins. Prof. Kikkawa and his group knocked-out efcab1 in zebrafish, encoding calaxin, and found that the mutant zebrafish have situs inversus due to the irregular ciliary beating of Kuppfer’s vesicle cilia (Sasaki et al. 2019), and also analyzed a complex between dynein microtubule binding domain and microtubule using high-resolution cryo-EM. Form these findings, he proposed a bound-unbound switching mechanism of the motor protein.
The next speaker was Dr. Shang-Te Danny Hsu from Academia Sinica, who talked about cryo-EM analysis of a feline coronavirus spike protein. Feline infectious peritonitis virus (FIPV) is an alphacoronavirus that causes nearly 100% mortality rate without effective treatments. Dr. Hsu and his colleagues have solved a 3.3 Å cryo-EM structure of an FIPV spike protein, responsible for host recognition and viral entry (Yang et al. 2020). In his talk, he presented an insight inspired by the EM structure for a better molecular understanding of the pathogenesis of FIP.
The third speaker was Dr. Kuen-Phon Wu from Academia Sinica, who talked about cryo-EM studies of bacterial glutamine synthetase. Bacterial glutamine synthetase (GS) is a dodecameric enzyme which responds in cellular glutamine biosynthesis as well as nitrogen fixation. Dr. Wu and his colleagues have determined the structures of GS in the catalytic cascade by cryo-EM at 2.6 Å resolution. He presented the new structures and a possible mechanism of recruiting magnesium, consuming ATP, and converting glutamine from glutamate.
The fourth speaker was Prof. Jun-ichi Kishikawa from Kyoto Sangyo University, who talked about single-particle analysis of membrane-embedded domain Vo of V-ATPase. V-ATPases are composed of hydrophilic V1 and membrane-embedded Vo (Fig. 1). The atomic structures of the V1 part and a rotational mechanism inspired by the structures have already been elucidated under the condition of the Vo structure missing (Arai et al. 2013). The group of Profs. Kishikawa and Yokoyama have solved the cryo-EM structures of a whole V-ATPase complex from Thermus thermophilus at low to middle resolution (Nakanishi et al. 2018). Recently, they have succeeded in solving the cryo-EM structure of the Vo part at a higher resolution. In his talk, he presented a new structure of Vo and proposed that the Vo part adopts an autoinhibited form when V1 dissociates from Vo.
The fifth speaker was prof. Kazuhiro Abe from Nagoya University, who talked about the transport mechanism of the gastric proton pump, H+,K+-ATPase, which is responsible for acidifying the gastric juice up to pH 1 and thus an important drug target for treating gastric acid-related diseases. Prof. Abe and his colleagues have solved the crystal structures of the H+,K+-ATPase in two different conformations (Abe et al. 2018). In his talk, he presented these cryo-EM structures and discussed the reasons why the pump is able to fulfill the requirements for the generation of a million-fold proton gradient across the membrane.
The last speaker was Prof. Hui-Chih Hung from National Chung-Hsing University, who talked about p53 function regulated by PAD4 through protein citrullination. Peptidylarginine deiminase 4 (PAD4)-catalyzed citrullination has been shown to play an emerging role as a potential cancer therapeutic target (Liu et al. 2017). Prof. Hung and her group have elucidated that p53 protein can be citrullinated by PAD4 in vitro and in the cell. Structural and functional analysis demonstrated that citrullination has detrimental effects on p53 tetramerization and thus impairs their DNA-binding ability. In her talk, she presented direct evidence of the PAD4-p53 interaction and proposed a potential mechanism of how PAD4 involves in p53 target gene regulation by citrullinating p53.
Compliance with ethical standards
Conflict of interest
The author declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Footnotes
IUPAB Biophysical Reviews Special Issue on the Biophysical Society of Japan
Session Commentary describing the topics and speakers selected for Session 2SDP
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abe K, Irie K, Nakanishi H, Suzuki H, Fujiyoshi Y. Crystal structures of the gastric proton pump. Nature. 2018;556:214–218. doi: 10.1038/s41586-018-0003-8. [DOI] [PubMed] [Google Scholar]
- Arai S, Saijo S, Suzuki K, Mizutani K, Kakinuma Y, Ishizuka-Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Iwata S, Yamato I, Murata T. Rotation mechanism of Enterococcus hirae V1-ATPase based on asymmetric crystal structures. Nature. 2013;493:703–707. doi: 10.1038/nature11778. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lee CY, Huang YN, Chen HY, Liu GY, Hung HC. Probing the roles of calcium-binding sites during the folding of human peptidylarginine deiminase 4. Sci Rep. 2017;25:2429. doi: 10.1038/s41598-017-02677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A, Kishikawa JI, Tamakoshi M, Mitsuoka K, Yokoyama K. Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus. Nat Commun. 2018;9(1):89. doi: 10.1038/s41467-017-02553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Shiba K, Nakamura A, Kawano N, Satouh Y, Yamaguchi H, Morikawa M, Shibata D, Yanase R, Jokura K, Nomura M, Miyado M, Takada S, Ueno H, Nonaka S, Baba T, Ikawa M, Kikkawa M, Miyado K, Inaba K. Calaxin is required for cilia-driven determination of vertebrate laterality. Commun Biol. 2019;2:226. doi: 10.1038/s42003-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Tzu-Jing, Chang Yen-Chen, Ko Tzu-Ping, Draczkowski Piotr, Chien Yu-Chun, Chang Yuan-Chih, Wu Kuen-Phon, Khoo Kay-Hooi, Chang Hui-Wen, Hsu Shang-Te Danny. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proceedings of the National Academy of Sciences. 2020;117(3):1438–1446. doi: 10.1073/pnas.1908898117. [DOI] [PMC free article] [PubMed] [Google Scholar]