Abstract
Herpes simplex virus (HSV) infections can cause considerable morbidity. Currently, nucleoside analogues such as acyclovir are widely used for treatment. However, HSV infections resistant to these drugs are a clinical problem among immunocompromised patients. To provide more efficient therapy and to counteract resistance, a different class of antiviral compounds has been developed. Pritelivir, a helicase primase inhibitor, represents a promising candidate for improved therapy. Here, we established an HSV-1 infection model on microneedle-pretreated human skin ex vivo. We identified HSV-1–specific histological changes (e.g., cytopathic effects, multinucleated giant cells), down-regulation of nectin-1, nuclear translocation of NF-κB (p65), interferon regulatory factor 3 (IRF3), and signaling of the IFN-inducible protein MxA. Accordingly, this model was used to test the potency of pritelivir compared with the standard drug acyclovir. We discovered that both drugs had a comparable efficacy for inhibiting HSV-1 replication, suggesting that pritelivir could be an alternative therapeutic agent for patients infected with acyclovir-resistant strains. To our knowledge, we present a previously unreported ex vivo HSV-1 infection model with abdominal human skin to test antiviral drugs, thus bridging the gap between in vitro and in vivo drug screening and providing a valuable preclinical platform for HSV research.
Introduction
Herpes simplex virus-1 (HSV-1), the prototype of the α-herpesvirus family, infects 60%–80% of people worldwide (Cunningham et al., 2006). Upon primary infection via the skin and mucocutaneous regions, HSV can reach sensory neurons where latent infection is established (Rahn et al., 2015). HSV-1 primary infections in humans are typically asymptomatic and are therefore difficult to study, but when symptomatic, replication of HSV-1 causes lesions around the oral mucosa (i.e., cold sores) (Birkmann and Zimmermann, 2016). Normally, skin forms an effective barrier to infection, but skin abrasions, burns, or atopic diseases are conditions favoring HSV susceptibility (Shenoy et al., 2015; Whitley and Roizman, 2001). HSV is a leading cause of encephalitis, genital ulcerative diseases (Wilson et al., 2009), corneal morbidity, and blindness in humans and can trigger keratitis (Remeijer et al., 2004). Transmission of virus during birth in newborns and in immunocompromised individuals can be life threatening (Krawczyk et al., 2013). Several animal models have been developed, leading to a better understanding of multiple aspects of HSV biology, pathogenesis, disease, and immunity. The species most commonly studied are mice, guinea pigs, rats, and rabbits, but models resembling human physiological conditions during HSV infection are limited (Kollias et al., 2015).
For a successful invasion and infection of cells, particular surface receptors such as HVEM, 3-O-sulfated heparin sulfate, and nectin-1 have been described as essential (Akhtar and Shukla, 2009; Rahn et al., 2015). Five viral glycoproteins have been implicated in the viral entry process: gB, gC, gD, gH, and gL. Release of viral DNA into the nucleus allows initiation of transcription, replication of viral DNA, and assembly of progeny capsid within the host by the sequential expression of three gene classes (immediate-early, delayed-early, late genes) (Akhtar and Shukla, 2009). HSV-derived cytosolic nucleic acids trigger innate immune response genes in host cells, including type I IFN and proinflammatory genes (IL-6, IL-1β, tumor necrosis factor-α), which are known to require the IRF3 and NF-κB pathways, respectively (Abe and Barber, 2014).
To inhibit viral replication, viral DNA-targeting chemotherapeutics such as nucleoside analogues (e.g., acyclovir, penciclovir, valacyclovir, and famciclovir) are widely used either as episodic therapy for a short period or as daily suppressive therapy for months or years, even though latent virus cannot be eradicated (Biswas et al., 2014). Nucleoside analogues inhibit viral DNA polymerase after activation by viral thymidine kinase. HSV resistance to nucleoside analogues has been reported as being due to mutations in viral thymidine kinase or polymerase resulting in a loss or alterations in substrate specificity (Strasfeld and Chou, 2010), and this is recognized as a clinical problem among immunocompromised patients (Biswas et al., 2014; Krawczyk et al., 2013). Although in nucleoside analogue-resistant cases thymidine kinase-independent drugs such as the viral DNA polymerase inhibitors foscarnet and cidofovir can be administered, these compounds frequently have severe toxic effects, particularly in patients with comorbidities (Kim et al., 2015). To fulfill the request for effective alternatives to nucleoside analogue inhibitors and to counteract resistance, several compounds have been screened for their antiviral activity (Biswas et al., 2014). Moreover, potential alternative therapeutic vaccines are under investigation, but limited protections to viral shedding and lesion development have been shown so far (Birkmann and Zimmermann, 2016). Among all developing drugs, only helicase-primase inhibitors, targeting the HSV helicase primase complex, fulfill specific, small-molecule attempts. Furthermore, helicase-primase inhibitors (i) activate in the absence of viral or cellular enzymes such as thymidine kinase, (ii) are equally effective against HSV-1 and HSV-2, (iii) are orally bioavailable, (iv) exhibit higher potency than therapeutic vaccines, and (v) lack crossresistance to nucleosides (Birkmann and Zimmermann, 2016). Pritelivir (PTV, BAY-57-1293, AIC316) is a member of the group of helicase-primase inhibitors and shows a superior pharmacological profile compared with standard HSV treatment (Betz et al., 2002; Biswas et al., 2014; Crute et al., 2002; Kleymann et al., 2002). Recently, it has successfully finished a phase 2 clinical trial (Wald et al., 2014, 2016).
Currently, the drug efficacy screening of unreported compounds is performed in monolayer cell culture based-tests, followed by animal model testing and clinical trials. Only about 10% of the compounds progress successfully through expensive clinical development, because data obtained from monolayers and animal models do not reflect the human microenvironment and associated responses (Hopkins, 2008; Kola, 2008; Mullard, 2016). To lower the cost of failed compounds, the dismissal of ineffective and/or toxic compounds should happen as early as possible, ideally before animal testing. Therefore, it is essential to develop ex vivo human models that can more realistically mimic the in vivo cell behavior of cells and provide more predictable results for in vivo tests.
Here, we describe a human ex vivo HSV-1 infection model showing virus-mediated changes at the cellular level. Moreover, we were able to investigate HSV-1 targeted proteins and the downstream signaling pathway, which is practically impossible to test directly in human skin in vivo. Furthermore, we showed effective viral inhibition using the investigational drug pritelivir (Wald et al., 2014, 2016) and the standard drug acyclovir in the HSV-1 ex vivo model.
Results and Discussion
Efficient HSV-1 infection of microneedle-pretreated human abdominal skin
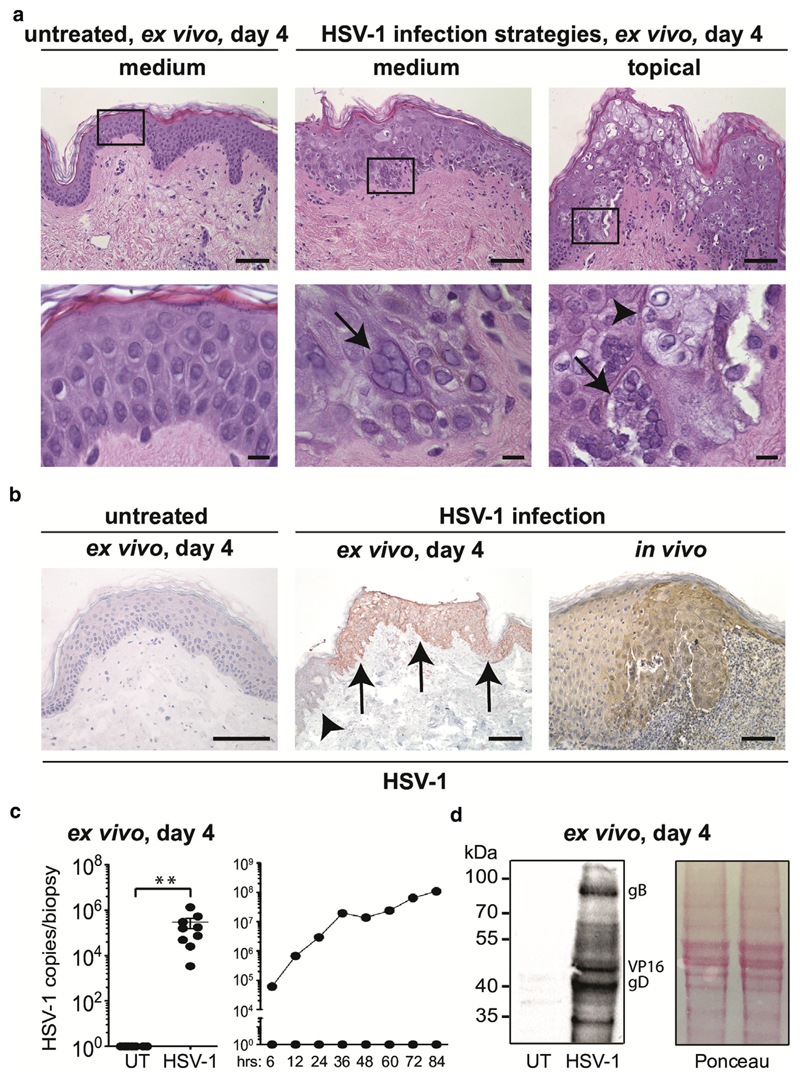
The inability to mimic the complexity of human skin using primary cells and/or cell lines poses significant limitations for studying host/pathogen interactions. Because of the lack of a human skin model resembling primary HSV-1 infection (Kim et al., 2015) and controversial findings in mouse skin (Rahn et al., 2015), we aimed to establish an ex vivo human skin model permissive to HSV-1 infection. To warrant efficient infection, dermatome-cut 0.5-mm–thick skin was treated with a derma roller, representing an established skin-needling procedure widely used to form micro injuries for anti-aging therapy (Singh and Yadav, 2016). Subsequently, the microneedle-pretreated skin was left untreated (i.e., uninfected), or HSV-1 was either administered into the medium or applied onto the top of the epidermis, cultured for 4 days, and analyzed. Examination of hematoxylin and eosin-stained skin sections exhibited no or modest (discrete spongiosis) morphological changes in untreated skin. In contrast, HSV-1–treated skin, irrespective of the administration route, showed viral-specific alterations such as multinucleated giant cells and cytopathic effects, predominantly in the epidermis (Figure 1a). These structural changes in keratinocytes after HSV-1 invasion are histological hallmarks for HSV diagnosis (Johannessen and Ogilvie, 2012). A similar infection pattern was achieved with both viral application routes with microneedle-pretreated skin. The phenomenon of spreading infection in particular skin regions is most likely due to the successful penetration and interaction of the virus with host cells after microneedling of the skin, which bypasses the stratum corneum via the holes, thereby allowing efficient virus infection in many parts of the skin. Without pretreatment, infection was adversely detected at the edge of the tissue (data not shown). Because we observed more infected area when virus was added to the medium after 4 days of culture compared with topical treatment (data not shown), we decided to use this application route for all further experiments.
Figure 1. Efficient HSV-1 infection in human skin explants.
(a) The upper panel shows hematoxylin and eosin staining of paraffin sections obtained from microneedle-pretreated, 4 day–cultured untreated skin (left) and HSV-1–treated 4 day-cultured skin (HSV-1 either applied into medium [middle] or topically [left]). Scale bars = 50 μm. The lower panel illustrates bigger magnifications (black box). Arrows denote multinucleated giant cells. Necroptotic changes are indicated with an arrowhead. Scale bars = 15 μm. (b) Immunohistochemistry staining of untreated (left) and HSV-1–treated skin (middle: infected area marked by arrows, uninfected area marked by an arrowhead) with an anti-HSV-1 serum. Data are representative from six donors. HSV-1–infected patient skin (right) was used as a positive control. Scale bars = 50 μm. (c) HSV-1 copies/biopsy sample in untreated (i.e., uninfected) and HSV-1–treated groups were analyzed using RT-PCR (left). Results are expressed as mean ± standard error of the mean from nine independent experiments, and donors and were analyzed with a Student t test (Wilcoxon matched-pairs signed rank test). **P < 0.01. HSV-1 replication kinetics in UT and HSV-1–treated skin were examined by measuring copies/biopsy sample with RT-PCR at indicated time points (right). Shown is one representative donor out of two with similar results. (d) Representative Western blot shows expression of HSV-1–specific glyocoproteins in HSV-1–treated skin but not in untreated skin using an anti-HSV-1 serum (n = 3). Ponceau S staining was included as loading control. HSV, herpes simplex virus; RT-PCR, real-time PCR; UT, untreated.
Immunohistochemical staining of paraffin-embedded skin sections showed a positive staining pattern with an anti-HSV-1 serum in HSV-1–treated but not untreated skin (Figure 1b). Specificity of the anti-HSV-1 serum was further confirmed on skin paraffin sections obtained from HSV-1–infected patients (Figure 1b). Isotype control staining results were always negative (data not shown). DNA quantification by real-time PCR (RT-PCR) showed comparable and significantly higher copies/biopsy sample in 4-day–cultured HSV-1–treated skin samples than in untreated control samples (Figure 1c). Analysis of HSV-1 replication kinetics in the skin over the course of infection showed an increase of copies/biopsy sample during a 4-day culture period in HSV-1–treated (i.e., infected) but not untreated (i.e., uninfected) skin (Figure 1c). Effective infection was confirmed with Western blot analysis, which detected HSV-1–specific glycoproteins in HSV-1–treated but not in untreated skin lysates (Figure 1d). Primary keratinocytes were used as a positive control (see Supplementary Figure S1a and b online) and confirmed results of previous in vitro studies (Montgomery et al., 1996). Here, we show successful HSV-1 infection in human abdominal skin with formation of syncytia and replication of virus at the protein level in situ.
HSV-1 entry receptor and downstream signaling pathway in human skin
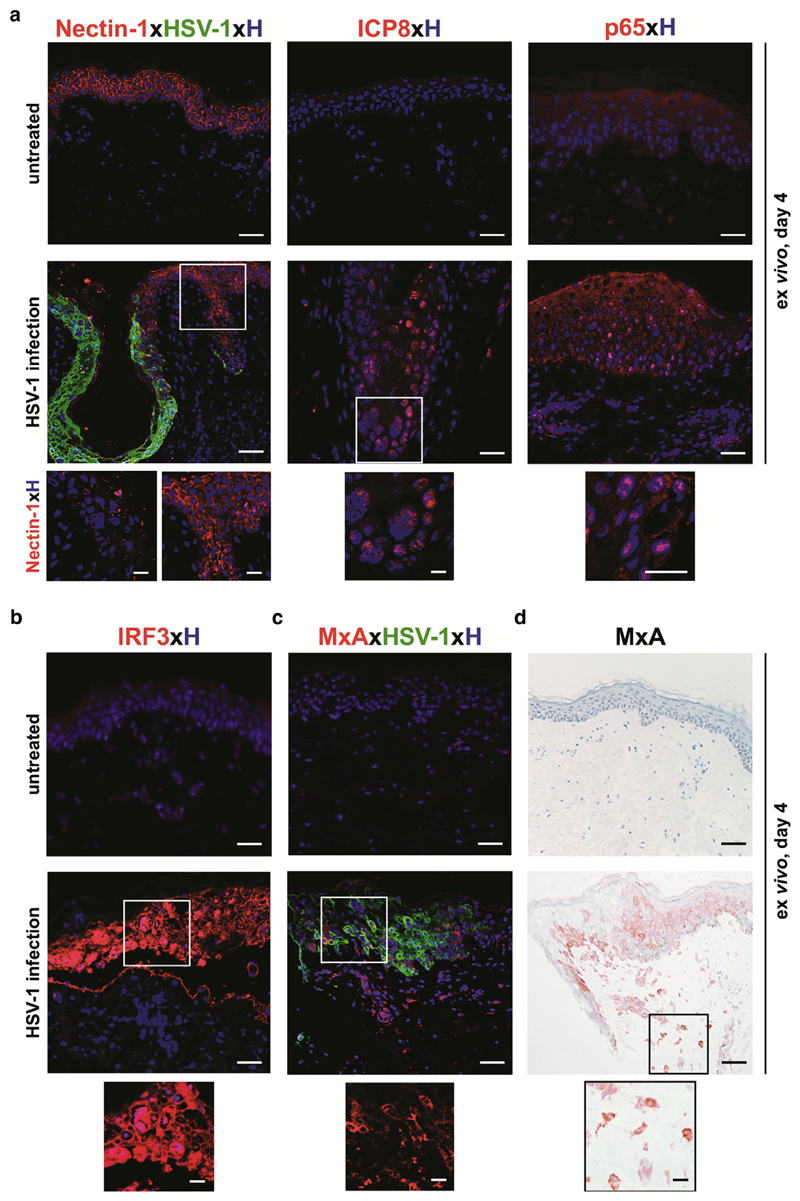
For a productive infection, HSV-1 glycoproteins must interact with various cell surface receptors such as nectin-1, HVEM, and 3-O-sulfated heparin sulfate (Rahn et al., 2015). In our model, we investigated nectin-1, because it has been reported to be an epithelial cell adhesion molecule serving as the major receptor for HSV-1 on keratinocytes (Petermann et al., 2015). Double immunofluorescence staining of paraffin sections showed prominent nectin-1 staining in the epidermis and no staining with HSV-1 serum in untreated skin. In contrast, a massive down-regulation of nectin-1 expression was observed in HSV-1–infected epidermal regions, whereas in the same section the uninfected epidermal area was comparable to untreated controls (Figure 2a). Our data are in line with results obtained with murine primary fibroblasts showing that HSV-1 infection mediates a rapid down-regulation of nectin-1, which correlated with virus endocytosis (Petermann et al., 2015).
Figure 2. HSV-1 successfully takes over the cellular system in skin cells.
(a) Immunofluorescence double labeling shows nectin-1+HSV-1– epidermal cells in untreated, 4-day–cultured skin. Upon HSV-1 treatment, nectin-1 expression is down-regulated in infected HSV-1+ epidermal areas only (inserts). Nuclear translocation of ICP8 and p65 appears in HSV-1–infected skin compared with untreated skin. Representative images with Hoechst nuclear staining are shown from three donors. (b) Immunofluorescence labeling indicates IRF-3 expression and translocation to the nucleus in HSV-1+ epidermal cells in HSV-1–treated but not untreated skin at day 4 of culture. No specific cytoplasmic IRF3 staining of any skin cells is observed in untreated skin. A representative merged image with Hoechst nuclear staining is shown from three donors. (c) Immunofluorescence double labeling shows MxA+HSV-1+ cells in the epidermis and MxA+HSV-1− cells in the dermis underneath the HSV-1–infected epidermal region, whereas the uninfected skin is negative. A representative merged image with Hoechst nuclear staining is shown from three donors. Insert shows single labeling of MxA-positive cells. (d) Immunohistochemical staining of 4-day–cultured HSV-1–treated and untreated skin paraffin sections show a similar MxA protein expression pattern (red-brown) compared with immunofluorescence. The insert indicates MxA+ cells in the dermis. Shown is one representative staining out of three independent experiments. Scale bars = 50 μm. Scale bars in inserts = 10 μm. HSV, herpes simplex virus.
During productive infection, HSV-1 efficiently redirects the host transcriptional machinery to express its own genes by modulating the NF-κB pathway (Amici et al., 2006; Lin et al., 2004). To ascertain productive infection in our model, we stained untreated and HSV-1–treated skin sections with antibodies directed against HSV-1 single-stranded DNA-binding protein (ICP8), NF-κB (p65), and IRF3. Antibodies were standardized with primary keratinocytes and freshly isolated, highly purified Langerhans cells, constituting the main cell types in the epidermis at the primary infection site. Nuclear translocations of ICP8, p65, and IRF3 were identified in the majority of HSV-1–treated keratinocytes but only in 10% of Langerhans cells (see Supplementary Figure S1c and d). The subsequent assessment of their translocation in our ex vivo model uncovered a comparable translocation pattern for ICP8 and p65 in HSV-1–infected skin (Figure 2a). In contrast, untreated skin tested negative for ICP8 expression and showed a typical cytoplasmic p65 staining (Figure 2a). Thus, down-regulation of nectin-1 indicates successful entry of HSV-1. Abundant nuclear ICP8 expression correlates with HSV-1 replication inside the cell, as reported previously (Tolun et al., 2013). Therefore, the observed nuclear expression of ICP8 in our model confirms viral DNA replication. p65 nuclear translocation endorses successful takeover of the cellular machinery by HSV-1 in our model.
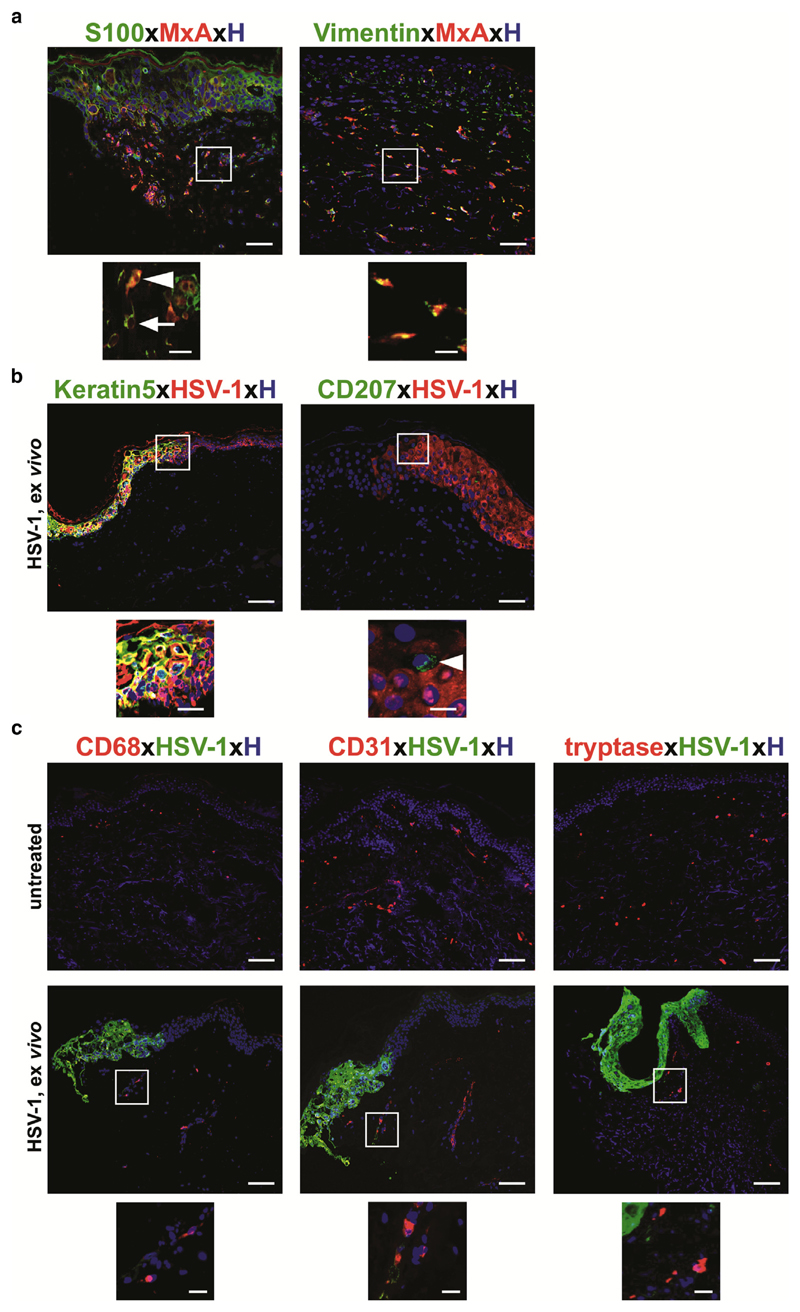
A virus-infected cell elicits a cellular response aimed at limiting virus replication and spread. The hallmark of this response is IRF3 nuclear translocation (Noyce et al., 2009) and the production and secretion of IFNs (Lin et al., 2004). Thus, we performed intracellular staining with an antibody directed against IRF3 and the IFN types I and III-inducible protein MxA as a sensitive readout for an IFN response. Immunofluorescence staining of HSV-1–treated skin showed expression and nuclear translocation of IRF3 only in HSV-1–infected areas (Figure 2b), whereas MxA-positive cells were present in HSV-1–infected epidermal layers and also in HSV-1–negative dermal areas located underneath in the same section (Figure 2c). This MxA staining pattern was confirmed by immunohistochemistry (Figure 2d). Untreated skin was negative for both IRF3 (Figure 2b) and MxA (Figure 2c and d). To uncover the nature of HSV-1–MxA+ dermal cells, paraffin sections were stained with an antibody directed against S100 protein, which is expressed by several cell types (e.g., neuronal crest, macrophages, keratinocytes, Langerhans cells, etc.). Only a few, but not all, MxA+ cells co-expressed S100 (Figure 3a). Furthermore, all MxA+ dermal cells co-expressed vimentin, which is found in various nonepithelial cells, especially mesenchymal cells (Figure 3a). Our data are in line with a report showing that MxA is strongly regulated by IFN during virus infection in human lung adenocarcinoma cells (Ku et al., 2011). Here, we confirm and extend MxA expression on HSV-1–infected epidermal cells and also uninfected mesenchymal cells. The role of mesenchymal cells during infection remains to be explored.
Figure 3. Epidermal keratinocytes are a primary portal for entry and spread of HSV-1 infection.
(a) Immunofluorescence double labeling shows a few MxA+S100+ cells (arrowhead), but most MxA+ dermal cells do not express this marker (arrow). All MxA+ cells express vimentin in HSV-1–treated paraffin sections. Inserts represent higher magnification of indicated staining. (b, c) Immunofluorescence double labeling indicates that HSV-1+ cells co-express keratin 5 but not CD207 (arrowhead). HSV-1+ cells do not express CD68, CD31, and tryptase. Representative merged images with Hoechst nuclear staining are shown from three donors. Scale bars = 50 μm; scale bars in inserts = 10 μm. HSV, herpes simplex virus.
We next investigated the nature of the cell types potentially involved in our HSV-1 infection model with cell-specific markers (e.g., keratinocytes [keratin-5], Langerhans cells [CD207], macrophages [CD68], endothelial cells [CD31], mast cells [tryptase]). We found that keratinocytes were the main cell type entangled in HSV-1 infection, whereas results for all other cells were negative (Figure 3b and c). Our findings extend a previous observation in human single cells in vitro showing that epidermal keratinocytes are the primary portal for entry and spread of HSV infection (Nicola et al., 2005). Although HSV-1 positivity in Langerhans cells and in other dermal cells have been shown by several groups (Bedoui et al., 2009; Kim et al., 2015) and are in line with our in vitro findings (see Supplementary Figure S1d), this is in contrast to our ex vivo observations, in which we never observed HSV-1–infected Langerhans cells, dermal CD207/Langerin+ dendritic cells, and fibroblasts. Explanations for this discrepancy could be that our time point of investigation at day 4 was too late compared with the observations at day 1 by Kim and colleagues (2015), and this remains to be addressed. Another possibility might be the different sources of human skin used for the explant models. We used abdominal skin and a microneedling procedure, whereas Kim and coworkers (2015) used intact inner foreskin, which lacks a stratum corneum and has a lower degree of keratinization (Ganor and Bomsel, 2011). In addition, in contrast to other groups, we used an unlabeled antibody to detect viral glycoproteins in infected cells. Even though we are aware that a 4-day culture period is a limiting factor for testing any adaptive response in this model, it is a unique tool for the analysis of cellular factors and pathways during the initiation phase of HSV-1 skin infection.
Acyclovir and pritelivir have a similar efficacy to prevent HSV-1 replication in human skin
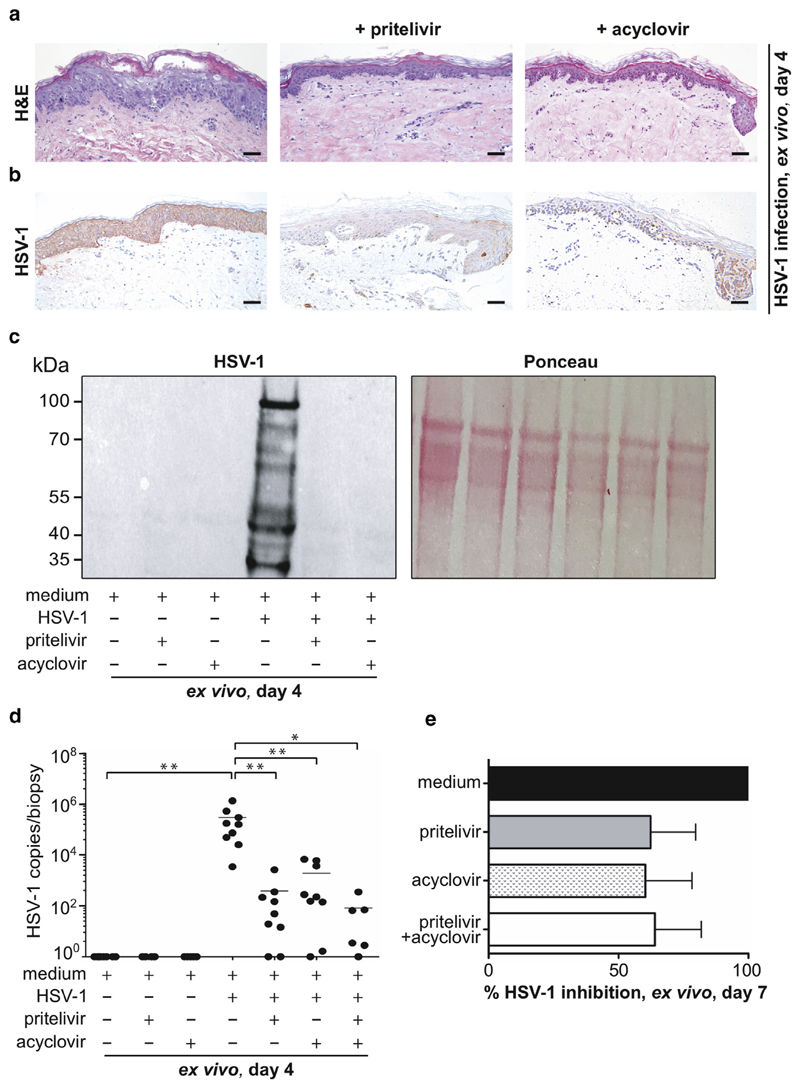
For almost five decades, the most common prescribed drug for the treatment of HSV-1 infection has been acyclovir. Recently, a compound called pritelivir has shown similar potential as acyclovir in a phase 2 clinical trial in HSV-2–infected patients (Wald et al., 2014, 2016). Here, we assessed the efficacy of both drugs at the cellular level and in our established skin explant model. Beforehand, pritelivir was evaluated for cytotoxicity on primary keratinocytes. None of the drug concentrations (range = 0.05–15 μmol/L) tested induced significant toxicities compared with the control (see Supplementary Figure S2a online). Next, microneedle-pretreated human skin was cultured for 4 days with HSV-1, as described (Figure 1a, middle panel) and, in addition, in the presence of the reported clinical maximum serum concentration (i.e., Cmax) of pritelivir (3.3 μmol/L) (Biswas et al., 2014) and acyclovir (43.6 μmol/L). Examination of hematoxylin and eosin-stained skin sections showed typical HSV-1–specific changes in HSV-1–treated skin without drugs, whereas no morphological changes were found in HSV-1–treated skin in the presence of either drug (Figure 4a). In line with this observation were data obtained using immunohistochemistry, because the signs of infection in the presence of either drug were drastically reduced compared with HSV-1–infected skin (Figure 4b). Effective infection was confirmed with Western blotting showing HSV-1–specific glycoproteins in virus-infected skin but not in the presence of either drug with or without HSV-1 (Figure 4c). Infection efficacy was evaluated using quantitative RT-PCR and showed a significant and relatively similar decrease in the HSV-1 viral copies/biopsy sample upon either drug treatment (Figure 4d). Again, similar observations were obtained in primary keratinocytes (see Supplementary Figure S2b and c). The combination of both drugs showed an augmented, although not statistically significant, effect upon HSV-1 infection compared with the single drug treatment (Figure 4d). These results are in line with mouse data showing that a combination of pritelivir and acyclovir has an additive effect against HSV-2 infection (Quenelle et al., 2018).
Figure 4. Pritelivir inhibits HSV-1 replication in human skin.
(a) H&E-stained skin sections show typical HSV-1–specific changes after 4 days of HSV-1 treatment (left), whereas no morphological changes are observed in the presence of pritelivir (middle) or acyclovir (right). (b) Immunohistochemical staining of paraffin sections with an anti HSV-1 serum shows a striking inhibition of HSV-1 infection with pritelivir (middle) and acyclovir (right) compared with the HSV-1–treated group (left). Shown is one representative from six donors. Scale bars = 50 μm. (c) Representative Western blot shows expression of typical HSV-1–specific glyoproteins in HSV-1–infected skin but not in untreated or drug-treated (pritelivir: 3.3 μmol/L, acyclovir: 43.6 μmol/L) skin when analyzed at day 4 of culture. Ponceau S staining is included as loading control. (d) HSV-1 copies/biopsy sample of indicated groups at day 4 of culture were analyzed by RT-PCR. Results are expressed as mean ± standard error of the mean from six to nine independent experiments and donors. *P < 0.01, **P < 0.001. + indicates HSV-1 treated, and − indicates untreated. (e) Skin was infected for 4 days and then shifted to indicated treatment for another 3 days with concentrations as indicated in (Figure 4d). Viral copies were determined with RT-PCR, and percent HSV-1 inhibition is indicated. Results expressed are mean ± standard error of the mean from three independent experiments with three different donors. H&E, hematoxylin and eosin; HSV, herpes simplex virus; RT-PCR, real-time PCR.
To mimic patient conditions during primary infection contrasting the prophylactic approach described, skin was first infected with HSV-1 for 4 days. Infected skin was further cultured with virus-free cell culture medium for 3 days in the presence or absence of either drug to measure their therapeutic potentials. RT-PCR showed that pritelivir and acyclovir partly inhibited further replication (62.3% and 60.3%, respectively) compared with non–drug-treated, HSV-1–infected control (100%) (Figure 4e). This suggests that both drugs have a similar potency to treat infection in our model at day 7. No remarkable additive effect was observed when both drugs were combined (64%). In future experiments, we plan to establish an acyclovir-resistant virus model.
To our knowledge, we established a previously unreported ex vivo model to study HSV infections in human skin. We decided to use abdominal skin because most often, HSV-1 primary infections cause sores inside the mouth, but this infection can recur on any part of the body containing all skin layers including the stratum corneum, the most superficial layer of skin. Thus, the location of recurrent infection is comparable to abdominal skin but is different from human foreskin, which was used in the study by Kim et al. (2015), which lacks the developed stratum corneum. Furthermore, a number of different widespread and disseminated viral infections can occur in patients with atopic dermatitis, a chronic, relapsing, inflammatory skin disease. Eczema herpeticum is the most common member of this group caused by one of the herpesviruses and can develop on any part of the body. To mimic virus infection and to study its mechanism, the use of abdominal skin was therefore physiologically most relevant. Our results clearly show that acyclovir and pritelivir have a similar potential to inhibit viral replication not only in primary keratinocytes but, most importantly, also in an ex vivo human skin explant model, providing further evidence that pritelivir may be an alternative candidate for HSV treatment. Even though we are aware that our model has some limitations (e.g., lacks influx of biochemicals [complement proteins from the circulation] and other immune cells (natural killer cells, neutrophils), adaptive responses cannot be tested, no innervation, limited culture periods], it will allow screening of unreported compounds to overcome the drug resistance problem, especially in immunocompromised individuals. Another key advantage of the presented model is the comparative assessment of infected and uninfected areas in the same skin sample and the ability to study cell-cell interactions in infected areas, which is of utmost importance for investigating virus-associated pathomechanisms and is impossible to study in monolayer cultures or directly in humans. Furthermore, because human and rodent skin differs significantly, a human skin model is of high relevance because it provides perhaps the laboratory model closest to the in vivo environment in terms of biological complexity and fidelity to human physiology. It is ethically advantageous, because it allows experiments in human tissue that would otherwise be impossible in living individuals. Furthermore, it is a simple, fast, and cost-effective tool for decreasing large-scale and expensive animal testing. Because there are increasing restrictions in Europe for using animals for testing compound properties and in line with the 3Rs (reduction, refinement, and replacement of animal models), our model represents an ideal preclinical platform.
Materials and Methods
Skin samples
Abdominal skin from anonymous healthy female and male donors (age range = 20–65 years) was obtained during plastic surgery procedures. A skin biopsy sample from a patient with disseminated HSV infection with compatible histological features was used as positive control for immune staining. The study was approved by the ethics committee of the Medical University of Vienna and was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all participants.
Preparation of HSV-1 virus
Wild-type HSV-1 virus was isolated from clinical specimens. The HSV-1–producing HepG2 cell line was used to expand the viral copies and to prepare the stock, as described (Kim et al., 2015). Cells were incubated in MEMα supplemented with 5% fetal bovine serum and 5% nonessential amino acid (all from Gibco, Thermo Fisher Scientific, Waltham, MA). Supernatant was collected after 2–3 days by centrifugation at 20,000g for 10 minutes and stored at −80°C until use (1.79 × 108 copies/ml stock). Virus titer was determined as described below. HSV-1 specificity was confirmed with RT-PCR, Western blotting, and immunohistochemistry.
Isolation and culture of primary keratinocytes and Langerhans cells
Dermatome-cut (0.5-mm) skin was incubated in 1.2 U/ml Dispase II in phosphate buffered saline (Roche Diagnostics, Indianapolis, IN), overnight at 4 °C. The next day, epidermis was separated mechanically and digested with 2.5 μg/ml of bovine trypsin and 0.05 U/ml DNase I (both from Sigma-Aldrich) for 30 minutes at 37 °C. Keratinocytes were cultured in a serum-free keratinocyte growth medium (KGM2; Lonza, Basel, Switzerland), as reported previously (Mildner et al., 2006). Two methods were applied for the separation of epidermal Langerhans cells, essentially as described (Tajpara et al., 2018).
Microneedle-pretreatment of abdominal human skin for HSV-1 infection in vitro
Dermatome-cut skin (4 cm2) was treated 20 times with a derma roller (SR150; Skin Radiance, London, UK) by the same operator to reduce variability. The derma roller device contained 1,200 needles of 1.5 mm in length, allowing penetration through the epidermis into the dermis, and generated 250 holes per square centimeter. Subsequently, 8-mm punch biopsy samples were obtained from dermal roller-treated skin and normal skin and floated for 4 days in 2 ml DMEM (Gibco, Thermo Fisher Scientific) in six-well plates supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Thermo Fisher Scientific). Skin was divided into two groups: (i) without stimulation (medium control) and (ii) infected with HSV-1 (stock). HSV-1 was applied using two different strategies: (i) 1.79 × 105 HSV-1 copies in 10 μl were added onto the top, or (ii) 8.95 × 106 HSV-1 copies in 50 μl were added into 1 ml culture medium. After 4 days of culture, skin explants were harvested for paraffin embedding and further analysis.
Drug treatment of primary cells and human skin explants
Primary keratinocytes were cultured in six-well plates (Costar, Cambridge, MA) to 70% confluence and subsequently incubated with 15 μl of HSV-1 (stock) or left untreated (medium control) for 24 hours and afterward analyzed by RT-PCR and Western blotting. In certain experiments, keratinocytes were incubated with HSV-1 and selected concentrations of the drugs acyclovir and pritelivir (Biovision, Milpitas, CA), as indicated in Supplementary Figures S1 and S2, or left untreated for 24 hours. Supernatants were collected, and viral copies were determined with RT-PCR. Keratinocytes were lysed for Western blotting.
Microneedle-pretreated skin biopsy samples (8 mm) were cultured with or without HSV-1 as described and in the absence or presence of acyclovir and/or pritelivir at concentrations specified in the figures. At 4 days of culture, skin explants were harvested for paraffin embedding, RT-PCR, and Western blotting. For certain experiments, 4-day HSV-1–treated skin was transferred to HSV-1–free conditions and cultured with medium, pritelivir, acyclovir, or a combination of both drugs for 3 days at concentrations specified in Figure 4. At day 7, skin explants were harvested for RT-PCR and percent inhibition was calculated.
Histochemistry and immunohistochemistry
A standard hematoxylin and eosin staining technique was used for histological analyses of paraffin-embedded (5-μm) skin sections. For immunohistochemistry, paraffin sections were deparaffinized with xylene and rehydrated with ethanol. Antigen unmasking was performed by boiling the slides in citrate-buffer (pH = 6, Dako, Santa Clara, CA) in a microwave for 5 minutes. After blocking the sections with 10% normal horse serum for 1 hour, sections were incubated with an unconjugated rabbit anti-human anti-HSV-1 serum (1:300; B01141, Dako) overnight at 4 °C. Subsequently, sections were incubated with a biotin-conjugated anti-rabbit IgG for 60 minutes at room temperature with the Elite rabbit IgG Vectastain kit (Vector Laboratories, Burlingame, CA). Biotinylated antibodies were detected with horseradish peroxidase-streptavidin, and staining was visualized with amino-ethyl-carbazole (Dako). Finally, sections were counterstained with hematoxylin (Merck, Darmstadt, Germany), mounted with Aquatex (Merck), and examined with a conventional bright field microscope (Olympus AX70; Olympus, Tokyo, Japan). Rabbit serum (Vector Laboratories) was used as isotype control.
Immunofluorescence
Paraffin sections (5 μm) were stained overnight at 4 °C with the following unconjugated primary antibodies/serum for surface and intracellular staining: HSV-1 (1:300), S100 (Z0311, 1:500), CD68 (EBM11, 1:100), and CD31 (JC70A, 1:100) (all from Dako); ICP8 (11E2, 1:500), tryptase (1:1,000), IRF3 (EP2419Y, 1:1,000), and vimentin (ab45939, 1:500) (all from Abcam, Cambridge, UK); p65 (27F9.G4, 1:1,000; Rockland Immunological Montgomery, PA); nectin-1 (610835, 1:200; R&D Systems); and keratin 5 (EP1601Y, 1:100, Thermo Fisher Scientific). Primary antibodies were detected with the corresponding species-specific goat anti-mouse Alexa Fluor 488 or goat anti-rabbit Alexa Fluor 488 or 546 antibodies (1:500, Thermo Fisher Scientific). Some sections were further stained for 2 hours at 37 °C with FITC-labeled anti-CD207 (929F3.01, 1:200; Dendritics, Lyon, France) or FITC-conjugated anti-MxA (383-7D4, 1:50; kindly provided by Otto Majdic, Medical University of Vienna). Isotype-matched controls were included at the same concentrations. Nuclei were stained with Hoechst (0.5 μg/ml) for 1 minute at room temperature (Sigma-Aldrich). The slides were washed with phosphate buffered saline and mounted with Fluroprep (BioMerieux, Marcy l’Etoile, France) for fluorescence analysis. Images were recorded using a confocal laser-scanning microscope (CLSM 410; Carl Zeiss, Jena, Germany) equipped with four lasers emitting light at 405, 488, 543, and 633 nm.
Nuclear translocation in primary cells upon HSV-1 infection was stained as follows. Primary keratinocytes cultured in six-well plates to 70% confluency on cover slips and freshly sorted Langerhans cells placed in round-bottom, 96-well plates were cultured without (medium control) or with HSV-1 for selected time points, as indicated in the figures. Keratinocytes were fixed with 4% paraformaldehyde for 15 minutes at 37 °C, whereas LCs were placed on poly-l-lysine–coated adhesion slides (Paul Marienfeld, Lauda-Königshofen, Germany) and fixed with acetone for 10 minutes at 4 °C. Fixed cells were stained with antibodies directed against ICP8, p65, and IRF3. Primary antibodies were detected with the corresponding species-specific goat anti-mouse Alexa Fluor 488 or goat anti-rabbit Alexa Fluor 488 or 546 antibodies. Langerhans cells were further stained for 2 hours at 37 °C with an anti-CD207 antibody.
Toxicity assay
The toxicity of pritelivir in primary human keratinocytes was evaluated with the cytotoxicity assay EZ4U (Biomedica GmbH, Eching, Germany) and was performed according to the manufacturer’s instructions. DMSO was used as a positive control.
Western blot
All samples were lysed in SDS–PAGE loading buffer, sonicated, centrifuged, and denatured with 0.1 mol/L dl-dithiothreitol (Sigma-Aldrich) before loading. SDS-PAGE was conducted on 8%–18% gradient gels (GE Amersham Pharmacia Biotech, Munich, Germany), and proteins were electrotransferred onto nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were incubated with the anti-HSV-1 serum (1:300, Dako) overnight at 4 °C. After washing, the blots were incubated with horseradish peroxidase-coupled sheep anti-rabbit secondary antibody (Amersham, Little Chalfont, UK) for 1 hour at room temperature, and reaction products were detected by chemiluminescence with the ImmunStar Western C Substrate kit (Bio-Rad) according to the manufacturer’s instructions. Ponceau S staining was included as loading control.
DNA isolation
Replica prepTMg (DNA) tissue miniprep kit (Promega, Madison, WI) was used to isolate DNA and was performed according to the manufacturer’s instructions and isolated DNA was stored at −80 °C until further analyses. Viral DNA was extracted with a High Pure Viral Nucleic Acid Kit (Roche), and amplification of DNA was performed with the RealStar HSV PCR Kit (Altona Diagnostics, Hamburg, Germany) on a Roche Light Cycler 480. Few HSV-1 copies were detected in some control samples after 4 days of culture and were most probably due to PCR carry over or cross-contamination during sample preparation and analysis.
Statistical analysis
Data are given as the mean, and error bars represent the standard error of the mean. Nonparametric paired and unpaired group comparisons (Wilcoxon signed-rank test, Mann-Whitney U test, and t test) were used when appropriate (GraphPad Software, La Jolla, CA). A P-value of less than 0.05 was considered significant. Statistical testing was performed using SPSS 15.0 software (IBM SPSS, Chicago, IL).
Supplementary Material
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2018.08.034.
Acknowledgments
The Austrian Science Fund (FWF) DK W1248-B30 supported this work. We thank S. Krönes-Hackl and G. Fischer (Department of Laboratory Medicine, Division of Clinical Virology, Medical University of Vienna, Vienna, Austria) for excellent technical help.
Abbreviations
- HSV
herpes simplex virus
- RT-PCR
real-time PCR
Footnotes
ORCID
Michael Mildner: http://orcid.org/000-0002-6892-925X
Adelheid Elbe-Bürger: http://orcid.org/0000-0003-2461-0367
Conflict of Interest
The authors state on conflict of interest.
Author contributions: PT co-designed, conducted, and analyzed the experiments and wrote the paper. RS performed RT-PCR experiments. MM, CS, and AE-B designed the experiments and analyzed data. MV, JM, and TP-K provided important resources. CS and MM edited the manuscript. AE-B secured funding, supervised the project, and wrote the paper. All authors had final approval of the submitted version.
References
- Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J Virol. 2014;88:5328–41. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar J, Shukla D. Viral entry mechanisms: cellular and viral mediators of herpes simplex virus entry. FEBS J. 2009;276:7228–36. doi: 10.1111/j.1742-4658.2009.07402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amici C, Rossi A, Costanzo A, Ciafrè S, Marinari B, Balsamo M, et al. Herpes simplex virus disrupts NF-κB regulation by blocking its recruitment on the IκBα promoter and directing the factor on viral genes. J Biol Chem. 2006;281:7110–7. doi: 10.1074/jbc.M512366200. [DOI] [PubMed] [Google Scholar]
- Bedoui S, Whitney PG, Waithman J, Eidsmo L, Wakim L, Caminschi I, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10:488–95. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- Betz UAK, Fischer R, Kleymann G, Hendrix M, Rübsamen-Waigmann H. Potent in vivo antiviral activity of the herpes simplex virus primase-helicase inhibitor BAY 57-1293. Antimicrob Agents Chemother. 2002;46:1766–72. doi: 10.1128/AAC.46.6.1766-1772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkmann A, Zimmermann H. HSV antivirals—current and future treatment options. Curr Opin Virol. 2016;18:9–13. doi: 10.1016/j.coviro.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Biswas S, Sukla S, Goldner T, Field HJ, Kropeit D, Paulsen D, et al. Pharmacokinetics-pharmacodynamics of the helicase-primase inhibitor pritelivir following treatment of wild-type or pritelivir-resistant virus infection in a murine herpes simplex virus 1 infection model. Antimicrob Agents Chemother. 2014;58:3843–52. doi: 10.1128/AAC.02641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute JJ, Grygon CA, Hargrave KD, Simoneau B, Faucher AM, Bolger G, et al. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat Med. 2002;8:386–91. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- Cunningham AL, Diefenbach RJ, Miranda-Saksena M, Bosnjak L, Kim M, Jones C, et al. The cycle of human herpes simplex virus infection: virus transport and immune control. J Infect Dis. 2006;194(Suppl.1):S11–8. doi: 10.1086/505359. [DOI] [PubMed] [Google Scholar]
- Ganor Y, Bomsel M. HIV-1 transmission in the male genital tract. Am J Reprod Immunol. 2011;65:284–91. doi: 10.1111/j.1600-0897.2010.00933.x. [DOI] [PubMed] [Google Scholar]
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Johannessen I, Ogilvie MM. Herpesviruses. In: Greenwood D, Barer M, Slack R, Irving W, editors. Medical Microbiology. 18th ed. London, UK: Churchill Livingston; 2012. pp. 419–45. [Google Scholar]
- Kim M, Truong NR, James V, Bosnjak L, Sandgren KJ, Harman AN, et al. Relay of herpes simplex virus between Langerhans cells and dermal dendritic cells in human skin. PLoS Pathog. 2015;11(4):e1004812. doi: 10.1371/journal.ppat.1004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleymann G, Fischer R, Betz UAK, Hendrix M, Bender W, Schneider U, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat Med. 2002;8:392–8. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- Kola I. The state of innovation in drug development. Clin Pharmacol Ther. 2008;83:227–30. doi: 10.1038/sj.clpt.6100479. [DOI] [PubMed] [Google Scholar]
- Kollias CM, Huneke RB, Wigdahl B, Jennings SR. Animal models of herpes simplex virus immunity and pathogenesis. J Neurovirol. 2015;21:8–23. doi: 10.1007/s13365-014-0302-2. [DOI] [PubMed] [Google Scholar]
- Krawczyk A, Arndt MAE, Grosse-Hovest L, Weichert W, Giebel B, Dittmer U, et al. Overcoming drug-resistant herpes simplex virus (HSV) infection by a humanized antibody. Proc Natl Acad Sci USA. 2013;110:6760–5. doi: 10.1073/pnas.1220019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CC, Che XB, Reichelt M, Rajamani J, Schaap-Nutt A, Huang KJ, et al. Herpes simplex virus-1 induces expression of a novel MxA isoform that enhances viral replication. Immunol Cell Biol. 2011;89:173–82. doi: 10.1038/icb.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol. 2004;78:1675–84. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner M, Ballaun C, Stichenwirth M, Bauer R, Gmeiner R, Buchberger M, et al. Gene silencing in a human organotypic skin model. Biochem Biophys Res Commun. 2006;348:76–82. doi: 10.1016/j.bbrc.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–36. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- Mullard A. Parsing clinical success rates. Nat Rev Drug Discov. 2016;15:447. doi: 10.1038/nrd.2016.136. [DOI] [PubMed] [Google Scholar]
- Nicola AV, Hou J, Major EO, Straus SE. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol. 2005;79:7609–16. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce RS, Collins SE, Mossman KL. Differential modification of interferon regulatory factor 3 following virus particle entry. J Virol. 2009;83:4013–22. doi: 10.1128/JVI.02069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann P, Rahn E, Thier K, Hsu M-J, Rixon FJ, Kopp SJ, et al. Role of nectin-1 and herpesvirus entry mediator as cellular receptors for herpes simplex virus 1 on primary murine dermal fibroblasts. J Virol. 2015;89:9407–16. doi: 10.1128/JVI.01415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenelle DC, Birkmann A, Goldner T, Pfaff T, Zimmermann H, Bonsmann S, et al. Efficacy of pritelivir and acyclovir in the treatment of herpes simplex virus infections in a mouse model of herpes simplex encephalitis. Antiviral Res. 2018;149:1–6. doi: 10.1016/j.antiviral.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn E, Petermann P, Thier K, Bloch W, Morgner J, Wickström SA, et al. In-vasion of herpes simplex virus type 1 into murine epidermis: an ex vivo infection study. J Invest Dermatol. 2015;135:3009–16. doi: 10.1038/jid.2015.290. [DOI] [PubMed] [Google Scholar]
- Remeijer L, Osterhaus ADME, Verjans GMGM. Human herpes simplex virus keratitis: the pathogenesis revisited. Ocul Immunol Inflamm. 2004;12:255–85. doi: 10.1080/092739490500363. [DOI] [PubMed] [Google Scholar]
- Shenoy R, Mostow E, Cain G. Eczema herpeticum in a wrestler. Clin J Sport Med. 2015;25(1):e18–9. doi: 10.1097/JSM.0000000000000097. [DOI] [PubMed] [Google Scholar]
- Singh A, Yadav S. Microneedling: advances and widening horizons. Indian Dermatol Online J. 2016;7(4):244. doi: 10.4103/2229-5178.185468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasfeld L, Chou S. Antiviral drug resistance: mechanisms and clinical implications. Infect Dis Clin North Am. 2010;24:413–37. doi: 10.1016/j.idc.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajpara P, Schuster C, Schön E, Kienzl P, Vierhapper M, Mildner M, et al. Epicutaneous administration of the pattern recognition receptor agonist polyinosinice–polycytidylic acid activates the MDA5/MAVS pathway in Langerhans cells. FASEB J. 2018;32:4132–44. doi: 10.1096/fj.201701090R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolun G, Makhov AM, Ludtke SJ, Griffith JD. Details of ssDNA annealing revealed by an HSV-1 ICP8-ssDNA binary complex. Nucleic Acids Res. 2013;41:5927–37. doi: 10.1093/nar/gkt266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald A, Corey L, Timmler B, Magaret A, Warren T, Tyring S, et al. Heli-case–primase inhibitor pritelivir for HSV-2 infection. N Engl J Med. 2014;370:201–10. doi: 10.1056/NEJMoa1301150. [DOI] [PubMed] [Google Scholar]
- Wald A, Timmler B, Magaret A, Warren T, Tyring S, Johnston C, et al. Effect of pritelivir compared with valacyclovir on genital HSV-2 shedding in patients with frequent recurrences: a randomized clinical trial. JAMA. 2016;316:2495–503. doi: 10.1001/jama.2016.18189. [DOI] [PubMed] [Google Scholar]
- Whitley RJ, Roizman B. Herpes simplex virus infections. The Lancet. 2001;357:1513–8. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- Wilson SS, Fakioglu E, Herold BC. Novel approaches in fighting herpes simplex virus infections. Expert Rev Anti Infect Ther. 2009;7:559–68. doi: 10.1586/eri.09.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.