Abstract
Clathrin forms diverse lattice and cage structures that change size and shape rapidly in response to the needs of eukaryotic cells during clathrin-mediated endocytosis and intracellular trafficking. We present the cryo-EM structure and molecular model of assembled porcine clathrin, providing new insights into interactions that stabilise key elements of the clathrin lattice, namely, between adjacent heavy chains, at the light chain-heavy chain interface and within the trimerisation domain. Furthermore, we report cryo-EM maps for five different clathrin cage architectures. Fitting structural models to three of these maps shows that their assembly requires only a limited range of triskelion leg conformations, yet inherent flexibility is required to maintain contacts. Analysis of the protein-protein interfaces shows remarkable conservation of contact sites despite architectural variation. These data reveal a universal mode of clathrin assembly that allows variable cage architecture and adaptation of coated vesicle size and shape during clathrin-mediated vesicular trafficking or endocytosis.
Introduction
Endocytosis enables material to be absorbed via specific ligand-receptor interactions through the assembly of specialised protein coats around vesicles formed from the plasma membrane 1,2. In the case of clathrin-mediated endocytosis (CME), three-legged clathrin structures called triskelia form a latticed scaffold around the outside of a vesicle derived from the plasma membrane and coordinate binding of a network of adaptor proteins, which together drive cargo selection, vesicle formation and detachment from the membrane. Clathrin-coated vesicles have been seen to emerge directly from flat clathrin lattices indicating that clathrin assemblies adapt to changes in membrane shape at endocytic sites3. Avinoam et al 4 provided evidence that these changes may be enabled by the rapid exchange of clathrin triskelia5 with the membrane-bound clathrin coat. The multiple shapes adopted by those assemblies observed in cells are also seen with purified clathrin, which forms cages with different architectures 6. The ability of clathrin to form diverse structures is inherently determined by its molecular structure but how, has remained unclear.
Clathrin’s role in endocytosis is mediated through engagement with adaptor proteins, most notably the heterotetrameric complex AP-2, which interacts with clathrin through a ‘clathrin box’ motif on an extended linker region within its β2-adaptin subunit and through a binding site on the β2-appendage domain 7,8,9, which interacts with clathrin’s ankle domain 10. AP-1, a homologue of AP-2, engages with clathrin during intracellular trafficking. A series of elegant studies leading to structures of the AP-1 11,12–14 and AP-2 15–18 core domains have made transformative advances but, despite these, the nature of the interaction of these key complexes with assembled clathrin is not fully understood. While NMR and X-ray studies of the clathrin terminal domain bound to β-arrestin 2 19 and adaptor binding motifs 20–23 revealed multiple adaptor protein binding sites on the clathrin terminal domain, understanding how such interactions are coordinated within the context of a growing clathrin coat requires a molecular level understanding of the clathrin scaffold with which they must engage. This highlights the importance of obtaining high resolution structural information of clathrin in its assembled form.
Clathrin can be purified from endogenous clathrin-coated vesicles and reconstituted into cage structures reminiscent of the clathrin polyhedral lattices seen in cells. The symmetry adopted by some of these cages led to their exploitation for single particle structural studies by cryo-EM as early as 1986 24,25. Further, clathrin cryo-EM structures have revealed the arrangement of clathrin triskelia within a cage 26–28, and the location of bound auxilin and Hsc70 29–32. X-ray structures of the clathrin proximal, terminal and linker domains provided atomic resolution information for these individual domains 33,34 and a crystallographic study 35 of clathrin hubs revealed coordinated changes in light and heavy chain conformation suggesting that the light chain could regulate assembly by influencing changes in knee conformation. In 2004, a 7.9 Å cryo-EM map of assembled clathrin provided an alpha carbon model of a hexagonal barrel cage 28 that revealed the location of the clathrin light chains and a helical tripod structure at the trimerization domain.
Here we present a higher resolution map and molecular model for assembled clathrin. Details of the interactions made by the tripod of helices identified previously 28 are now much clearer, experimentally proving that it adopts a coiled-coil structure. Density for the key light chain tryptophans is now visible, enabling the light chain interaction with the proximal domain to be further defined. Additional elements of the heavy chain secondary structure can be visualised with confidence and energetic analysis based on our molecular model has revealed interaction sites that are of potential importance for assembly. Furthermore, structural analysis of three different clathrin cage architectures reveals how these different architectures can arise as a result of flexibility at defined positions on the clathrin leg combined with remarkable conservation of contact sites between the heavy chain legs. Thus clathrin adopts a universal mode of assembly that allows variable cage geometry and may facilitate rapid adaptation of coated vesicle size and shape during clathrin mediated vesicular trafficking or endocytosis.
Results
Multiple clathrin cage architectures
We determined structures of five clathrin cage architectures (Fig. 1a) from 12,785 particles selected from cryo-electron microscopy images of endogenous clathrin assembled in the presence of the clathrin-binding domain of β2-adaptin (Supplementary Note 1a). We found that incorporating β2-adaptin into the cages promoted formation of more regular cage structures, as reported previously 9. To address the challenge that the multiple cage types in the sample presented for structural analysis, cages were identified using a library of ten cage architectures built in silico (Fig. 1b) that had been proposed by Schein and Sands-Kidner to be the most likely stable structures to be formed for cages with fewer than 60 vertices 36. A supervised and subsequent unsupervised structure classification scheme determined the five cage architectures, which could be refined to nominal resolutions of 24 – 9.1 Å (Supplementary Notes 1c-e, 2, 3 and Table 1).
Figure 1. Identification of multiple clathrin cage architectures.
(a) Cryo-EM maps of five clathrin cage architectures with resolutions between 24 – 9.1 Å. (b) Library of cage architectures with 20 – 38 vertices 36 used for particle classification. (c) Cryo-EM map of the mini coat architecture (left) with four triskelia highlighted to show quaternary level interactions. A triskelion is shown in isolation (middle) and in the context of neighbouring heavy chains (right). Structural features are coloured according to the domain structure of the clathrin heavy and light chains defined in Fig. 2a.
Table 1. Cryo-EM data collection, refinement and validation statistics.
Whole cage and consensus hub substructure statistics are shown. See Supplementary Tables 1 and 2 for additional information. *Datasets with different total doses were combined. The maximum dose is shown.
| 28 mini coat cage (EMD-0114) |
32 sweet potato cage (EMD-0115) |
36 barrel cage (EMD-0116) |
36 tennis cage (EMD-0118) |
37 big apple cage (EMD-0120) |
Consensus hub (EMD-0126, PDB 6SCT) |
||
|---|---|---|---|---|---|---|---|
| Data collection and processing | |||||||
| Magnification | 82,111 | 82,111 | 82,111 | 82,111 | 82,111 | 82,111 | |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | |
| Electron exposure (e–/Å2) | 69* | 69* | 69* | 69* | 69* | 69* | |
| Defocus range (μm) | 1.8 – 2.8 | 1.8 – 2.8 | 1.8 – 2.8 | 1.8 – 2.8 | 1.8 – 2.8 | 1.8 – 2.8 | |
| Pixel size (Å) | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | 1.71 | |
| Symmetry imposed | T | D3 | D6 | D2 | C1 | C3 | |
| Initial particle images (no.) | 12,785 | 12,785 | 12,785 | 12,785 | 12,785 | 12,785 | |
| Final particle images (no.) | 2,945 | 1,761 | 1,160 | 1,624 | 2,010 | 313,406 | |
| Map resolution (Å) | 9.07 | 23.7 | 12.2 | 13.8 | 23.7 | 4.69 | |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | |
| Map resolution range (Å) | 6.5 – 50.0 | 12.7–50.0 | 8.5–50.0 | 9.1–50.0 | 10.1–50.0 | 3.4 – 50.0 | |
| Refinement | |||||||
| Initial model used (PDB code) | 6SCT | 6SCT | 6SCT | 6SCT | 6SCT | 3IYV, 1B89 | |
| Model resolution (Å) | 4.59 | ||||||
| FSC threshold | 0.5 | ||||||
| Model resolution range (Å) | 4.3 – 6.7 | ||||||
| Map sharpening B factor (Å2) | -164.9 | ||||||
| Model composition | |||||||
| Nonhydrogen atoms | 40,680 | ||||||
| Protein residues | 4,836 | ||||||
| Ligands | – | ||||||
| B factors (Å2) | |||||||
| Protein | 77.4 | ||||||
| Ligand | – | ||||||
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.00 | ||||||
| Bond angles (°) | 0.76 | ||||||
| Validation | |||||||
| MolProbity score | 1.80 | ||||||
| Clashscore | 6.06 | ||||||
| Poor rotamers (%) | 0.30 | ||||||
| Ramachandran plot | |||||||
| Favored (%) | 92.4 | ||||||
| Allowed (%) | 7.47 | ||||||
| Disallowed (%) | 0.12 |
Mini coat cage and hub substructure
The most abundant clathrin architecture was the mini coat cage, which reached the highest resolution of 9.1 Å (Fig. 1c, Supplementary Note 3). We note that the terminal domain densities are weaker than the main cage density due to domain flexibility, and consequently adaptor binding was not resolved. The density around the mini coat is observed to be variable (Supplementary Video. 1) and presented an opportunity for further structural averaging. Thus, in order to improve the resolution further, we used subparticle extraction and refinement 37 to obtain the structure of the invariant mini coat hub, which encompasses the vertex of the cage. This yielded an improved map resolution of 5.1 Å. The subparticle extraction and refinement procedure was repeated for all five of the cage architectures, determining the invariant hub structure at resolutions of 7.8 – 5.1 Å (Supplementary Notes 1d-f, 3). All of the hub substructures were combined and refined to determine a single consensus hub structure across all determined cage types, with a global resolution of 4.7 Å, with local resolutions reaching 4.3 Å and a marked improvement in density quality (Fig. 2b-d and Supplementary Notes 3, 4). Such improvement indicates the invariance across all the hub substructures, even though they are from different cage structures. This improved map enabled us to build a model of the clathrin hub (Table S2). The model, built into the C3 averaged map, contains contiguous segments of three heavy chains (residues 635-1075, 809-1474, 1250-1629) and two light chains (residues 99-157 of one and 99-165,189-225 of the other). Together the heavy chain comprises heavy chain repeat (HCR) 1g through to HCR7j, including the trimerization domain (TxD) (Fig 2a and Table S3). Within these areas of the map local resolution measurements report resolutions ranging 5.9 – 4.3 Å and 6.7 – 4.4 Å for the heavy and light chain respectively with the worst density appearing in regions furthest from the hub. Bulky side chains from landmark aromatic residues are well resolved to model the main chain through the density and assign the register (Fig. 2c and Supplementary Fig. 1a). The model was validated using emRinger, map-vs-model measurements and per residue attribute plots (Supplementary Fig. 1a-b and Tables 1, S1 and S2). These report reasonable statistics 38 for a model at this resolution. Although there is variation in the quality of the map, due to the repeating structure of the hub (e.g. residues 1250-1474 are seen in two chains) good per residue statistics are found across the whole residue range modelled.
Figure 2. The consensus hub substructure from all and individual clathrin cage architectures.
(a) The domain structure of the clathrin heavy and light chains. (b) The consensus hub structure resulting from subparticle refinement of all cage types at 4.7 Å resolution. The domain structure is coloured according to (a) and the unsharpened density is shown as transparent. (c) The consensus map density and model between HCR6c-7c are shown as well as (d) the gold-standard FSC curves for the consensus hub structure and each cage type with its respective hub substructure.
Interface stabilisation analysis
Our model provides new insights into the interactions made by triskelion legs within a clathrin cage and our understanding of the cage assembly mechanism. A cage edge is formed from four separate triskelion legs and is composed of two antiparallel proximal domains that sit above two antiparallel distal domains (Fig. 1c and Supplementary Fig. 2). We were interested to identify areas of the structure that formed stable interactions and so performed an analysis of intermolecular interaction energies predicted by our model using Rosetta. Complementary analysis that accommodated uncertainty in rotamer position was made in a new application of the molecular docking programme, BUDE 39,40, where we were able to calculate interaction energies for all favourable rotamer positions in the model. Results from both Rosetta and BUDE calculations (Fig. 3 and Supplementary Fig. 1c-d) showed good agreement in energy scoring indicated by an overall cross-correlation of 0.80 and 0.81 for the heavy and light chains respectively. To illustrate the implications of these results for an entire cage, the Rosetta results obtained for the consensus hub model were mapped onto the minicoat cage map (Fig 3a). While the energy-per-residue profiles between BUDE analysis of the EM structure and the Rosetta scored and partially relaxed structure are similar, there are some differences. For example, while BUDE identifies the interaction of light chain tryptophan 105 with the heavy chain, this interaction is not evident in the Rosetta scoring, which is more sensitive to the conformational uncertainties associated with a map in the 4 Å resolution range. Nonetheless results from both approaches showed strong interaction energies between light chain tryptophans 127 and 138 and the proximal domain, and at the trimerization domain. This is consistent with biochemical studies that have shown that two light chain tryptophans (W105 and W127) are required for light chain binding to the heavy chain, 41 and demonstrated the stability of the trimerization domain interaction 42. There were also strong interaction energies at additional positions along the length of the cage edge. This led us to investigate our molecular model at these positions, as discussed in the following sections.
Figure 3. Clathrin cage stabilisation.
(a) Rosetta energy scores mapped onto the mini coat. Inset is an equivalent view to the left panel in (c). (b) Interaction energies between clathrin subunits predicted from analysis by Rosetta of the consensus hub structure for heavy chain (Top) and light chain (Bottom). BUDE interaction energies are shown in grey. (c) A view of the proximal-distal contacts made by heavy chains of the cage inner and outer shell is shown from the side (left) and the distal-distal contacts made by heavy chains on the coat inner shell are shown from below (right). See also Supplementary Fig. 2. The locations of the disease-related mutations P890L and L1074P are shown by gold and green spheres respectively.
Leg to leg interactions
Our analysis of interaction energy indicated several interaction ‘hotspots’ along the length of the assembled legs (Fig. 3). Of these, two are close to point mutations in the human clathrin gene which have been identified in patients with autosomal dominant mental retardation-56 (MRD56)43,44. These mutations, P890L and L1074P, fall close to interaction hotspots formed between residues 883-888 and 981-984 of the distal domain and between distal and proximal domain residues 1040-1046 and 1428-1433 (Fig. 3c and Supplementary Fig. 2 and Supplementary Table 4). This emphasises the importance of such stabilising interactions for the cellular function of clathrin. A number of studies have highlighted the potential significance of histidines in clathrin assembly, which in vitro is pH dependent 45-47. In our consensus hub structure, the only histidines involved in potential intermolecular contacts are 1279 and 1432, which are close to asparagine 853 and glutamate 1042 respectively on neighbouring heavy chain legs, and have favourable interaction energies in our Rosetta analysis. This does not rule out the possibility that other histidines 46, not identified in our structure as forming stabilising interactions, contribute to clathrin cage assembly via alternative mechanisms.
Trimerisation domain stabilisation
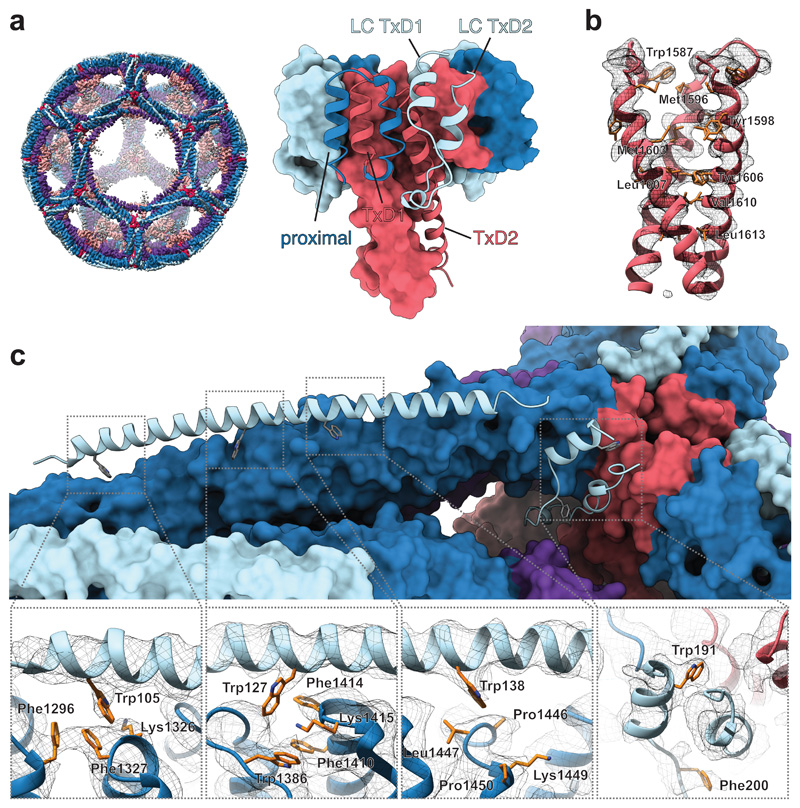
Our model of the assembled clathrin hub reveals the interactions that stabilise key elements of the clathrin lattice, between adjacent heavy chains, at the light chain-heavy chain interface and within the trimerisation domain (TxD). In the clathrin trimerisation domain strong interactions between the C-termini of three heavy chains 42 determine the triskelion structure. Biochemical studies have defined residues 1550-1675 as the smallest region capable of trimerisation 48 and further shown that the C-terminus of the light chain stabilises trimerisation 42. It has also previously been shown that the respective heavy chains of the TxD form a tripod of three helices 28 with two further C-terminal light chain helices embedded in the heavy chain tripod structure 35. Our structure reveals (Fig. 4a-b) that this helical tripod forms extensive coiled-coil contacts between residues 1606-1617 (within helix TxD2). Cysteines 1565, 1569 and 1573 have been implicated in hub assembly 49. In our model these residues face helix HCR7h-i1546-1561 and align along one side of helix HCR7j1564-1575 which interacts with one of the light chain C-terminal helices (LC1 TxD) that engage the hub (Supplementary Fig. 3). Their role in hub assembly may therefore be to stabilise the helices involved in creating the interface for the energetically-favoured light chain – heavy chain interaction (Fig. 3b and Supplementary Fig. 3).
Figure 4. Structural features of the trimerisation domain, coiled-coil and light chain interactions.
(a) Close up view of the trimerisation domain (TxD) composed of three heavy chains (TxD1 and TxD2) and three associated light chains (LC TxD1 and LC TxD2). A whole mini coat is shown for reference. (b) Molecular model and density for the TxD helical tripod, showing coiled-coil interactions between the three helices. (c) Detail of light chain – heavy chain interactions. Panels show the molecular model and consensus hub map density for interactions involving four key tryptophans. At HCR7f the light chain becomes disordered (LC166-189). The light chain becomes ordered again at LC TxD 1 and 2 which bind at the trimerisation domain (TxD). The newly determined sequence register for LC TxD helices 1 and 2 places them between two adjacent TxD heavy chains in trans. Colouring as for Fig. 2a.
Light chain association at the proximal and trimerisation domain
Our structure reveals in detail how the light chain associates with the heavy chain and how the light chain C-terminal helices engage with the trimerisation domain. Light chains of 23-26 kDa are tightly associated with heavy chains requiring strong chemical denaturants such as sodium thiocyanate to remove them 50. They have been implicated in the timing and productivity of vesicle formation 51, in regulation of GPCR endocytosis 52, clathrin disassembly 53, and negative regulation of association of huntingtin interacting proteins (HIP1 and HIP1R) with actin 54. Previous studies showed that the light chain forms a helix that binds to the proximal domain of the heavy chain 28 and crystallographic studies have suggested the presence of further light chain helical structure at the trimerisation domain 35. In our map, this proximal binding light chain helix is well-resolved, enabling fitting of the continuous density from residues 99 to 157, spanning 11 helical repeats of the heavy chain proximal domain from HCR6b to HCR7f (Fig. 4c). The sequence register of this area of our map and model also agrees closely with the X-ray structure of residues 1210-1517 34 of the proximal domain (Supplementary Fig. 1e). Our model explains yeast two-hybrid studies 41 that have previously demonstrated the importance of Trp 105 and Trp 127 for light chain binding and their stabilising role is highlighted by energetic analysis (Fig. 3). Mutation of Trp 105 to arginine abolishes LC-HC binding but is rescued by mutation of lysine to glutamate at 1326 41. In our model, Trp 105 forms an aromatic stacking interaction with Phe 1327 and Phe 1296. In the yeast-2-hybrid study, binding was lost upon mutation of Trp 127 to arginine but rescued by mutation of Lys 1415 to glutamate 41. We show that LC Trp 127 binds in a hydrophobic pocket between HCR6h-HCR6i created by Phe 1410, Phe 1414 and Trp 1386, further adjacent to Lys 1415. We modelled the mutations resulting from yeast two-hybrid studies, using the most common rotamers. In each case the rescue mutation forms a plausible salt bridge with the originally disruptive mutation, with the charged groups for the pairs Arg 105/Glu 1326 and Arg 127/Glu 1415 within 3 Å of one another (Supplementary Fig. 3c-d). Between residues 158 and 189 the light chain becomes disordered before rebinding at the trimerisation domain in a helix-loop-helix arrangement. The further two helices (LC TxD1 and TxD2) in the helix-loop-helix arrangement stably associate with two TxD domains in trans, bridging adjacent legs that join to make the triskelion vertex (Fig. 4c and Supplemental Fig. 3a).
Cage structural modelling and analysis
Clathrin is notable for forming a wide variety of lattice types, including multiple cage architectures, flat lattices 3 and even tubular structures 55. In order to find out how triskelion legs adapt to form varying cage architectures, we locally fitted triskelion leg segments from our consensus hub structure into the whole cage maps determined in this study. Three cages, the 28 mini coat, 36 barrel and 36 tennis ball (Fig. 5) were at sufficient resolution to allow docking of the segments 1629-1281, 1280-1131 and 1130-840 and 839-635 for each leg. The independent local fitting of each cage type provides an unbiased view of the variable leg conformations and angles across three different cage types (Supplemental Movie 2–3). We found across cage types, as expected, that there were variable leg conformations (Supplementary Fig. 4a). These were characterised by consistent angular differences of ~18° in the proximal to distal joint region, ~12° in the distal to ankle region, and more substantial angular variation of 7 – 17° between the distal joint and distal domain. We note that the heavy chain helices from the N-terminus to residue 965 are twisted relative to previously published alpha carbon models (Supplementary Fig. 4b-c). This indicates that a different ankle-distal surface is presented to the inside of the cage which may have implications for understanding adaptor protein binding.
Figure 5. Contacts between triskelion legs for different cage architectures.
The panels show the pattern of intermolecular contacts for molecular models fitted into the mini coat, hexagonal barrel and tennis ball cage structures. Intermolecular contacts for the consensus hub model are also shown. Aligned, individually modelled heavy chain legs for each cage structure are shown, with cylinders depicting each domain. See also Supplementary Fig. 6.
It might be expected that the distribution of the leg angles would correlate with the local cage geometry that a leg contributes to, i.e. whether a leg segment associates with a hexagon or a pentagon. In order to address this question, we assigned each leg a geometric signature describing its local geometric context within the cage architecture and plotted the leg angles for each individual conformation (Supplementary Fig. 5). We found that legs related by symmetry in the mini coat and barrel cages had, as expected, comparable conformations. However, where some legs shared the same geometric signature but were not related by global symmetry, as frequently found in the tennis ball, this was not the case. This shows that legs can fit within a particular geometric context with a variety of conformations and suggests that individual leg conformations may result more from longer range influences from other legs rather than simply the local geometric environment.
Conserved interaction patterns between cage types
In order to determine how universal the leg-leg contacts were in the formation of different clathrin structures we determined contact maps for the complete cage maps of the mini coat, hexagonal barrel and tennis ball structures based on the individual fitting of triskelion legs into these maps discussed above (Fig. 5). The envelope provided by the lower resolution whole cage maps provides information on the path of a particular triskelion leg, which we observed to vary in whole cages. Such changes in the path of a leg might be presumed to result in differences in the contacts made between legs in a whole cage in comparison to the consensus hub structure. To investigate this, we determined contact maps reporting contact pairs between alpha carbons on different chains that were within 6 Å of one another for individual legs within whole cage models. At the resolutions of the whole cage maps (Supplemental Notes 1-3) we do not expect to be able to define precise molecular contacts, however changes in the pattern of intermolecular contacts could indicate broader changes in interaction between legs in whole cages. Interestingly we see no obvious qualitative change in the pattern of these contacts across cage types nor in comparison to the consensus hub structure contact map (Fig. 5). Thus while angular changes, which may result from small movements within the hub, enable a leg to adapt to different geometries, they do not significantly alter the pattern of intermolecular contacts that are formed. We do note intriguing differences in the distributions of contacts between heavy chain legs of different geometries (Supplementary Fig. 6) but higher resolution structures of these whole cages would be required for further interpretation. Overall our data suggest that leg-leg contacts are well-preserved between cages with different architectures and leg conformations.
Discussion
Our map and molecular model for assembled clathrin in combination with analysis of five distinct cage architectures provide new insights into the assembly mechanism of clathrin coats. The pattern of interactions we observe, combined with a limited range of leg conformations, suggests that gentle flexing of the legs between established contact points allows clathrin to adapt its conformation to form the architecture required to support membrane and adaptor dynamics, without the requirement for new contacts specific to a particular cage architecture. This extends previous observations on the contribution of variations in leg conformation to the hexagonal barrel cage structure 27,28,56. The series of interaction points that we have identified at intervals along the clathrin legs suggests the strength of the leg-leg interaction would depend on the degree of alignment of these multiple binding sites. Thus we can speculate that, where binding sites are aligned this would result in a stable lattice which may nonetheless be easily destabilised by factors that alter that alignment. This scenario is consistent with the rapid exchange of triskelia observed during clathrin-coated vesicle formation 4,5,57. Our data thus reveal a universal mode of clathrin assembly that supports its role in rapidly changing and morphologically varied cellular coat structures.
For rapid exchange of triskelia to take place, clathrin-adaptor interactions must also be considered. Clathrin is recruited to the growing coated vesicle at the plasma membrane by adaptor proteins, primarily AP-2 which, through specific binding to receptor internalisation motifs, enables selection of cargo for inclusion in the vesicle. AP-2 recruits further adaptor proteins, through its appendage domains, which can bring additional cargo to the vesicle 58 or influence the size or shape of the resulting coated vesicle 59,60. Aguet et al 60 showed that removing the α–appendage domain from AP-2 resulted in clathrin-coated structures that lacked curvature suggesting that, in cells, interaction between AP-2 and certain adaptor proteins is required to generate a curved clathrin lattice. This contrasts with experiments in vitro 61 in which clathrin polymerisation on liposomes incubated with a clathrin binding epsin domain drove membrane curvature sufficiently to form clathrin-coated buds. This highlights the fine balance of competing interactions involved in coated vesicle formation. In allowing diverse cage architectures to be formed using relatively small changes in leg conformation and common sites of interaction, the structure of clathrin presents a neutral framework amenable to adaptation by changing conditions.
The cage architectures examined in this study are relatively small compared to many of the structures likely to occur in cells. However, along with other larger (less symmetrical) cage arrangements, the tennis ball structure has been observed to enclose a membrane in purified coated vesicles 62. Since the hub structure does not significantly vary across cage architecture, it seems possible that the interaction patterns observed in this study are relevant for other cage types formed from pentagons and hexagons. Whether these interactions are universal for other types of clathrin lattice, for example flat 3 or cylindrical lattices 55 or coats with heptagonal3,62 or square faces63 remains to be determined.
Methods
Buffer compositions
Polymerisation buffer. 100 mM MES pH 6.4, 1.5 mM MgCl2, 0.2 mM EGTA, 0.02 w/v NaN3. Depolymerisation buffer. 20 mM TEA pH 8.0, 1 mM EDTA, 1 mM DTT, 0.02 w/v NaN3. Tris buffer. 1 M Tris pH 7.1, 1 mM EDTA, 1 mM DTT, 0.02 w/v NaN3. HKM buffer. 25 mM HEPES pH 7.2, 125 mM K Ac, 5 mM Mg Ac, 0.02 w/v NaN3. Ficoll/Sucrose buffer. 6.3 w/v Ficoll PM 70, 6.3 w/v sucrose in HKM pH 7.2. Saturated ammonium sulphate. Excess ammonium sulphate dissolved in 10 mM Tris pH 7, 0.1 mM EDTA. Prescission buffer. 50 mM tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1mM DTT, 0.02 w/v NaN3.
Protein purification and expression
Endogenous clathrin coated vesicles were extracted from Sus scrofa brains and clathrin purified from them as triskelia using previously described methods 64. The initial assembly for harvesting cages was performed by dialysis into polymerisation buffer pH 6.2 and subsequent ultracentrifugation with concentration by resuspension of the pellet into a small volume of polymerisation buffer. All subsequent uses of polymerisation buffer utilised a pH of 6.4. Clathrin concentration was assayed by A280 of triskelia to avoid effects from light scattering.
The GST β2-adaptin616-951 plasmid was a kind gift from Steve Royle, University of Warwick 65. β2-adaptin616-951 was expressed as a GST fusion protein in an E. coli BL21 strain and purified using GSH resin (GE Healthcare), the GST tag was subsequently removed by cleavage using a commercially available GST fusion 3C protease (Prescission, GE Healthcare) overnight at 4 °C in Prescission buffer. Cleavage enzyme was removed by GSH resin and the cleaved protein collected from the flow through after which it was concentrated and exchanged into Tris buffer on vivaspin columns (Sartorius).
Complex preparation
The clathrin cage β2-adaptin complex was made by reconstitution in depolymerisation buffer (pH 8.0) at 4 °C, at 3.0 μM clathrin with a 6-fold molar excess of β2-adaptin and dialysis overnight into polymerisation buffer at pH 6.4. Clathrin-β2 cages were harvested by centrifugation at 230,000 g for 30 mins and concentrated 10-fold by pellet resuspension into a small volume of polymerisation buffer pH 6.4.
Negative stain transmission electron microscopy
4 μl of 1 μM assembled clathrin cages was applied to a glow-discharged formvar carbon 300-mesh copper grid (Agar Scientific) and allowed to adsorb for 1 minute. The sample was removed by blotting and 4 μl of 2% w/v uranyl acetate (UA) rapidly applied to the grid. This was incubated at room temperature for 1 minute to stain the sample after which the UA removed by blotting. Grids were air dried at room temperature before transfer to the microscope. Samples were imaged using a JEOL 2010F FEG transmission electron microscope with a Gatan Ultrascan 2000™ camera, at an accelerating voltage of 200kV.
Cryo-electron microscopy
Samples of clathrin-β2 cages were inspected by negative stain transmission electron microscopy (TEM) before freezing in vitreous ice for cryo-electron microscopy (cryo-EM). The concentration of clathrin for cryo-EM grid preparation was between 15-30 μM and determined by inspection of particle distribution in negative stain prior to freezing. 3 μl of clathrin-β2 cages were applied to glow-discharged 300 mesh copper Quantifoil R1.2/1.3 grids and blotted at ambient temperature and humidity for 3 seconds before plunging into an ethane/propane (80/20%) mix liquefied and cooled by liquid nitrogen using a hand freezing device (built by Birkbeck mechanical workshop, University of London). Cryo-EM micrographs were collected automatically in movie mode and acquired in four datasets using a Titan Krios (MRC-LMB) operated at 300 kV each equipped with a Falcon II detector. Using EPU for data acquisition of each dataset, a total dose of 42 – 69 e-/Å2 were collected over 3 seconds at dose rates of between 1 – 1.2 e-/Å2/s and a magnified pixel size of 1.705 Å/px using a 1.5 μm beam and 70 μm C2 aperture to ensure illumination of the carbon support with one image acquired per hole. Micrographs were targeted for collection between 1.4 – 3.2 μm defocus.
Cage library construction
A list was made of those cage species that are geometrically `probable' conformations, according to the `head-to-tail dihedral angle discrepancy exclusion' rule that has been proposed by Schein and co-workers 36, and such that the number of vertices was less than or equal to 38. The software CaGe 66,67 http://www.math.uni-bielefeld.de/~CaGe http://caagt.ugent.be/CaGe} was used to generate a set of labelled coordinates in three dimensions and a tabulation of vertex connectivity that identifies the edges of the polyhedron. These coordinates were then manipulated in R 68 so that the centroid was positioned at the origin; the fullerene's axes of symmetry were correctly orientated for application of symmetrical averaging at a later stage (see Data processing); alternate `handed' versions of the (chiral) 32- and 38-vertex fullerenes were obtained by a reflection; the coordinates were scaled to give a distance between connected vertices of 185 Å 26; and the coordinate set was augmented by points at a spacing of 1 Å along each edge. Hence, a library of 10 species of fullerene or cage architecture were compiled. The R library `bio3d' 69 was used to record each coordinate set in PDB format, which was then converted to an MRC volume suitable for use as a reference for classification using the program e2pdb2mrc.py from the EMAN2 package 70. Coordinates were converted to a volume at a low pass filtered resolution of 60 Å.
Data processing
The movies were motion corrected using MotionCor2 71 to produce motion corrected summed micrographs, with and without dose weighting. The contrast transfer function of the micrographs was estimated from the non-dose weighted micrographs using gctf v1.06 72 using the validation and EPA functions. 12,785 particles were manually picked from the motion corrected micrographs using e2boxer.py from the Eman2 package. Non dose weighted particles at a binned pixel size of 10.2 Å/px were extracted and subjected to reference-free 2D classification over 28 iterations in Relion. High quality 2D classes were selected for further classification. The selected subset of particles contained 9,500 particles and was used in a supervised asymmetric 3D classification into 10 structural classes (see Cage library construction sections). The 38 big apple reference contains 38 triskelia however the resulting 3D classification volume contained only 37 triskelia indicative of the robustness against reference bias in this particular analysis. The volumes from the classification and associated particles were used for subsequent classification and refinement. Reconstructions with and without imposing symmetry were found to correlate, further indicating a lack of reference and symmetry bias.
Further image processing was performed using Relion-2.1 73. Phase flipped particles were extracted from the dose weighted micrographs. The extracted box size was 500 px at a pixel size of 1.705 Å to include cages up to a size of 850 Å. Each cage architecture was refined unbinned without imposing symmetry (Table 1) from the supervised 3D classification subsets and volumes low pass filtered to 40 Å. A mask was generated from the C1 reconstruction at 3σ, extended and softened by 4 and 9 pixels. This mask was used to refine the respective structures imposing symmetry whilst employing solvent flattening and a Gaussian noise background. The resolutions of each reconstruction were estimated using the gold-standard FSC measurement within a mask created from the refinement volume at 3σ, expanded by 4 pixels and softened by 9 pixels (3σ/e4/s9). The MTF of the Falcon II camera operated at 300 KeV was applied and the B-factor of the map automatically calculated when the resolution exceeded 10 Å 74. The resolution of each reconstruction was found to correlate with the number of asymmetric units utilised in the refinement (Supplementary Note 1d). Further unsupervised 3D classification of each structure was performed into 3 classes with a regularisation parameter T of 4, no imposed symmetry and no mask to ensure no reference bias was present in the particle subsets (Supplementary Note 2).
Localised subparticle extraction and reconstruction
Relion localised reconstruction python scripts 37 and in-house written scripts were utilised to extract trimerisation hub subparticles from the whole cage particles. The hub subparticles were then refined and reconstructed independently to obtain the highest resolution hub structure. This is described in detail in the supplemental methods. A structure of the hub containing residues 581-1180, 700-1550, 1100-1630 was generated from 3IYV and fitted into the hub of the 9.1 Å 28 mini coat volume. This model was fitted at each of the hub vertexes of each cage and used to define a vector in UCSF Chimera describing the location of the hub in the respective C1 whole cage reconstruction. The hubs were extracted and recentred as new subparticles in 256 px boxes. The subparticles were extracted from whole cage particles boxed in at 750 px at 1.705 Å without phase flipping to include all signal delocalised due to the defocus 74 applied during data collection. This was repeated for each cage architecture and thus in each case the subparticle extraction expanded each whole cage dataset to n times the original particle number where n is equal to the number of hubs in the respective cage architecture. Reconstructions of the newly extracted hub subparticles were made to serve as references for refinements. Refinements were first conducted in C1 without masking, and further refinements made using C3 symmetry, masking applied from a 3σ extended 4px and softened 9px mask (3σ/e6/s9), with solvent flattening and a Gaussian noise background. Due to the special case where protein density extends outside of the refinement mask and box, for the hub subparticles it was necessary to apply a soft spherical mask (320 Å diameter, 10 Å softened) to the unfiltered half maps before resolution estimation, to remove protein density outside the sphere of refinement. Gold-standard resolution estimation of each hub subparticle then followed and estimated the reconstructions to be between 5.1 and 7.8 Å. Each hub subparticle particle set was combined to create a consensus refinement of the hub structure in every cage architecture that had been determined. This structure was refined as previously described and the map used for subsequent final model building and optimisation (see model building). Global resolution was measured as described by gold-standard FSC (3σ/e6/s11) and found the reconstruction to have converged at a resolution of 4.7 Å, with a measured map B-factor of -165 Å2. Local resolution estimations were made using Relion revealing resolutions extending to 4.3 Å in the areas that were modelled.
Protocol for localised subparticle extraction and reconstruction
The invariant hub structures at the vertices of all cages types present an opportunity for symmetry averaging. However the formal symmetry of these cages does not describe the location of every hub due to the presence of additional local symmetry and potentially cage deformation. We used localised reconstruction 37 as mentioned in the previous section to identify the locations of all repeating hub subunits within each cage structure and adopted a generalised protocol for this as follows:
Complete C1 refinement of whole cage complex
Generate and fit a hub PDB to the repeating subparticle of the whole complex
Use localrec_create_cmm.sh to create a vector defining the origin of the map and the centre of mass each PDB fitted to the subparticle
Use localrec_create_subparticles.sh and relion_localized_reconstruction.py to localise the position of the subparticle in the C1 reconstruction and by reference to the Relion star file, the subparticle in each original particle image. Subparticles will be reboxed and extracted with new origins.
Use localrec_create_substructures.sh and relion_localized_reconstruction.py to reconstruct the substructure volume representing each subparticle
Join all subparticle star files to process in 2D/3D classification, and refinement.
Use localrec_create_subtration_masks.sh to generate softened volumes minus the subparticle of interest for partial signal subtraction.
Use localrec_create_subparticles.sh to generate the same localized subparticles with and without signal subtraction.
Use localrec_create_substructures.sh to reconstruct the substructure volume representing each partial signal subtracted subparticle
Join all star files for the partially signal subtracted subparticles and process in 2D and 3D classification, and refinement.
Model building
Density for bulky side chains was visible, and comparison of the relevant map density to an X-ray structure of residues 1210-1517 of the proximal domain showed a good agreement in residue register 34. The crystal structure of the proximal clathrin heavy chain (1B89) fitted to the hub substructure was thus used to obtain the initial register of the map. The remaining helices were built as ideal polyalanine helices in Coot 75. Register was assigned manually in Coot using landmark residues, secondary structure prediction and comparison to the existing Cα cryo-EM model of clathrin (3IYV). Loops were initially modelled using phenix.fit_loops. The light chain modelled as isoform B (Accession: F1S398_PIG) was assigned sequence register by landmark tryptophan residues in the proximal region (W105/127/138). The TxD associated helix-loop-helix sequence register was assigned by density consistent with the landmark tryptophan residue (W191) as well as a C-terminal phenylalanine (F200). For clarity and consistency with the literature, we have referred to light chain residues using the bovine numbering. Further modelling proceeded iteratively between Coot 75, O 76 and phenix.real_space_refine 77 aiming to maintain idealised geometry and sequence register. Map-to-model comparison in phenix.mtriage validated that no overfitting was present in the structures. Atomic displacement factor refinement was used to calculate the residue B-factors and EMringer to score side chains. Model geometry was validated using MolProbity 78. All map and model statistics are detailed in Table 1.
Interface energy analysis with BUDE and Rosetta
Interface energy analysis was performed on the cryo-EM consensus hub structure using Rosetta and the docking program Bristol University Docking Engine (BUDE). Rosetta (v3.10) was used to calculate interface energy scores, reported in rosetta energy units (REU) using the residue_energy_breakdown application and the ref2015 scoring function 79,80. The structure was prepared for energy scoring by minimising the structure with the relax application 81,82 with all-atom constraints for five iterations and a final single iteration of backbone-only constrained relaxation. The RMSD between the starting and relaxed structure for C-alpha positions and all atoms was 0.22 and 1.48 Å respectively. A utility was written to sum the energy contribution of all pairwise attractive and repulsive intermolecular contacts identified by Rosetta for each residue in the consensus hub structure, which were subsequently summed over five residue windows for comparison to the analysis by BUDE. BUDE (v1.2.9) 39,40 was configured to report the theoretical free energy (ΔG/kJ/mol) of binding between two static input structures using the heavy_by-atom_2016-v1.bhff forcefield. BUDE is suitable for use with a medium resolution model because it uses soft-core atoms and distance-based functions (with 2–6 Å range beyond the sum of the atomic radii permitted) to describe hydrophobicity, hydrogen bonding and salt bridges and is therefore tolerant of model inaccuracies inevitable in the 4 Å resolution range. BUDE analysis was initially performed on the consensus hub structure. Then, to examine the influence of sidechain positioning, the interaction energy between pairs of sidechain residues across the interface was calculated as the average of all favourable rotamer-rotamer interactions using a customised version of BUDE. A utility was written to extract one chain from the consensus hub structure and assign the rest of the structure as the BUDE receptor. The extracted chain was split into five residue segments, at one-residue increments, and passed to BUDE as the ligand. This produced a sliding window of interaction energies along the interfaces of the clathrin structure. A good agreement in the energy scoring determined by BUDE and Rosetta is indicated by high cross correlations (Supplementary Fig. 1c-d). Energy minima in the Rosetta and BUDE energy scoring data were found using a five residue sliding window and using threshold criteria rejecting minima that are below 20% of the largest minima of the data (Table S4). We note that BUDE successfully reports realistic energy scoring without the necessity to modify the input structure.
Cage architecture model building
Molecular models for the whole cage maps were constructed in UCSF Chimera by building a complete triskelion (635-1629) from the consensus hub structure as follows. Firstly, the consensus hub model was fitted as a rigid body into each hub of the 28 mini coat, 36 barrel and 36 tennis ball. Each hub contains segments of 9 clathrin heavy chains (chains A,F,K: 1248-1629; chains B,G,L: 809-1474; chains C,H,M: 635-1075) and 6 clathrin light chains (chains D,I,N: 99-225; chains E,J,O). To construct a full triskelion the common portions of each of these chains were structurally superimposed keeping the TxD-proximal 1629-1281 region fixed to maintain the invariant hub. The fit of the triskelion from 1280-635 was then optimised by local model to map rigid body fitting of heavy chain segments. In summary the hub structure from 1629-1281 was maintained and three fragments of 1280-1131, 1130-840 and 839-635, were fitted to the map. These fragments were selected manually through inspection of the density and we note that an analysis of the alpha carbon models of clathrin heavy chains from the previously published 36 barrel 28 reveal bend points at 1281-1130 and 837-840, consistent with the choice of fragments for fitting in this study. The heavy chain modelling was repeated individually for each heavy chain in every triskelia of the 28 mini coat, 36 barrel and 36 tennis ball to produce molecular models that reflect the changing heavy chain conformations in the cages studied. Principal axes were defined through the regions 1629-1594 (TxD), 1550-1280 (proximal), 1230-1135 (distal junction), 1090-870 (distal), 815-635 (ankle) using UCSF Chimera. The angles between the regions were measured in every heavy chain for each cage type, using the minor principal axes. The proximal to TxD angle was found to be effectively invariant and as such only the remaining angles were plotted in python.
Contact maps
Contact maps were generated using UCSF Chimera ‘Find Clashes/Contacts’ to detect alpha carbons in proximity of one another (less than 6 Å). The points on each map represent a contact between residues on separate chains.
Map and model visualisation
Maps and models were visualised using UCSF Chimera 83.
Supplementary Material
Acknowledgements
CJS was funded by BBSRC grants BB/K003461/1 and BB/N008391/1 and KLM by BBSRC travel award BB/L018888/1. CJS was a Royal Society Leverhulme Trust Senior Research Fellow. YC was funded by National Institute of Health (NIH) awards R01GM098672 and P50GM082250. YC is a Howard Hughes Medical Institute investigator. RBS and AAI thank the EPSRC for support (EP/N013573/1). JRJ was funded by the EPSRC via the MOAC Doctoral Training Centre. MH was supported by the Medical Research Council Doctoral Training Partnership (MR/J003964/1). MB was funded by BBSRC (MIBTP) grant BB/J014532/1. We acknowledge the use of EM facilities at the MRC-LMB (Cambridge, UK), NeCEN and UCSF Mission Bay for data collection and in particular K. R. Vinothkumar for microscope operational assistance at the MRC-LMB. We acknowledge Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging centre (eBIC), proposals EM13142 and EM13909, funded by the Wellcome Trust, MRC and BBSRC. Sample preparation and development was supported by I. Hands-Portman, Warwick Life Sciences Imaging Suite (now Advanced Bioimaging Research Technology Platform), using equipment funded by the Wellcome Trust (grant no. 055663/Z/98/Z). We thank S. Royle, T. Burnley (CCP-EM), J. Huiskonen and S. Scheres for helpful discussions.
Footnotes
Reporting Summary statement. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability. BUDE is available under a free academic license from the developer Richard Sessions (http://www.bris.ac.uk/biochemistry/research/bude). All utilities and scripts are available on github (https://github.com/kylelmorris).
Data availability. Structural data have been deposited into the Worldwide Protein Data Bank (wwPDB), the Electron Microscopy Data Bank (EMDB) and EMPIAR84. EM electron density maps were deposited in the EMDB with accession numbers EMD-0114, 0115, 0116, 0118, 0120 for the 28 mini coat, 32 sweet potato, 36 D6 barrel, 36 tennis ball and 37 big apple respectively. The corresponding hub structure maps were deposited as EMD-0121, 0122, 0123, 0124, 0125 respectively. The consensus hub substructure map was deposited with the accession number EMD-0126. The atomic coordinates for the consensus hub were deposited with the PDB accession code 6SCT. Particle stacks associated with EMD-0114–0120 were deposited to EMPIAR as 10294. Particle stacks associated with EMD-0114–0120 without phase flipping and suitable for subparticle extraction were deposited to EMPIAR as 10295. Particle stacks associated with EMD-0121–0125 and EMD-0126 were deposited to EMPIAR as 10296. Other data are available upon reasonable request.
Author Contributions. C.J.S. conceived the overall project. C. J. S and Y. C. supervised research. K.L.M., J. R. J., M. H., S. W., M. J. B., A. A. I., R. B. S., A. D. C and C.J.S. performed research. K.L.M designed and developed the experimental analysis strategy, performed EM and image analysis, collected data, prepared the samples and purified the protein. J. R. J. constructed the cage library. K. L. M. and A. D. C. conducted modelling. R. B. S. and A. A. I. performed BUDE and Rosetta calculations. M.J.B assisted with protein preparation and M.H with data acquisition. S.W. assisted with data acquisition. J.P.A. assisted with data analysis. C. J. S. and K. L. M. wrote the initial manuscript with assistance and editing by all authors equally.
Competing interests. The authors declare no competing interests.
References
- 1.Brodsky FM. Diversity of clathrin function: new tricks for an old protein. Annual review of cell and developmental biology. 2012;28:309–336. doi: 10.1146/annurev-cellbio-101011-155716. [DOI] [PubMed] [Google Scholar]
- 2.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annual review of cell and developmental biology. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 3.Heuser J. Three-dimensional visualization of coated vesicle formation in fibroblasts. J Cell Biol. 1980;84:560–583. doi: 10.1083/jcb.84.3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avinoam O, Schorb M, Beese CJ, Briggs JAG, Kaksonen M. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 2015;348:1369–1372. doi: 10.1126/science.aaa9555. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, et al. Clathrin exchange during clathrin-mediated endocytosis. J Cell Biol. 2001;155:291–300. doi: 10.1083/jcb.200104085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowther RA, Finch JT, Pearse BM. On the structure of coated vesicles. Journal of molecular biology. 1976;103:785–798. doi: 10.1016/0022-2836(76)90209-6. [DOI] [PubMed] [Google Scholar]
- 7.Shih W, Gallusser A, Kirchhausen T. A clathrin-binding site in the hinge of the beta 2 chain of mammalian AP-2 complexes. The Journal of biological chemistry. 1995;270:31083–31090. doi: 10.1074/jbc.270.52.31083. [DOI] [PubMed] [Google Scholar]
- 8.Dell'Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 9.Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. Embo J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knuehl C, et al. Novel binding sites on clathrin and adaptors regulate distinct aspects of coat assembly. Traffic. 2006;7:1688–1700. doi: 10.1111/j.1600-0854.2006.00499.x. [DOI] [PubMed] [Google Scholar]
- 11.Ren X, Farias GG, Canagarajah BJ, Bonifacino JS, Hurley JH. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell. 2013;152:755–767. doi: 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen QT, Ren X, Zhang R, Lee IH, Hurley JH. HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science. 2015;350:aac5137. doi: 10.1126/science.aac5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldwein EE, et al. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci U S A. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris KL, et al. HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation. Cell. 2018;174:659–671 e614. doi: 10.1016/j.cell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular Architecture and Functional Model of the Endocytic AP2 Complex. Cell. 2002;109:523–535. doi: 10.1016/S0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 16.Kelly BT, et al. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson LP, et al. A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly BT, et al. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang DS, et al. Structure of an Arrestin2-Clathrin Complex Reveals a Novel Clathrin Binding Domain That Modulates Receptor Trafficking. The Journal of biological chemistry. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the β-propeller of clathrin. Proceedings of the National Academy of Sciences. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miele AE, Watson PJ, Evans PR, Traub LM, Owen DJ. Two distinct interaction motifs in amphiphysin bind two independent sites on the clathrin terminal domain [beta]-propeller. Nat Struct Mol Biol. 2004;11:242–248. doi: 10.1038/nsmb736. [DOI] [PubMed] [Google Scholar]
- 22.Zhuo Y, et al. Nuclear Magnetic Resonance Structural Mapping Reveals Promiscuous Interactions between Clathrin-Box Motif Sequences and the N-Terminal Domain of the Clathrin Heavy Chain. Biochemistry. 2015;54:2571–2580. doi: 10.1021/acs.biochem.5b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muenzner J, Traub LM, Kelly BT, Graham SC. Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin. Traffic (Copenhagen, Denmark) 2017;18:44–57. doi: 10.1111/tra.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigers GP, Crowther RA, Pearse BM. Three-dimensional structure of clathrin cages in ice. Embo J. 1986;5:529–534. doi: 10.1002/j.1460-2075.1986.tb04242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vigers GP, Crowther RA, Pearse BM. Location of the 100 kd-50 kd accessory proteins in clathrin coats. Embo J. 1986;5:2079–2085. doi: 10.1002/j.1460-2075.1986.tb04469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith CJ, Grigorieff N, Pearse BMF. Clathrin coats at 21 angstrom resolution: a cellular assembly designed to recycle multiple membrane receptors. Embo Journal. 1998;17:4943–4953. doi: 10.1093/emboj/17.17.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musacchio A, et al. Functional organization of clathrin in coats: Combining electron cryomicroscopy and x-ray crystallography. Molecular Cell. 1999;3:761–770. doi: 10.1016/s1097-2765(01)80008-3. [DOI] [PubMed] [Google Scholar]
- 28.Fotin A, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 29.Smith CJ, et al. Location of auxilin within a clathrin cage. Journal of molecular biology. 2004;336:461–471. doi: 10.1016/j.jmb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Fotin A, et al. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- 31.Heymann JB, et al. Visualization of the binding of Hsc70 ATPase to clathrin baskets: implications for an uncoating mechanism. The Journal of biological chemistry. 2005;280:7156–7161. doi: 10.1074/jbc.M411712200. [DOI] [PubMed] [Google Scholar]
- 32.Xing Y, et al. Structure of clathrin coat with bound Hsc70 and auxilin: mechanism of Hsc70-facilitated disassembly. Embo J. 2010;29:655–665. doi: 10.1038/emboj.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic Structure of Clathrin: A β Propeller Terminal Domain Joins an α Zigzag Linker. Cell. 1998;95:563–573. doi: 10.1016/S0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ybe JA, et al. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature. 1999;399:371–375. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- 35.Wilbur JD, et al. Conformation Switching of Clathrin Light Chain Regulates Clathrin Lattice Assembly. Developmental Cell. 2010;18:841–848. doi: 10.1016/j.devcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schein S, Sands-Kidner M. A geometric principle may guide self-assembly of fullerene cages from clathrin triskelia and from carbon atoms. Biophys J. 2008;94:958–976. doi: 10.1529/biophysj.107.110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ilca SL, et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nature Communications. 2015;6 doi: 10.1038/ncomms9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIntosh-Smith S, Wilson T, Ibarra AÁ, Crisp J, Sessions RB. Benchmarking Energy Efficiency, Power Costs and Carbon Emissions on Heterogeneous Systems. The Computer Journal. 2012;55:192–205. doi: 10.1093/comjnl/bxr091. [DOI] [Google Scholar]
- 40.Wood CW, et al. CCBuilder: an interactive web-based tool for building, designing and assessing coiled-coil protein assemblies. Bioinformatics. 2014;30:3029–3035. doi: 10.1093/bioinformatics/btu502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CY, et al. Clathrin light and heavy chain interface: alpha-helix binding superhelix loops via critical tryptophans. Embo Journal. 2002;21:6072–6082. doi: 10.1093/emboj/cdf594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ybe JA, et al. Light chain C-terminal region reinforces the stability of clathrin heavy chain trimers. Traffic. 2007;8:1101–1110. doi: 10.1111/j.1600-0854.2007.00597.x. [DOI] [PubMed] [Google Scholar]
- 43.DeMari J, et al. CLTC as a clinically novel gene associated with multiple malformations and developmental delay. Am J Med Genet A. 2016;170A:958–966. doi: 10.1002/ajmg.a.37506. [DOI] [PubMed] [Google Scholar]
- 44.Hamdan FF, et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am J Hum Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilbur JD, Hwang PK, Brodsky FM. New faces of the familiar clathrin lattice. Traffic. 2005;6:346–350. doi: 10.1111/j.1600-0854.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 46.Boecking T, et al. Key Interactions for Clathrin Coat Stability. Structure. 2014;22:819–829. doi: 10.1016/j.str.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crowther RA, Pearse BMF. Assembly and packing of clathrin into coats. Journal of Cell Biology. 1981;91:790–797. doi: 10.1083/jcb.91.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 49.Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4:850–856. doi: 10.1046/j.1600-0854.2003.00139.x. [DOI] [PubMed] [Google Scholar]
- 50.Winkler FK, Stanley KK. Clathrin heavy chain, light chain interactions. Embo J. 1983;2:1393–1400. doi: 10.1002/j.1460-2075.1983.tb01597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loerke D, et al. Cargo and Dynamin Regulate Clathrin-Coated Pit Maturation. Plos Biology. 2009;7:628–639. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferreira F, et al. Endocytosis of G Protein-Coupled Receptors Is Regulated by Clathrin Light Chain Phosphorylation. Current Biology. 2012;22:1361–1370. doi: 10.1016/j.cub.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 53.Young A, et al. Hsc70-induced Changes in Clathrin-Auxilin Cage Structure Suggest a Role for Clathrin Light Chains in Cage Disassembly. Traffic. 2013;14:987–996. doi: 10.1111/tra.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Molecular Biology of the Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elkhatib N, et al. Tubular clathrin/AP-2 lattices pinch collagen fibers to support 3D cell migration. Science. 2017;356 doi: 10.1126/science.aal4713. [DOI] [PubMed] [Google Scholar]
- 56.Kirchhausen T, Owen D, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6:a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, et al. Adaptor and clathrin exchange at the plasma membrane and trans-Golgi network. Mol Biol Cell. 2003;14:516–528. doi: 10.1091/mbc.e02-06-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 59.Morgan JR, et al. A role for the clathrin assembly domain of AP180 in synaptic vesicle endocytosis. J Neurosci. 1999;19:10201–10212. doi: 10.1523/JNEUROSCI.19-23-10201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- 62.Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T. Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. Journal of molecular biology. 2007;365:892–899. doi: 10.1016/j.jmb.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heymann JB, et al. Clathrin-coated vesicles from brain have small payloads: a cryo-electron tomographic study. J Struct Biol. 2013;184:43–51. doi: 10.1016/j.jsb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothnie A, Clarke AR, Kuzmic P, Cameron A, Smith CJ. A sequential mechanism for clathrin cage disassembly by 70-kDa heat-shock cognate protein (Hsc70) and auxilin. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6927–6932. doi: 10.1073/pnas.1018845108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hood FE, et al. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J Cell Biol. 2013;202:463–478. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinkmann G, F OD, Dress A, Harmuth T. CaGe – a virtual environment for studying some special classes of large molecules. MATCH Commun Math Comput Chem. 1997;36:233–237. [Google Scholar]
- 67.Brinkmann G, F OD, Lisken S, Peeters A, Van Cleemput N. CaGe - a Virtual Environment for Studying Some Special Classes of Plane Graphs - an Update. MATCH Commun Math Comput Chem. 2010;63:533–552. [Google Scholar]
- 68.R_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. http://www.R-project.org/ [Google Scholar]
- 69.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 70.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of molecular biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 75.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 77.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alford RF, et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park H, et al. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J Chem Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nivon LG, Moretti R, Baker D. A Pareto-optimal refinement method for protein design scaffolds. PLoS One. 2013;8:e59004. doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 84.Iudin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. EMPIAR: A public archive for raw electron microscopy image data. Nature Methods. 2016;13 doi: 10.1038/nmeth.3806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.