Abstract
Oncolytic herpes simplex viruses (HSVs), in clinical trials for the treatment of malignant gliomas, are assumed to be selective for tumor cells because their replication is strongly attenuated in quiescent cells, but not in cycling cells. Oncolytic selectivity is thought to occur because mutations in viral ICP6 (encoding a viral ribonucleotide reductase function) and/or γ34.5 function are respectively complemented by mammalian ribonucleotide reductase and GADD34, whose genes are expressed in cycling cells. However, it is estimated that only 5–15% of malignant glioma cells are in mitosis at any one time. Therefore, effective replication of HSV oncolytic viruses might be limited to a subpopulation of tumor cells, since at any one time the majority of tumor cells would not be cycling. However, we report that an HSV with defective ICP6 function replicates in quiescent cultured murine embryonic fibroblasts obtained from mice with homozygous p16 deletions. Furthermore, intracranial inoculation of this virus into the brains of p16−/− mice provides evidence of viral replication that does not occur when the virus is injected into the brains of wild-type mice. These approaches provide in vitro and in vivo evidence that ICP6-negative HSVs are ‘molecularly targeted,’ because they replicate in quiescent tumor cells carrying specific oncogene deletions, independent of cell cycle status.
Keywords: oncolytic virus, gene therapy, glioma, brain tumor, mechanism of replication, tumor-suppressor gene
Main
Oncolytic herpes simplex viruses (HSVs) have undergone extensive laboratory investigation (Kirn et al., 2001) and early clinical trials (Aghi and Martuza, 2005) as anticancer agents. The oncolytic HSVs that have undergone clinical trials have had mutations in HSV genes γ34.5 and/or ICP6. ICP6 encodes a viral ribonucleotide reductase (vRR) function that allows replication of wild-type HSV to occur even in quiescent cells, such as the neurons that are infected during encephalitis caused by wild-type HSV type 1 (HSV1; Goldstein and Weller, 1988). Without this function, HSV replication is severely curtailed in quiescent cells (Goldstein and Weller, 1988). However, cycling cells upregulate expression of S-phase specific genes such as mRR for their own nucleic acid metabolism, which complements the defective viral ICP6 function, thus allowing these mutant HSVs to replicate (Goldstein and Weller, 1988). This complementation provides a biologic rationale for employment of ICP6-defective HSV in oncolytic therapy, rationale based on their preferential replication in cycling versus quiescent cells.
However, if oncolytic HSVs only target the 5–15% of glioma cells replicating at any moment in time (Hoshino et al., 1989), than the majority of glioma cells will evade oncolytic HSV therapy, particularly because oncolytic HSVs are administered intratumorally rarely more than once, while other S-phase specific treatments like radiation and chemotherapy can be administered daily for an extended period of time, allowing targeting of cells that might enter the cell cycle later in the course of treatment. Knowing that the downstream effect of p16INK4a (hereafter termed p16), a tumor suppressor gene on human chromosome 9p21 deleted in 30% of glioblastomas (Kamiryo et al., 2002), is to prevent transcription factor E2F1 from transcribing S-phase genes such as mRR (Elledge et al., 1992), we hypothesized that mutations in p16 would complement ICP6-defective HSV replication, even if cells were in quiescence and not actively cycling, thereby enabling ICP6-defective HSV to overcome limited infectability in quiescent cells in a molecularly targeted fashion.
The yield of an infectious ICP6-defective HSV1 (hrR3) was determined for cultured human T98 glioma and fibroblast cells that were cycling, as well as for a quiescent population of each cell type in G0 that was isolated by fluorescence activated cell sorting (FACS). Overall, T98 cells produced significantly more hrR3 than fibroblasts, regardless of cell cycle status. More interestingly, the ratio of hrR3 yield in cycling versus noncycling cells was nearly threefold higher in normal fibroblasts than in T98 glioma cells (Figure 1). This suggested that the cellular machinery of quiescent T98 glioma cells complemented hrR3 replication to a higher degree than that of quiescent fibroblasts.
Figure 1.

hrR3 replication is greater in arrested p16-deleted cells than in arrested p16-expressing cells. (a) Recovery in plaque forming units (pfu) ml−1 of infectious KOS-derived vRR(ICP6, UL39)–LacZ+ hrR3 (from S Weller, University of Connecticut, Farmington, CT, USA) (Goldstein and Weller, 1988) 48 h after infecting (multiplicity of infection (MOI)=1.5) cultured human (U87 glioma, HF (human fibroblasts), T98 glioma, U373 glioma and U373-mRR cells; all obtained from American Type Culture Collection/ATCC, Manassas, VA, USA) and murine (MEFs (murine embryonic fibroblasts), obtained from R DePinho, Dan Farber Cancer Institute, Boston, MA, USA) cells that were either cycling or in G0/G1 (isolated by flow cytometry). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U ml−1 of penicillin, and 100 μg ml−1 of streptomycin at 37 °C and 5% CO2. U373-mRR were generated by lipofectamine (Invitrogen, Carlsbad, CA, USA) transfection of U373 with the neomycin resistance gene-containing plasmid pCDNA3.1 (Invitrogen), with the cDNA for the M2 subunit of mRR subcloned into the plasmid. The M2 subunit of mRR cDNA was generated by RT–PCR of mRNAs obtained from U373 cells using a 5′ primer from positions 189 to 211 of the M2 subunit coding sequence including the AUG site (5′-ATCCGGATCCACTATGCTCTCCCTCCGTGT-3′) and the 3′ primer from positions 1346 to 1368 including the UAA site (5′-GCTTAAGCTTATTTAGAAGTCAGCATCCAAG-3′), with the resulting PCR product first cloned into the TA cloning vector (Invitrogen). MEFs were isolated as described (Sharpless et al., 2001). For FACS analysis, 106 cells were trypsinized, centrifuged at 1000 g for 5 min, fixed by gradual addition of ice cold 70% ethanol (30 min at 4 °C) and washed with phosphate buffered saline (PBS). Cells were then treated with RNase (10 μg ml−1) for 30 min at 37 °C, washed once with PBS, resuspended and stained in 1 ml of 69 μM propidium iodide in 38 mM sodium citrate for 30 min at room temperature. The cell cycle phase distribution was determined by analytical DNA flow cytometry as described (Gray-Bablin et al., 1997). Alternatively, to isolate G0/G1 cells by DNA content, 109 cells were trypsinized, resuspended in PBS, and fluorescence activated cell sorting (FACS)-sorted. To measure viral yield, 105 cells were plated into 12-well plates in DMEM plus 10% fetal calf serum. The next day, cells were infected with hrR3 (MOI=1.5) for 48 h. Infected cells were scraped into the medium and subjected to three freeze-thaw cycles. Virus titers were determined by plaque assays on Vero cells. Standard deviations are shown. (b) Ratio of viral yield in cycling cells to viral yield in G0/G1 cells for the six different cell lines analysed in (a). The p16+/+ expressing cells (HF, MEF, p53− MEF, U373) exhibited significantly greater ratios than their p16−/− deleted counterparts (U87/T98 for HF, p16−/− MEF for MEF and for p53−/− MEF) or U373-mRR (P<0.05). Similar results (not shown) were obtained with murine fetal astrocytes of all p16/p53 phenotypes (obtained from R DePinho) and cells in G1 obtained by treating with 40 μM lovastatin (Merck, Rathway, NJ, USA) for 36 h, with lovastatin converted to its active form as described previously (Gray-Bablin et al., 1997).
T98 glioma cells possess both p16 and p53 mutations (Wang et al., 2006). To investigate the role of these two mutations in viral replication in cultured G0 cells, the same experiment was repeated in cycling and G0 U87 glioma cells, which express wild-type p53 but possess homozygous deletions in p16 (Wang et al., 2006). The ratio of hrR3 yield in cycling-to-G0 U87 glioma cells was nearly identical to that in cycling-to-G0 T98 glioma cells (Figure 1). This finding suggested that p53 mutations were not necessary to support hrR3 replication in arrested tumor cells and was in agreement with previous findings in cycling tumor cells (Yoon et al., 1999).
On the other hand, when the same experiment was repeated in U373 glioma cells, which are p16+/+ p53−/− (Komata et al., 2000), the ratio of hrR3 yield in cycling-to-G0 U373 cells was higher than that of human fibroblasts and over 10-fold higher than that of p16−/− p53+/+ U87 and p16−/− p53−/−T98 glioma cells (Figure 1). These findings raised the possibility that p16 mutations might contribute to the support of hrR3 replication seen in quiescent T98 and U87, but not in U373, glioma cells.
Because glioma and other tumor cells possess not only mutations in p16, but also a variety of genetic mutations that could affect viral or cellular replication (Farassati et al., 2001; Smith et al., 2006), the hrR3 yield was determined in cycling and FACS-isolated G0 murine embryonic fibroblasts (MEFs) and fetal astrocytes from wild-type as well as transgenic mice with homozygous deletions in p16 or p53. p16−/− MEFs and fetal astrocytes supported hrR3 replication even in G0, whereas viral replication was not maintained in G0 wild-type and p53-deleted MEFs and fetal astrocytes (Figure 1). Comparable results were obtained when pharmacologic agents such as lovastatin were used to induce growth arrest (data not shown). These findings indicate that deletions in p16, but not p53, will support productive replication of an ICP6-defective HSV in quiescent cells.
To confirm that loss of p16 function would result in higher levels of mammalian ribonucleotide reductase (mRR) gene expression, regardless of cell cycle status (Elledge et al., 1992), we measured the mRNA levels of the M2 subunit of mRR in cycling relative to noncycling cells using real-time reverse transcription (RT)–PCR. In p16-deleted cells (that is, U87, T98 and p16−/− MEFs), the ratio of mRR M2 subunit mRNA in cycling relative to noncycling cells was 0.9–1.3. However, in p16-expressing cells (that is, wild-type MEFs, human fibroblasts, p53−/− MEFs and U373), this ratio was much higher (2.5–5.8; Figure 2). The finding of decreased mRR expression in noncycling p16+/+ cells agreed with published data showing mRR expression in cycling cells, with expected downregulation in G0 or growth-arrested cells (Elledge et al., 1992). However, in p16−/− cells, mRR expression was maintained during G0, regardless of cell-cycle status, in agreement with the mechanism of p16-mediated suppression of mRR transcription (Elledge et al., 1992). We then stably transfected p16-expressing U373 glioma cells to overexpress the M2 subunit of mRR, creating a cell line, U373-mRR, which expressed 5.6-fold more mRR M2 subunit mRNA than U373 (data not shown). When infected with hrR3, the ratio of viral yield in cycling U373-mRR relative to noncycling U373-mRR was comparable to that seen in U87 or T98 cells and nearly threefold lower than that of U373 cells (Figure 1), suggesting that stably elevating the mRR M2 subunit expression throughout the cell cycle in U373 cells (Figure 2) enabled the support of viral replication independent of cell cycling status in a manner comparable to that seen in p16-deleted cells, such as T98 or U87 (Figure 1).
Figure 2.
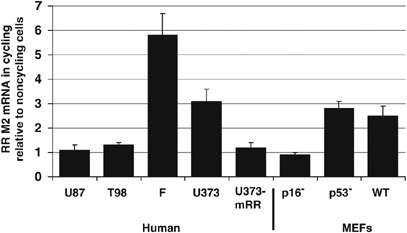
Ribonucleotide reductase expression is elevated in noncycling cells with p16 deletions. Reverse transcription (RT)–PCR shows the ratio of M2 subunit of mammalian ribonucleotide reductase (mRR) mRNA in cycling relative to noncycling cells was 0.9–1.3 in p16-deleted cells (U87, T98, and p16−/− MEFs). However, in p16-expressing cells (that is, wild-type MEFs, human fibroblasts, p53−/− MEFs and U373), this ratio was much higher (2.5–5.8). A high capacity cDNA archive kit (Applied Biosystems, Foster City, CA, USA) was used to generate cDNA from RNA isolated from cells using Trizol (Invitrogen) per protocol. Real-time RT–PCR was performed on an ABI Prism 7000 (Applied Biosystems) machine using human primer-probe combinations for RR subunit M2 (Applied Biosystems part nos. Hs00357247_g1 and Hs00168784_m1) and 18S rRNA (Applied Biosystems part no. 4308329) combined with TaqMan Master Mix (Applied Biosystems). Relative quantification was performed using 18S rRNA as an endogenous control. All reactions began with 10 min at 95 °C for AmpliTaq Gold activation, followed by 40 cycles at 95 °C for 15 s for denaturation and then 60 °C for 1 min for annealing/extension. Abbreviations used: F, fibroblasts; WT, wild-type; U373-mRR, U373 clone selected after stable transfection with plasmid constitutively expressing the M2 subunit of mammalian ribonucleotide reductase.
To determine the effect of p16 deletions on hrR3 replication in G0 cells in vivo, we studied the replication and toxicity of hrR3 and KOS, the wild-type HSV1 from which hrR3 is derived, after intracerebral inoculation in wild-type and p16−/− ‘knock-out’ mice. Cerebral viral titers were similar when wild-type HSV was inoculated into the brains of wild-type and p16−/− mice. However, p16−/− mice produced three orders of magnitude more hrR3 than wild-type mice after intracerebral inoculation (Figure 3a). The LD50 (viral dose that killed 50% of inoculated mice 14 days after inoculation) was two orders of magnitude less when hrR3 was inoculated into the brains of p16−/− mice compared to wild-type mice, but not significantly different when wild-type HSV was inoculated into the brains of wild-type or p16−/− mice (Figure 3b). Finally, the anatomic distribution of hrR3-mediated gene expression in neurons was much greater in the brains of p16−/− mice than in the brains of wild-type mice and the former also exhibited evidence of increased inflammatory and necrotic effects caused by the host immune response which likely arose due to significant viral replication in this treatment group (Figure 3c). Taken together, these findings confirm the validity of the hypothesis that the ICP6-defective HSV1 retains significant replicative ability in cells with defects in p16, even if the cells are postmitotic or growth arrested.
Figure 3.

hrR3 exhibits greater replication in brains of p16−/− transgenic mice than in brains of wild-type mice. (a) Plaque forming units (pfu) per gram brain tissue recovered 5 days after intracranial inoculation of wild-type FVB (Jackson Laboratories; Bar Harbor, ME, USA) and FVB-derived p16−/− mice (provided by R DePinho) with wild-type KOS (obtained from D Knipe, Harvard Medical School, Boston, MA, USA) and hrR3 viruses. Viral recovery was nearly 30-fold lower when hrR3 was inoculated into the brains of FVB mice than in the other three experimental groups (P<0.05 by Student's t-test). Mice (n=10 per treatment group) were anesthetized with ketamine/xylazine (75/15 mg kg−1 intraperitoneally) and immobilized in a stereotactic apparatus. Through a midline sagittal incision, a 1 mm skull burr hole was drilled l 1 mm anterior to and 2.5 mm right of the bregma. KOS and hrR3 virus (105–1010 pfu) in 2 μl virus buffer (150 mM NaCl, 20 mM Tris pH 7.5) were injected 4.5 mm below dura using Hamilton syringes. Mice were euthanized when they became lethargic, anorexic, dehydrated or distressed. Brains were cut in small pieces, suspended in a volume of PBS twice the tumor volume, homogenized manually, sonicated and centrifuged at 16 110 × g for 5 min at 4 °C. The supernatant was isolated and freeze thawed three times, and viral yield was determined by plaque assays on Vero cells as described above. Standard deviations are shown. (b) LD50, the viral dose in pfu required for death of 50% of mice 14 days after intracranial inoculation, was nearly 3-log higher when hrR3 was inoculated into FVB mice than in the other three experimental groups. The LD50 for each virus/mouse strain combination was calculated as follows: (1) the portion of 10 mice who survived 14 days after inoculating 4 viral doses in FVB or p16−/− mice (105, 107, 109, and 1010 pfu) was calculated; and (2) Finney's probit analysis method was used to calculate LD50 from these four data points (Finney, 1971). (c) hrR3 positivity in neurons, as determined by staining for the marker gene β-galactosidase, was far greater with significant associated necrosis and surrounding inflammatory infiltrate in the brains of p16−/− mice (upper panel) than in the brains of wild-type FVB mice (lower panel; × 60 magnification). Brains were frozen in liquid nitrogen-cooled N-methylbutane, and sectioned coronally by cryostat to 8 μm thickness. Slides underwent blocking of endogenous peroxidase (DAKO PAP kit, Dako, Copenhagen, Denmark), incubating with goat serum for 30 min at 4 °C to reduce nonspecific background staining, incubation with mouse-anti-β-galactosidase antibody (Promega Biosciences, Madison, WI, USA) overnight at 4 °C, incubation with rabbit anti-mouse peroxidase conjugated secondary antibody (Dako) for 2 h, incubation with 3′,3′-deamino-benzidine tetrahydrochloride per protocol (Dako) and hematoxylin staining. Scale bar represents 100 μm.
Oncolytic viruses such as engineered HSVs, have produced significant anticancer effects in preclinical models, but have not shown definite efficacy in clinical trials (Aghi and Martuza, 2005). Speculated reasons for this lack of efficacy include viral doses that are less than the maximum tolerated dose, mutations that render oncolytic HSV less potent, inefficient spatial or temporal distribution of virus (Wein et al., 2003) and anti-HSV immunity impeding viral replication (Ikeda et al., 1999). Another reason for limited efficacy has been assumed to be that oncolytic HSV will infect and lyse cells in the tumor that are actively dividing, but that quiescent tumor cells may not support viral replication. In the case of gliomas, 85–95% of tumor cells may not be actively cycling at any one time (Hoshino et al., 1989). If quiescent tumor cells cannot support oncolytic HSV replication, then oncolytic HSVs would be unable to treat tumors like gliomas with large noncycling fractions, particularly because oncolytic viruses are administered intratumorally once at surgery, unlike other S-phase specific adjuvant treatments such as chemotherapy or radiation which can be administered daily, enabling the targeting of cells that enter the cell cycle at different points in time. However, our results indicate that noncycling p16−/− cells will still support the replication of ICP6-defective HSVs. These findings imply that ICP6-defective oncolytic HSV represents a molecularly targeted therapy for tumors with defects in the p16 tumor suppressor pathway. They also imply that other types of herpes viruses and poxviruses, such as vaccinia virus (McCart et al., 2001), with genetic defects other than vRR, such as viral thymidine kinase, whose cellular ortholog (mammalian thymidine kinase) is also under transcriptional control of the p16 tumor suppressor pathway, may also replicate in quiescent cells with defects in this pathway.
Another type of targeting of a downstream effector of p16 is represented by the E1A-defective adenoviruses (Jiang et al., 2005). In addition, viruses, such as coronavirus (Wurdinger et al., 2005) and measles virus (Nakamura et al., 2005), can be engineered to be selectively taken up by cells that express cell surface receptors, such as mutant epidermal growth factor receptor, that are specifically found on tumors. Other viruses target cells with alterations in the ras mitogenic pathway. Examples of such viruses include reovirus (Norman et al., 2004), HSV-2 with a deletion in the viral ICP10 gene (Fu et al., 2006) and HSV-1 with a deletion in the viral γ34.5 gene (Smith et al., 2006). Other viruses target tumor cells' propensity to exhibit altered biochemical properties, such as vesicular stomatitis virus, which targets cells with altered interferon responsiveness (Stojdl et al., 2000) or adenovirus ONYX-015 which targets cells that shut down cellular protein synthesis after infection (O'Shea et al., 2005). One key distinction between the type of targeting we found in HSVs lacking ICP6 and the targeting of these other viruses is that the targeting of HSVs lacking ICP6 allows them to infect quiescent tumor cells carrying p16 deletions, which can represent 85–95% of the cells in a glioma at any given moment in time (Hoshino et al., 1989).
The experimental approach of comparing MEFs from p16−/− mice to MEFs from wild-type mice to validate the hypothesis that an ICP6-defective HSV will target nonreplicating cells with p16 deletions is superior to an approach employing tumor cells with or without apparently intact p16 gene loci because: (1) tumor cells possess many other genetic mutations that may also affect viral replication (Farassati et al., 2001; Smith et al., 2006) and (2) even tumor cells that appear to have intact p16 gene loci may have p16 gene inactivation by promoter methylation and/or inactivation of other components of the p16 tumor suppressor pathway (Costello et al., 1996; Ono et al., 1996; Ueki et al., 1996; Burns et al., 1998; Nakamura et al., 1998; Watanabe et al., 2001; Ohgaki and Kleihues, 2007).
As clinical trials continue to be carried out with such oncolytic viruses it will be important to determine retrospectively and prospectively if there is enhanced viral replication in patients whose tumors carry a particular mutation known to complement the viral defect. For the ICP6-defective HSV1 trials, determination of the status of the p16 tumor suppressor pathway in treated tumors may be crucial in order to identify patients with tumors sensitive to these agents.
Acknowledgements
This study was funded by an American Brain Tumor Association Fellowship to MA and by NIH PO1 CA69246, R01 NS41571, R01 CA85139 to EAC. RAD was supported by an ACS Research Professor Award and NIH U01 CA084628 and P01 CA095616.
References
- Aghi M, Martuza RL. Oncolytic viral therapies—the clinical experience. Oncogene. 2005;24:7802–7816. doi: 10.1038/sj.onc.1209037. [DOI] [PubMed] [Google Scholar]
- Burns KL, Ueki K, Jhung SL, Koh J, Louis DN. Molecular genetic correlates of p16, cdk4, and pRb immunohistochemistry in glioblastomas. J Neuropathol Exp Neurol. 1998;57:122–130. doi: 10.1097/00005072-199802000-00003. [DOI] [PubMed] [Google Scholar]
- Costello JF, Berger MS, Huang HS, Cavenee WK. Silencing of p16/CDKN2 expression in human gliomas by methylation and chromatin condensation. Cancer Res. 1996;56:2405–2410. [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci. 1992;17:119–123. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- Farassati F, Yang AD, Lee PW. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat Cell Biol. 2001;3:745–750. doi: 10.1038/35087061. [DOI] [PubMed] [Google Scholar]
- Finney DJ. Probit Analysis. Cambridge University Press: New York, NY; 1971. [Google Scholar]
- Fu X, Tao L, Cai R, Prigge J, Zhang X. A mutant type 2 herpes simplex virus deleted for the protein kinase domain of the ICP10 gene is a potent oncolytic virus. Mol Ther. 2006;13:882–890. doi: 10.1016/j.ymthe.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Goldstein DJ, Weller SK. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988;166:41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Gray-Bablin J, Rao S, Keyomarsi K. Lovastatin induction of cyclin-dependent kinase inhibitors in human breast cells occurs in a cell cycle-independent fashion. Cancer Res. 1997;57:604–609. [PubMed] [Google Scholar]
- Hoshino T, Prados M, Wilson CB, Cho KG, Lee KS, Davis RL. Prognostic implications of the bromodeoxyuridine labeling index of human gliomas. J Neurosurg. 1989;71:335–341. doi: 10.3171/jns.1989.71.3.0335. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ichikawa T, Wakimoto H, Silver JS, Deisboeck TS, Finkelstein D. Oncolytic virus therapy of multiple tumors in the brain requires suppression of innate and elicited antiviral responses. Nat Med. 1999;5:881–887. doi: 10.1038/11320. [DOI] [PubMed] [Google Scholar]
- Jiang H, Gomez-Manzano C, Alemany R, Medrano D, Alonso M, Bekele BN. Comparative effect of oncolytic adenoviruses with E1A-55 kDa or E1B-55 kDa deletions in malignant gliomas. Neoplasia. 2005;7:48–56. doi: 10.1593/neo.04391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiryo T, Tada K, Shiraishi S, Shinojima N, Nakamura H, Kochi M. Analysis of homozygous deletion of the p16 gene and correlation with survival in patients with glioblastoma multiforme. J Neurosurg. 2002;96:815–822. doi: 10.3171/jns.2002.96.5.0815. [DOI] [PubMed] [Google Scholar]
- Kirn D, Martuza RL, Zwiebel J. Replication-selective virotherapy for cancer: biological principles, risk management and future directions. Nat Med. 2001;7:781–787. doi: 10.1038/89901. [DOI] [PubMed] [Google Scholar]
- Komata T, Kondo Y, Koga S, Ko SC, Chung LW, Kondo S. Combination therapy of malignant glioma cells with 2-5A-antisense telomerase RNA and recombinant adenovirus p53. Gene Therapy. 2000;7:2071–2079. doi: 10.1038/sj.gt.3301327. [DOI] [PubMed] [Google Scholar]
- McCart JA, Ward JM, Lee J, Hu Y, Alexander HR, Libutti SK. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res. 2001;61:8751–8757. [PubMed] [Google Scholar]
- Nakamura M, Konishi N, Hiasa Y, Tsunoda S, Nakase H, Tsuzuki T. Frequent alterations of cell-cycle regulators in astrocytic tumors as detected by molecular genetic and immunohistochemical analyses. Brain Tumor Pathol. 1998;15:83–88. doi: 10.1007/BF02478888. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Peng KW, Harvey M, Greiner S, Lorimer IA, James CD. Rescue and propagation of fully retargeted oncolytic measles viruses. Nat Biotechnol. 2005;23:209–214. doi: 10.1038/nbt1060. [DOI] [PubMed] [Google Scholar]
- Norman KL, Hirasawa K, Yang AD, Shields MA, Lee PW. Reovirus oncolysis: the Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection. Proc Natl Acad Sci USA. 2004;101:11099–11104. doi: 10.1073/pnas.0404310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Tamiya T, Ichikawa T, Kunishio K, Matsumoto K, Furuta T. Malignant astrocytomas with homozygous CDKN2/p16 gene deletions have higher Ki-67 proliferation indices. J Neuropathol Exp Neurol. 1996;55:1026–1031. doi: 10.1097/00005072-199655100-00002. [DOI] [PubMed] [Google Scholar]
- O'Shea CC, Soria C, Bagus B, McCormick F. Heat shock phenocopies E1B-55 K late functions and selectively sensitizes refractory tumor cells to ONYX-015 oncolytic viral therapy. Cancer Cell. 2005;8:61–74. doi: 10.1016/j.ccr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Smith KD, Mezhir JJ, Bickenbach K, Veerapong J, Charron J, Posner MC. Activated MEK suppresses activation of PKR and enables efficient replication and in vivo oncolysis by Deltagamma(1)34.5 mutants of herpes simplex virus 1. J Virol. 2006;80:1110–1120. doi: 10.1128/JVI.80.3.1110-1120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;6:821–825. doi: 10.1038/77558. [DOI] [PubMed] [Google Scholar]
- Ueki K, Ono Y, Henson JW, Efird JT, Von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- Wang CC, Liao YP, Mischel PS, Iwamoto KS, Cacalano NA, McBride WH. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006;66:6756–6762. doi: 10.1158/0008-5472.CAN-06-0185. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation and homozygous deletion of the p14ARF and p16INK4a genes in oligodendrogliomas. Acta Neuropathol (Berl) 2001;101:185–189. doi: 10.1007/s004010000343. [DOI] [PubMed] [Google Scholar]
- Wein LM, Wu JT, Kirn DH. Validation and analysis of a mathematical model of a replication-competent oncolytic virus for cancer treatment: implications for virus design and delivery. Cancer Res. 2003;63:1317–1324. [PubMed] [Google Scholar]
- Wurdinger T, Verheije MH, Raaben M, Bosch BJ, De Haan CA, Van Beusechem VW. Targeting non-human coronaviruses to human cancer cells using a bispecific single-chain antibody. Gene Therapy. 2005;12:1394–1404. doi: 10.1038/sj.gt.3302535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SS, Carroll NM, Chiocca EA, Tanabe KK. Influence of p53 on herpes simplex virus type 1 vectors for cancer gene therapy. J Gastrointest Surg. 1999;3:34–48. doi: 10.1016/S1091-255X(99)80005-5. [DOI] [PubMed] [Google Scholar]


