ABSTRACT
It is crucial to understand the molecular mechanisms involved in epileptogenesis. This study aims to investigate the role of lncRNA NEAT1, miR-129-5p and Notch signaling pathway in epilepsy. In this research, temporal lobe tissues were collected from patients with epilepsy and healthy controls. The CTX-TNA cells were treated with IL-1β to establish as epilepsy cell model, which were then manipulated the expression level of NEAT1, miR-129-5p and Notch1 to investigate their roles in the epilepsy progression. The expression levels of RNA and protein in temporal lobe tissues and epilepsy cell model were determined by RT-qPCR, western blotting or ELISA, respectively. MTT assay was utilized to analyze the cell viability. Dual-luciferase reporter assay was used to explore the interaction relationship between lncRNA NEAT1, miR-129-5p and Notch1. Silencing NEAT1 significantly reduced the expression levels of IL-6, COX-2 and TNF-α in epilepsy cell model. The overexpression of NEAT1 suppressed the expression level of miR-129-5p. Inhibiting miR-129-5p significantly increased the expression of IL-6, COX-2, TNF-α and Notch1. Furthermore, the expression levels of IL-6, COX-2 and TNF-α were increased after overexpressing Notch1 in miR-129-5p mimics-treated cells. The expression levels of Notch1, JAG1, and HES1 were decreased after transfecting with sh-NEAT1. However, compared with sh-NEAT1 group, the expression levels of Notch1, JAG1, HES1, IL-6 and TNF-α were reversed by miR-129-5p inhibition or Notch1 overexpression. The present study verified that lncRNA NEAT1 affected inflammatory response of epilepsy by suppressing miR-129-5p and further regulating Notch signaling pathway in IL-1β-induced epilepsy cell model.
Abbreviations: CNS: Central nervous system; lncRNAs: Long noncoding RNAs; NEAT1: Nuclear-enriched abundant transcript 1; miRNAs: MicroRNAs; ATCC: American Type Culture Collection; DMEM: Dulbecco’s Modified Eagle Medium; FBS: Fetal bovine serum; ELISA: Enzyme-linked immunosorbent assay; RT-qPCR: Reverse transcription-quantitative polymerase chain reaction; SD: Standard deviation; ANOVA: Analysis of variance; LPS: Ligand lipopolysaccharide; GLO1: Glyoxalase I
KEYWORDS: LncRNA NEAT1, MiR-129-5p, Notch signaling pathway, Epilepsy
1. Introduction
Epilepsy is a general chronic neurologic disorder disease, which affects over 50 million people worldwide [1]. Epilepsy is triggered by over discharge due to the neuronal abnormality in a certain specific area of the brain [2]. The typical clinical features are the recurrent disturbance of consciousness and the generalized or localized muscle spasms and convulsions [3]. A recent study reports that inflammatory dysregulation is the pathological feature in the central nervous system (CNS) diseases, including epilepsy [4]. Astrocytes are richly present in the epileptogenic brain, which is one of the most crucial type of glial cells leading to the neuroinflammatory response [5]. Astrocytes can generate a variety of pro-inflammatory mediators, including chemokines and cytokines, which result in activation of the innate and adaptive immune response [6]. The candidate pathogenic factors and mechanisms of epilepsy remain unclarified though numerous researches contributing to study the mechanisms of epileptogenesis [7]. Exploring the molecular mechanism of astrocyte inflammation in epilepsy is helpful to explain the development of epilepsy.
Studies have shown that the development and function of the nervous system are modulated by long non-coding RNAs (lncRNAs), and dysregulated lncRNAs may contribute to disorder of the function of brain [8]. LncRNAs are involved in the complex epigenetic regulation of the nervous system [7]. Donato et al. demonstrated that non-coding RNAs targeting host genes involved in several biochemical pathways which are related to compromised response to synaptic impairment of retinal neurotransmission, impairment of the interphotoreceptor matrix and, blood-retina barrier [9]. As previously described, overexpression of lncRNA TUSC7 inhibited microglial activation and the expression of inflammatory factors in microglia cells by regulating miR-449a/PPAR-γ [10]. Jang et al. [11] reported that the dysregulated lncRNAs might be potential therapeutic targets for the regulation of epilepsy in rat model. It was reported that lncRNA H19 contributed to the activation of hippocampal astrocytes and microglia cells, as well as the inflammatory response in epileptic rats [12]. NEAT1 (nuclear-enriched abundant transcript 1) is the lncRNA that exhibits high level in the brain [13]. Feifei Zhang et al. [14] found that the expressions of inflammatory chemokines and cytokines were regulated by NEAT1 in systemic lupus erythematosus via the MAPK pathway. This research indicated that lncRNA NEAT1 was one of the lncRNAs associated with inflammation, but there were few studies on NEAT1 in epilepsy and its regulation mechanism is still unclear.
It has been demonstrated that astrocytes also produce a series of microRNAs (miRNAs), which can regulate the inflammatory pathways related to various neurological disorders, including epilepsy. Marek Rajman et al. [15] found that inhibition of miR-129-5p in vitro prevented synaptic contraction and reduced the severity of seizures in vivo. Studies also showed that miR-129 could regulate Notch signaling pathway in the pathogenesis of many cancers, but the effect of Notch signaling pathway on inflammation in epilepsy is rarely reported [16,17]. The Notch signaling pathway is related to a variety of diseases in the nervous, immune and cardiovascular systems, which is an evolutionarily highly conserved pathway [18]. Yushuang Li et al. [19] indicated that neurogenesis during epileptogenesis was enhanced by HIF-1α-Notch signaling pathway and neurogenesis was reduced once this pathway was blocked in acute epilepsy.
In the present study, we found the expression level of NEAT1 was markedly increased in the hippocampal neurons of epilepsy patients. Thus, we investigated the role of NEAT1 in epilepsy, as well as examined the function of miR-129-5p and Notch signaling pathway in epilepsy model in vitro. We also reveal the regulation mechanism of NEAT1, miR-129-5p and Notch signaling pathway in the occurrence of epilepsy and provide a potentially viable new direction and target for epilepsy clinical treatment.
2. Materials and methods
2.1. Preparation of tissues
Surgical resection specimens from clinical patients were performed from Xiangya Hospital (Changsha, Hunan, China) in patients undergoing neurological surgery, including six patients with epilepsy and six healthy controls. Patients with epilepsy had a age of 21 ~ 45 years and included 2 males and 4 females. Epilepsy was diagnosed according to standard proposed by the International League Against Epilepsy (ILAE). No patients were received antiepileptic drugs before surgery. Healthy controls were acquired from patients who required craniocerebral surgery resulting from the severe traumatic brain injury. Healthy controls had an age of 18 ~ 45 years and included 3 males and 3 females. None of the patients in the control group had a history of seizures or epilepsy. Normal temporal lobe tissues and temporal lobe tissues in epilepsy patients were collected from these samples. Tissue collection and manipulation were authorized by the Ethics Committee of Xiangya Hospital. Informed consents were collected from all subjects.
2.2. Cell culture
The CTX-TNA astrocyte cell line was obtained from the American Type Culture Collection (ATCC), which is derived from the cerebral cortex of healthy newborn SD rats and retained the typical features of type I astrocytes. The cells were cultured using DMEM (Dulbecco’s Modified Eagle Medium, Gibco, Carlsbad, CA, USA) complete medium with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, USA), 100 U/ml penicillin and streptomycin and treated with 10 ng/mL of IL-1β (PeproTech, USA) for 24 h.
2.3. Plasmid construction and cell transfection
The pcDNA3.1-NEAT1 and pcDNA3.1-Notch1 vectors were established by Invitrogen (USA). The shRNA against NEAT1 plasmid (sh-NEAT1) were devised and synthesized by Vigenebio (Maryland, USA). MiR-129-5p mimics, miR-129-5p inhibitor and their negative controls (NC) were synthesized by GenePharma Co. Ltd. (Shanghai, China). The CTX-TNA cell line was seeded in six-well plates with the density of 1 × 106 cells per well, then cells were disposed with IL-1β (10 ng/mL) for 24 h, which was established as the cell model of epilepsy. Cell transfections were carried out in CTX-TNA cells with Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. After 48 h of transfection, RT-qPCR analysis or western blot analysis was used to verify transfection efficiency.
2.4. Total RNA isolation and RT-qPCR
Total RNA was isolated from temporal lobe tissues and normal control tissues as well as CTX-TNA cells using Trizol reagent (Invitrogen) according to the standard procedures. After reverse transcription of total RNA by using a cDNA Reverse Transcription Kit (Applied Biosystems), we obtained the cDNA. The expression levels of lncRNA NEAT1, miR-129-5p, Notch1, JAG1, HES1 and inflammatory cytokines IL-6, COX-2, TNF-α were measured using Applied Biosystems 7500 Real Time PCR System (Applied Biosystems) with a SYBR Green QPCR Master Mix (Invitrogen). The results were normalized to the expression levels of GAPDH or U6 snRNA using the 2−ΔΔCt method for quantification. Primers were devised and produced by Sangon Biotech (Shanghai, China).
2.5. Western blot analysis
The details of western blot assay were performed as previously described [20]. In brief, CTX-TNA cells were lysed with RIPA buffer (Sigma-Aldrich). The protein concentrations were determined using BCA protein quantitation kit (Thermo Fisher Scientific, Waltham, MA, USA). Thirty micrograms proteins were subjected to SDS-PAGE and then transferred to PVDF membranes (Roche, Basel, Switzerland). Membranes were blocked with 5% nonfat milk for 1 h, and incubated with primary antibodies against Notch1 (Abcam, 1:500), JAG1 (Abcam, 1:500), HES1 (Abcam, 1:500) and GAPDH (Abcam, 1:1000) overnight at 4°C. The membranes were washed three times with wash buffer prior to incubation with horseradish peroxidase-conjugated secondary antibody (Abcam, 1:2000) for 1 h at room temperature. The protein bands were visualized using Immobilon Western Chemiluminescent HRP substrate (Millipore, Burlington, MA, USA). The proteins were quantified using Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
2.6. MTT assay
The cell viability was evaluated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. CTX-TNA cells (4 × 103 cells/well) were plated in 96-well plates for growth of 24 h. The medium was removed, and MTT solution was added to the culture medium and incubated at 37°C for 4 h. Then, optical density (OD) values were measured at 490 nm using a Multlskan Mk3 microplate reader (Thermo Fisher, Waltham, MA, USA) [21].
2.7. Enzyme-linked immunosorbent assay (ELISA)
The expression levels of COX-2, TNF-α and IL-6 in the tissues and cell culture supernatant were measured according to the manufacturer’s instructions by ELISA kits (Thermo Fisher Scientific, USA). A spectrophotometer was used to read the absorbance of each well at 450 nm, and the contents of each well were calculated using a standard curve.
2.8. Dual-luciferase reporter assay
The interaction relationships between NEAT1 and miR-129-5p, Notch1 and miR-129-5p were detected by using dual-luciferase reporter assay. To construct dual-luciferase reporter plasmids, the predicted binding sequence of miR-129-5p in lncRNA NEAT1 (NEAT1-WT) or Notch1 3ʹUTR (Notch1-WT) and their mutated sequence (NEAT1-MUT and Notch1-MUT) were separately cloned into pmirGLO vector (Promega, Madison, WI, USA). For luciferase assay, CTX-TNA cells were transfected with above constructs and co-transfected with mimics NC or miR-129-5p mimics (GenePharma, Shanghai, China). After incubated for 48 h at 37°C, cells were harvested, and then Firefly and Renilla luciferase activities were measured using the Dual-Glo® Luciferase Assay System (Promega) according to the manufacturer’s direction. Measurements were performed on a microplate reader. The relative firefly luciferase activity was calculated by normalizing to renilla luciferase activity [22].
2.9. Statistical analysis
All experiments were repeated at least for three times, which referred to biological replicates. Data were analyzed with Prism 6.0 (GraphPad Software, Inc.) and were shown as mean ± SD (standard deviation). Comparison between two groups was performed using the Student’s t test. Comparison among three or more groups was conducted using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. p< 0.05 was considered statistically significant.
3. Results
3.1. NEAT1 expression is increased in epilepsy patients
To explore the role of NEAT1 in epilepsy patients, we detected the expression level of NEAT1 in the temporal lobe tissues obtained from six epilepsy patients and six normal controls. RT-qPCR assay was utilized to detect the expression level of NEAT1 (Figure 1(a)). Compared with the normal controls, the epilepsy patients group showed increased NEAT1 level. And the expression levels of IL-6, IL-1β and TNF-α were measured by ELISA. As shown in Figure 1(b–d), compared with normal tissues, epilepsy patients’ tissues had high levels of IL-6, IL-1β and TNF-α, this indicated that inflammatory response may be involved in the progression of epilepsy.
Figure 1.
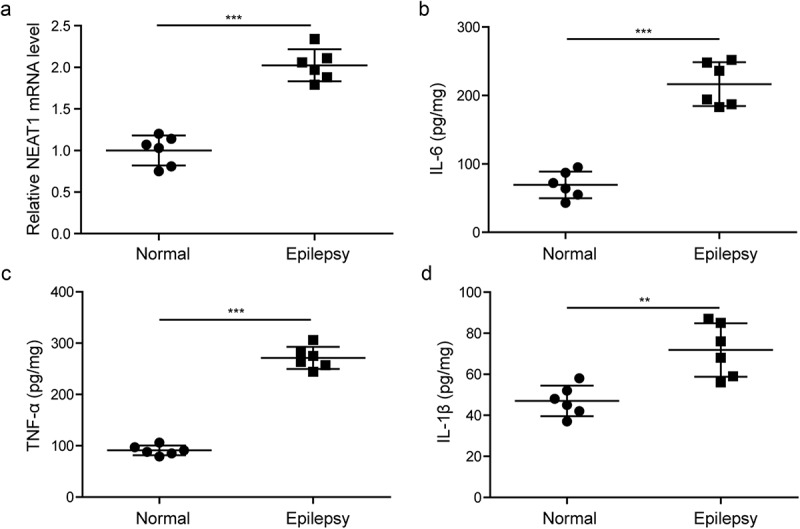
NEAT1, IL-6, IL-1β and TNF-α expression levels are increased in epilepsy patients.
(a). Altered expression of lncRNA NEAT1 was measured by RT-qPCR in temporal lobe tissues of epilepsy patients and normal controls. (b). Protein level of IL-6 detected by ELISA in temporal lobe tissues of epilepsy patients and normal controls. (c). Protein level of TNF-α detected by ELISA in temporal lobe tissues of epilepsy patients and normal controls. (d). Protein level of IL-1β detected by ELISA in temporal lobe tissues of epilepsy patients and normal controls. n = 6, **p < 0.01 and ***p < 0.001.
3.2. NEAT1 regulates the expression level of inflammatory factors and cell viability
To prove the function of NEAT1 in the production of inflammatory cytokines, we silenced NEAT1 via sh-NEAT1 and overexpressed NEAT1 with pcDNA3.1-NEAT1 vector in CTX-TNA astrocyte cells. As indicated in Figure 2(a), the mRNA levels of IL-6, COX-2 and TNF-α were measured by RT-qPCR, silencing NEAT1 significantly attenuated mRNA levels of IL-6, COX-2 and TNF-α. However, overexpressing NEAT1 significantly increased the mRNA levels of IL-6, COX-2 and TNF-α. These results indicated that NEAT1 could regulate the expression of inflammatory factors in astrocytes. To further explore the role of NEAT1 in cell viability, we performed the MTT assay. Compared with negative control groups, the cell viability of the pcDNA3.1-NEAT1-transfected cells was significantly lessened, while the cell viability of sh-NEAT1-transfected cells was significantly increased (Figure 2(b)). We concluded that NEAT1 increased the expression levels of inflammatory factors (IL-6, COX-2 and TNF-α), also inhibited cell viability in epilepsy.
Figure 2.
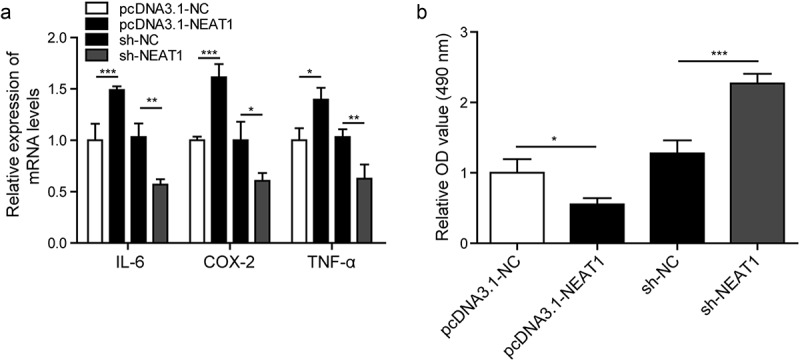
NEAT1 regulates the expression level of inflammatory factors and cell viability.
(a). Altered expression levels of IL-6, COX-2 and TNF-α measured by RT-qPCR. (b). Cell viability measured by MTT assay. *p < 0.05, **p < 0.01 and ***p < 0.001.
3.3. NEAT1 targets the expression of miR-129-5p
Researches indicated that NEAT1 was associated with inflammation, but there were few studies on NEAT1 in epilepsy and its regulation mechanism is still unclear [11]. Firstly, we determined the expression level of miR-129-5p in the model cells and normal astrocytes, as shown in Figure 3(a), the expression of miR-129-5p was decreased in the model cells compared with normal astrocytes. Next, we used bioinformatics software to predict the binding sites of NEAT1 and miR-129-5p, which was indicated in Figure 3(b). In Figure 3(c), in the experiment of transfecting the recombinant vector plasmid NEAT1-WT, the luciferase activity of the miR-129-5p mimics group was significantly decreased compared with the mimics NC group. In the experiment of transfecting the recombinant vector plasmid NEAT1-MUT, there was no difference in the expression of luciferase activity in each group. We also utilized RT-qPCR to study the expression level of miR-129-5p in after silencing or overexpression NEAT1, which is shown in Figure 3(d). There expression level of miR-129-5p was induced in the sh-NEAT1 group, and was decreased in pcDNA3.1-NEAT1 group. These results indicated that the differentially expressed level of miR-129-5p was regulated by NEAT1, which was involved in the progression of epilepsy.
Figure 3.
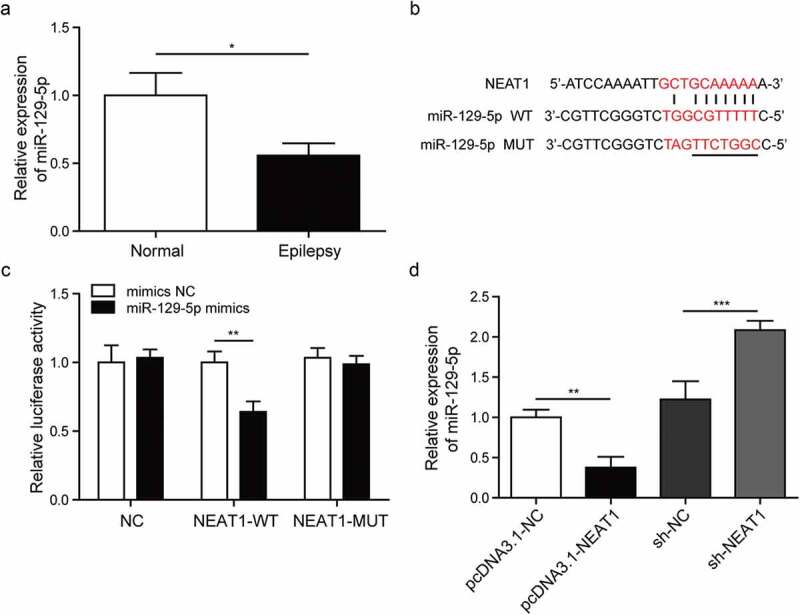
NEAT1 targets the expression of miR-129-5p.
(a). Detection of miR-129-5p expression in model cells and normal cells by RT-qPCR. (b). Analysis of binding sites of NEAT1 and miR-129-5p using bioinformatics software. (c). Analysis of the relationship between NEAT1 and miR-129-5p in astrocytes by dual-luciferase reporter assay. (d). RT-qPCR was used to detect the expression of miR-129-5p after silencing or overexpression NEAT1 in model cells. *p < 0.05, **p < 0.01 and ***p < 0.001.
3.4. MiR-129-5p affects the expression of inflammatory cytokines and cell viability in astrocytes
To indicate the function of miR-129-5p in inflammatory cytokines production, we silenced miR-129-5p with miR-129-5p inhibitor and overexpressed miR-129-5p with miR-129-5p mimics in CTX-TNA cells. In Figure 4(a), the mRNA levels of IL-6, COX-2 and TNF-α were measured by RT-qPCR, silencing miR-129-5p significantly increased the expression of IL-6, COX-2 and TNF-α. However, miR-129-5p mimics significantly decreased the expression levels of IL-6, COX-2 and TNF-α. These results indicated that miR-129-5p could regulate the expression levels of inflammatory factors in astrocytes. After culture for 48 h, the cell viability was dramatically reduced by miR-129-5p inhibitor, however, it was significantly increased by miR-129-5p mimics (Figure 4(b)).
Figure 4.
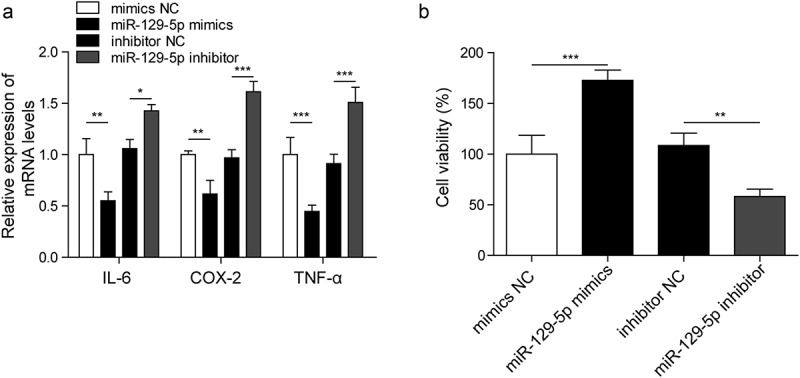
MiR-129-5p affects the expression of inflammatory cytokines in astrocytes and cell viability.
(a). Detection of inflammatory factors IL-6, COX-2 and TNF-α by RT-qPCR. (b). Cell viability measured by MTT assay. *p < 0.05, **p < 0.01 and ***p < 0.001.
3.5. MiR-129-5p regulates the notch signaling pathway
Studies have shown that miR-129 can regulate Notch signaling pathway in the pathogenesis of many cancers [16,17], we explored the relationship between miR-129-5p and Notch1 in epilepsy. Firstly, we explored the mRNA level of Notch1 in the model cells and normal astrocytes using RT-qPCR, the expression of Notch1 was dramatically increased in epilepsy compared with normal astrocytes, as shown in Figure 5(a). Next, binding sites of miR-129-5p and Notch1 were predicted by bioinformatics software, which is shown in Figure 5(b). The luciferase activity of the miR-129-5p mimics group was dramatically decreased compared with the mimics NC group in wild-type cells, whereas it did not affect the luciferase activity of mutated Notch1 (Figure 5(c)). We also utilized RT-qPCR to study the expression level of Notch1 in miR-129-5p mimics and miR-129-5p inhibitor groups, which is shown in Figure 5(d). The expression level of Notch1 in the miR-129-5p mimics group was decreased compared with mimics NC group, while it was increased in the miR-129-5p inhibitor group compared with inhibitor NC group. These results indicated that miR-129-5p can regulate the pathway of Notch1.
Figure 5.
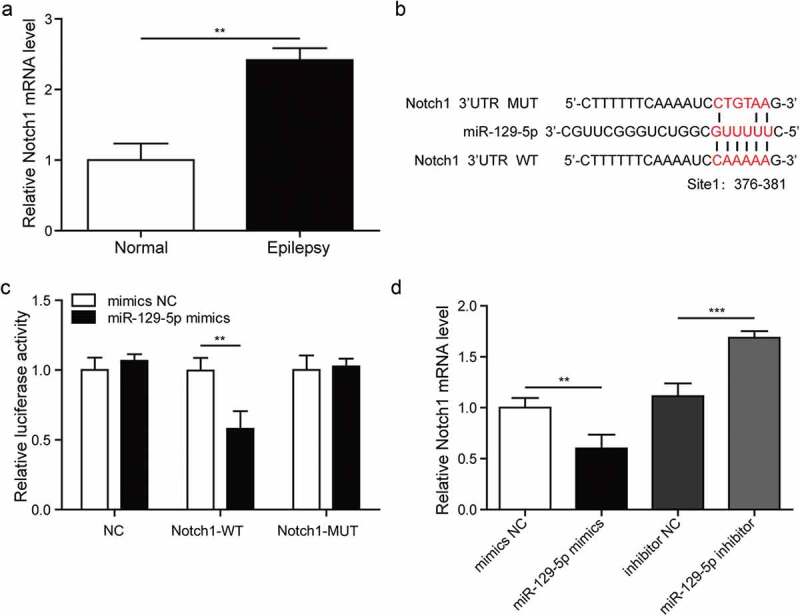
MiR-129-5p regulates the Notch signaling pathway.
(a). Detection of Notch1 expression in model cells and normal cells by RT-qPCR. (b). Analysis of binding sites of miR-129-5p and Notch1 using bioinformatics software. (c). Analysis of the relationship between miR-129-5p and Notch1 in astrocytes by dual-luciferase reporter assay. (d). The expression of Notch1 in mimic NC, miR-129-5p mimics, inhibitor NC and miR-129-5p inhibitor groups measured by RT-qPCR. **p < 0.01 and ***p < 0.001.
3.6. MiR-129-5p regulates inflammatory response via the notch signaling pathway
Then, we explored the relationship between miR-129-5p, Notch signaling pathway and inflammatory factors in epilepsy. The CTX-TNA cells were treated with IL-1β to set as epilepsy cell model, and they were transfected with miR-129-5p mimics or miR-129-5p inhibitor, and divided into four groups: mimics NC, miR-129-5p mimics and inhibitor NC, miR-129-5p inhibitor. Firstly, we explored the mRNA levels of JAG1 and HES1, and the protein levels of Notch1, JAG1 and HES1 in the astrocytes with differentially expressed miR-129-5p using RT-qPCR and western blotting, respectively (Figure 6(a,b)). It showed that miR-129-5p mimics down-regulated mRNA expression levels of JAG1 and HES1, while miR-129-5p inhibitor increased the expression levels of JAG1 and HES1 (Figure 6(a)). In Figure 6(b), miR-129-5p mimics decreased the protein levels of JAG1, HES1 and Notch1, but these protein levels were increased in the miR-129-5p inhibitor group. Next, we utilized RT-qPCR to determine the expression levels of IL-6, COX-2 and TNF-α in mimics NC, miR-129-5p mimics and miR-129-5p mimics+pcDNA3.1-Notch1 groups, which are shown in Figure 6(c). The expression levels of IL-6, COX-2 and TNF-α in the miR-129-5p mimics group were decreased, while they were increased in miR-129-5p mimics+pcDNA3.1-Notch1 group. To further study the role of miR-129-5p and Notch signaling pathway in cell viability, we performed the MTT assay. Our data showed that the cell viability was increased by miR-129-5p mimics as compared with the mimics NC, while was decreased in miR-129-5p mimics+pcDNA3.1-Notch1 group compared with miR-129-5p mimics group (Figure 6(d)). Taken together, these results indicated that miR-129-5p affected the inflammatory response of epilepsy through Notch1 signaling pathway, which further affected cell viability.
Figure 6.
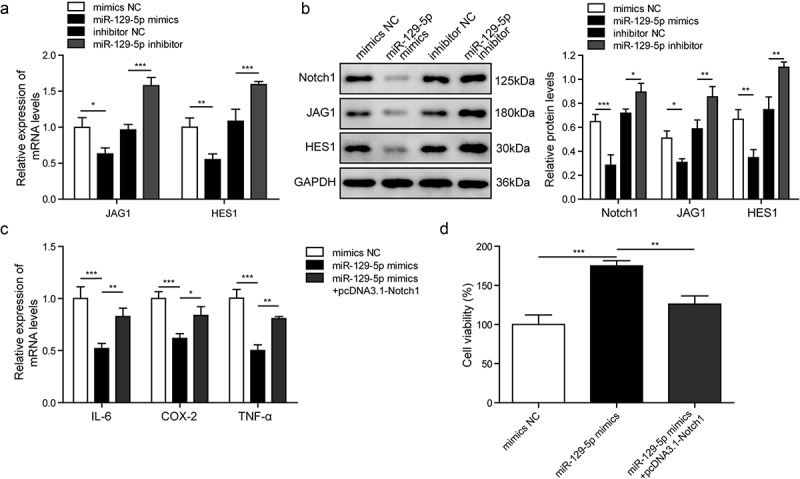
MiR-129-5p regulates cell inflammation via the Notch pathway.
(a). The expression levels of JAG1 and HES1 measured by RT-qPCR. (b). The protein expression of Notch1, JAG1, HES1 measured by Western blotting. (c). Detection of inflammatory factors IL-6, COX-2 and TNF-α by RT-qPCR in mimics NC, miR-129-5p mimics and miR-129-5p mimics +pcDNA3.1-Notch1 groups. (d). Cell viability measured by MTT assay. *p < 0.05, **p < 0.01 and ***p < 0.001.
3.7. NEAT1 regulates inflammatory response via miR-129-5p affecting notch signaling pathway
The CTX-TNA cells were treated with IL-1β to set as epilepsy model in vitro, and they were divided into four groups: sh-NC, sh-NEAT1, sh-NEAT1+ miR-129-5p inhibitor and sh-NEAT1+ pcDNA3.1-Notch1. In Figure 7(a), the mRNA level of Notch1 was measured by RT-qPCR, the expression level of Notch1 in sh-NEAT1 group was decreased compared with sh-NC group. However, the expression level of Notch1 was increased after miR-129-5p inhibition or Notch1 overexpression. This suggested that NEAT1 regulated Notch signaling pathway by targeting miR-129-5p. The protein levels of Notch1, JAG1 and HES1 in the epileptic astrocytes with different treatments were tested using western blotting. The protein levels of JAG1, HES1 and Notch1 were decreased in sh-NEAT1 group, while were increased in sh-NEAT1+ miR-129-5p inhibitor or sh-NEAT1+ pcDNA3.1-Notch1 groups compared with sh-NEAT1 group (Figure 7(b)). And the protein levels of IL-6 and TNF-α were further measured by ELISA. The expression levels of IL-6 and TNF-α in sh-NEAT1 group were decreased. However, compared with sh-NEAT1 group, the expression levels of IL-6 and TNF-α were increased in sh-NEAT1+ miR-129-5p inhibitor or sh-NEAT1+ pcDNA3.1-Notch1 groups (Figure 7(c)). To further study the role of NEAT1, miR-129-5p and Notch signaling pathway in cell viability, we performed the MTT assay. Our data showed that, following the culture for 48 h, the cell viability was clearly increased in sh-NEAT1-transfected cell as compared with sh-NC group. Nevertheless, the cell viability was decreased in sh-NEAT1+ miR-129-5p inhibitor or sh-NEAT1+ pcDNA3.1-Notch1 groups (Figure 7(d)). It indicated that NEAT1 affected the Notch signaling pathway via miR-129-5p, which in turn affected the cell viability of astrocytes in epilepsy.
Figure 7.
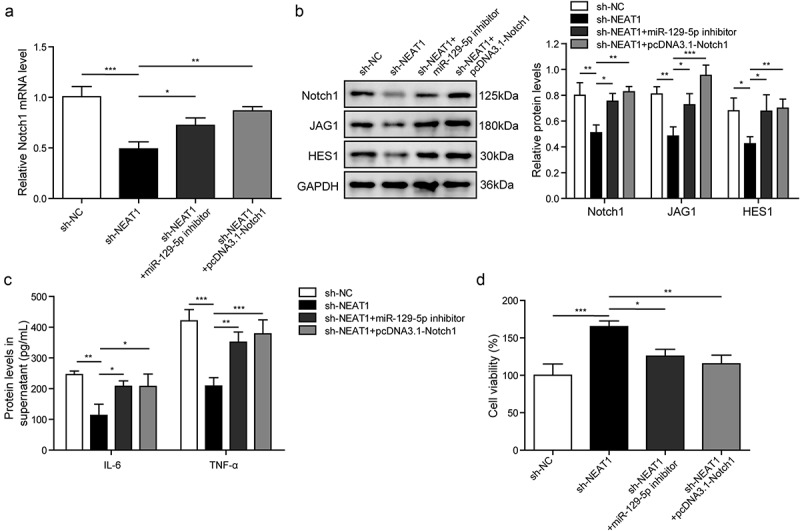
NEAT1 regulates inflammatory response via miR-129-5p affecting Notch pathway.
(a). The mRNA level of Notch1 measured by RT-qPCR. (b). The protein expression of Notch1, JAG1, HES1 measured by Western blotting. (c). Detection of inflammatory factors IL-6 and TNF-α by ELISA. D. Cell viability measured by MTT assay. *p < 0.05, **p < 0.01 and ***p < 0.001.
4. Discussion
Epilepsy is caused by aberrantly hypersynchronous electrical discharges in brain neurons, which is clinically characterized with symptom of transient dysfunction in CNS [23]. Repeated episodes generally result in damage or even necrosis of brain neurons, which leads to the proliferation of astrocytes and the plasticity of brain function and in turn irritates seizures. The abovementioned circulatory plight is the main cause of epilepsy, which places a heavy burden on families and society [24]. Peripheral inflammation is associated with epilepsy susceptibility or epilepsy, and inflammation can exacerbate seizure frequency in patients with epilepsy [25,26]. Animal models have also shown similar association, in many epilepsy models, peripheral injection of the toll-like receptor 4 ligand lipopolysaccharide (LPS) [27] can accelerate epileptogenesis or enhance epilepsy-induced damage. This study investigated the expression of lncRNA NEAT1, miR-129-5p and Notch signaling pathway in epilepsy, and explored the interaction and regulatory relationships between them. We found that lncRNA NEAT1 regulated the Notch signaling pathway by inhibiting the expression of miR-129-5p, thereby inhibiting inflammatory response caused by epilepsy, and providing a new strategy for the treatment of epilepsy.
It has been noted that the disorder of lncRNA is related to the process of epilepsy. Microarray analysis showed that 279 lncRNAs were significantly misregulated in the epilepsy (kainite) mouse model induced by rutin, providing new insights into the potential treatment based on lncRNAs [28]. The lncRNA NEAT1 is critical for formation of mysterious subnuclear domains called paraspeckles, which are present in all mammalian cells. The role of NEAT1 played in many kinds of cancers was fully reported, but the function of NEAT1 in epilepsy was poorly understood. Pavel Katsel et al. [29] reported that NEAT1 was related to the oligodendrocytes function including myelination and dysregulation of NEAT1 had particular correlation with schizophrenia. Zhang et al. [30] suggested that NEAT1 might represent a therapeutic target for inflammasome-associated diseases, by enhancing the activation of NLRP3 and NLRC4 inflammasomes and promoting pyroptosis in vivo. Lei Wu et al. [31] reported that Notch signaling played a key role in neuroinflammation and inflammation-related neuronal damage in epilepsy. In view of the NEAT1 has been proved to modulate human neuronal activity, we performed this study to investigate its role in epilepsy. We firstly explored the expression level of NEAT1 and found that it was increased in temporal lobe tissues of epilepsy patients. This suggested that the NEAT1 was not normally expressed in epilepsy patients. To further research the relationship between NEAT1 and inflammatory factors, we utilized RT-qPCR to examine the expression levels of IL-6, COX-2 and TNF-α and used MTT assay to study the cell viability. Silencing NEAT1 significantly attenuated mRNA levels of IL-6, COX-2 and TNF-α. We concluded that NEAT1 increased the expression levels of inflammatory factors (IL-6, COX-2 and TNF-α), thus inhibited cell viability in epilepsy.
MiRNAs are widely expressed in the vertebrate brains and participates in a variety of neurodegenerative diseases through gene regulation [32,33]. For example, miR-124 is specifically expressed in the brain and is an important regulator of neuronal differentiation and nervous system development, and has been shown to inhibit epileptic seizures and regulate proteins-associated with epileptic development [34]. Torabi et al. reported that miR-455-5p acted as an anti-inflammatory role in multiple sclerosis [35]. Donato et al. discovered new regulative functions of miRNAs in retinitis pigmentosa [36]. MiR-129-5p was commonly reported to function in various cancers by modulating cell proliferation, invasion and migration [37–39]. It was initially found that the miR-129-5p was implicated in neurologic diseases after the identification of its expression in the ventral hippocampus, and was firstly discovered as a new target of NEAT1 [40]. In our results, we found that miR-129-5p was downregulated in epilepsy, and NEAT1 silencing increased the expression level of miR-129-5p, which showed that NEAT1 negatively regulate the expression of miR-129-5p. The results of dual-luciferase reporter assay and prediction of binding sites using bioinformatics software also indicated that there was interaction between miR-129-5p and NEAT1. Silencing miR-129-5p significantly increased the expression of IL-6, COX-2 and TNF-α, and the viability of cells was dramatically reduced by miR-129-5p inhibitor. Also, the expression levels of Notch1, JAG1, HES1, IL-6 and TNF-α and cell viability of astrocytes caused by sh-NEAT1 were reversed by miR-129-5p inhibitor. Thus, we indicated that miR-129-5p might be involved in the mechanism of inflammation and cell viability regulated by NEAT1 in epilepsy.
Notch signal pathway modulates the differentiation and proliferation of neurons [41] and the migration and excitability of mature neurons in the CNS [42,43]. Furthermore, some evidence reported that Notch signaling was involved in the release of proinflammatory cytokines in CNS [44,45]. The Notch signaling pathway was closely related to immune cells and the inflammatory response [46]. Notch could enhance neuronal sensitivity to apoptosis via modulating microglial activation and leukocyte infltration in an ischemic stroke model [47]. In addition, Notch signaling induced microglial-mediated brain inflammation and led to neuronal degeneration in cerebral ischemia [48]. In our study, the expression level of Notch1 in the miR-129-5p mimics group was decreased, while it was increased in the miR-129-5p inhibitor group. Results also indicated that the expression of Notch1, JAG1 and HES1 were significantly elevated after inhibiting miR-129-5p, which indicated that miR-129-5p could affect Notch signaling pathway. Furthermore, the expression levels of IL-6, COX-2 and TNF-α in the miR-129-5p mimics group were decreased, while were rescued in miR-129-5p mimics+pcDNA3.1-Notch1 group. Cell viability was increased by miR-129-5p mimics, while was decreased in miR-129-5p mimics+pcDNA3.1-Notch1 group. These results demonstrated that miR-129-5p inhibited the inflammatory response and cell viability by regulating Notch signaling pathway.
Although the novel findings have been validated in epilepsy cell model, the results of this study may be limited due to lack of animal experiments, which may be improved in the future study. Besides, the role of glyoxalase I (GLO1) in epilepsy inflammation pathway has caught our attention. Recent studies indicated that the increased expression of GLO1 could result in epileptic seizures, for example, Tao et al. reported that the GLO1 SNPs were significantly associated with epilepsy [49]. Recently, GLO1 was analyzed in relation to an important neurodegenerative pathology, also involving inflammation pathway, that was retinitis pigmentosa [50]. Thus, the relationship between GLO1 and epilepsy inflammation pathway may be one of our research objectives in the future. Furthermore, in order to extend the knowledge on non-coding RNAs in relationship with epilepsy, we will perform the whole transcriptomic experiment in epilepsy patients according to the experiment reference workflow [51].
In conclusion, the present study verified that lncRNA NEAT1 affected inflammatory response and cell viability by suppressing miR-129-5p and further regulating Notch signaling pathway in IL-1β-induced epilepsy model in vitro. Specifically, this study clarified the mechanisms of inflammatory response in epilepsy and provided a hopeful strategy for the treatment of epilepsy.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Camfield P, Camfield C.. Regression in children with epilepsy. Neurosci Biobehav Rev. 2019;96:210–218. [DOI] [PubMed] [Google Scholar]
- [2].Huang WS, Zhu L.. MiR-134 expression and changes in inflammatory cytokines of rats with epileptic seizures. Eur Rev Med Pharmacol Sci. 2018;22(11):3479–3484. [DOI] [PubMed] [Google Scholar]
- [3].Wang HK, Yan H, Wang K, et al. Dynamic regulation effect of long non-coding RNA-UCA1 on NF-kB in hippocampus of epilepsy rats. Eur Rev Med Pharmacol Sci. 2017;21(13):3113–3119. [PubMed] [Google Scholar]
- [4].Aronica E, Ravizza T, Zurolo E, et al. Astrocyte immune responses in epilepsy. Glia. 2012;60(8):1258–1268. [DOI] [PubMed] [Google Scholar]
- [5].Annamaria V, Jacqueline F, Tamas B, et al. The role of inflammation in epilepsy. Nat Rev Neurol. 2011;7(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Burmeister AR, Johnson MB, Marriott I. Murine astrocytes are responsive to the pro-inflammatory effects of IL-20. Neurosci Lett. 2019;708:134334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li X, Giri V, Cui Y, et al. LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR-21-5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem Biophys Res Commun. 2019;512(1):79–86. [DOI] [PubMed] [Google Scholar]
- [8].Xiao W, Cao Y, Long H, et al. Genome-Wide DNA methylation patterns analysis of noncoding RNAs in temporal lobe epilepsy patients. Mol Neurobiol. 2018;55(1):793–803. [DOI] [PubMed] [Google Scholar]
- [9].Donato L, Scimone C, Rinaldi C, et al. Non-coding RNAome of RPE cells under oxidative stress suggests unknown regulative aspects of Retinitis pigmentosa etiopathogenesis. Sci Rep. 2018;8(1):16638. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [10].Yu Y, Zhu M, Zhao Y, et al. Overexpression of TUSC7 inhibits the inflammation caused by microglia activation via regulating miR-449a/PPAR-gamma. Biochem Biophys Res Commun. 2018;503(2):1020–1026. [DOI] [PubMed] [Google Scholar]
- [11].Jang Y, Moon J, Lee ST, et al. Dysregulated long non-coding RNAs in the temporal lobe epilepsy mouse model. Seizure. 2018;58:110–119. [DOI] [PubMed] [Google Scholar]
- [12].Han CL, Ge M, Liu YP, et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J Neuroinflammation. 2018;15(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. [DOI] [PubMed] [Google Scholar]
- [15].Rajman M, Metge F, Fiore R, et al. A microRNA-129-5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J. 2017;36(12):1770–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen X, Zhang Y, Shi Y, et al. MiR-129 triggers autophagic flux by regulating a novel Notch-1/E2F7/Beclin-1 axis to impair the viability of human malignant glioma cells. Oncotarget. 2016;7(8):9222–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiao G, Li X, Li G, et al. MiR-129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem-like cells. Oncotarget. 2017;8(61):103261–103273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yin J, Hu H, Li X, et al. Inhibition of Notch signaling pathway attenuates sympathetic hyperinnervation together with the augmentation of M2 macrophages in rats post-myocardial infarction. Am J Physiol Cell Physiol. 2016;310(1):C41–53. [DOI] [PubMed] [Google Scholar]
- [19].Li Y, Wu L, Yu M, et al. HIF-1alpha is Critical for the Activation of Notch Signaling in Neurogenesis During Acute Epilepsy. Neuroscience. 2018;394:206–219. [DOI] [PubMed] [Google Scholar]
- [20].Geng JF, Liu X, Zhao HB, et al. LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int J Biochem Cell Biol. 2018;99:133–139. [DOI] [PubMed] [Google Scholar]
- [21].Ji S, Zheng Z, Liu S, et al. Resveratrol promotes oxidative stress to drive DLC1 mediated cellular senescence in cancer cells. Exp Cell Res. 2018;370(2):292–302. [DOI] [PubMed] [Google Scholar]
- [22].Donato L, Scimone C, Rinaldi C, et al. Stargardt phenotype associated with two ELOVL4 promoter variants and ELOVL4 downregulation: new possible perspective to etiopathogenesis? Invest Ophthalmol Vis Sci. 2018;59(2):843–857. [DOI] [PubMed] [Google Scholar]
- [23].McCormick DA, Contreras D. On the cellular and network bases of epileptic seizures. Annu Rev Physiol. 2001;63:815–846. [DOI] [PubMed] [Google Scholar]
- [24].Grillo E. The circular dilemma of seizure-induced brain injury. Brain. 2014;137(Pt 11):e305. [DOI] [PubMed] [Google Scholar]
- [25].Tellez-Zenteno JF, Matijevic S, Wiebe S. Somatic comorbidity of epilepsy in the general population in Canada. Epilepsia. 2005;46(12):1955–1962. [DOI] [PubMed] [Google Scholar]
- [26].Scheid R, Teich N. Neurologic manifestations of ulcerative colitis. Eur J Neurol. 2010;14(5):483–493. [DOI] [PubMed] [Google Scholar]
- [27].Auvin S, Porta N, Nehlig A, et al. Inflammation in rat pups subjected to short hyperthermic seizures enhances brain long-term excitability. Epilepsy Res. 2009;86(2–3):124–130. [DOI] [PubMed] [Google Scholar]
- [28].Lee DY, Moon J, Lee ST, et al. Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem Biophys Res Commun. 2015;462(4):433–440. [DOI] [PubMed] [Google Scholar]
- [29].Katsel P, Roussos P. The expression of long noncoding RNA NEAT1 is reduced in schizophrenia and modulates oligodendrocytes transcription. NPJ Schizophr. 2019;5(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang P, Cao L, Zhou R. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10(1):1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu L, Li Y, Yu M, et al. Notch signaling regulates microglial activation and inflammatory reactions in a rat model of temporal lobe epilepsy. Neurochem Res. 2018;43(6):1269–1282. [DOI] [PubMed] [Google Scholar]
- [32].Karnati HK, Panigrahi MK, Gutti RK, et al. miRNAs: key players in neurodegenerative disorders and epilepsy. J Alzheimers Dis. 2015;48(3):563–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang W, Wang X, Chen L, et al. The microRNA miR-124 suppresses seizure activity and regulates CREB1 activity. Expert Rev Mol Med. 2016;18:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Torabi S, Tamaddon M, Asadolahi M, et al. miR-455-5p downregulation promotes inflammation pathways in the relapse phase of relapsing-remitting multiple sclerosis disease. Immunogenetics. 2019;71(2):87–95. [DOI] [PubMed] [Google Scholar]
- [36].Donato L, Bramanti P, Scimone C, et al. miRNA expression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio. 2018;8(2):219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96(1):19–25. [DOI] [PubMed] [Google Scholar]
- [38].Lu X, Ma J, Chu J, et al. MiR-129-5p sensitizes the response of Her-2 positive breast cancer to trastuzumab by reducing Rps6. Cell Physiol Biochem. 2017;44(6):2346–2356. [DOI] [PubMed] [Google Scholar]
- [39].Gu LP, Jin S, Xu RC, et al. Long non-coding RNA PCAT-1 promotes tumor progression by inhibiting miR-129-5p in human ovarian cancer. Arch Med Sci. 2019;15(2):513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang H, Cai Y, Zheng L, et al. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J Cell Physiol. 2018;233(10):6638–6648. [DOI] [PubMed] [Google Scholar]
- [41].Breunig JJ, Silbereis J, Vaccarino FM, et al. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc Natl Acad Sci U S A. 2007;104(51):20558–20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ables JL, Breunig JJ, Eisch AJ, et al. Not(ch) just development: notch signalling in the adult brain. Nat Rev Neurosci. 2011;12(5):269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sha L, Wu X, Yao Y, et al. Notch signaling activation promotes seizure activity in temporal lobe epilepsy. Mol Neurobiol. 2014;49(2):633–644. [DOI] [PubMed] [Google Scholar]
- [44].Yao L, Kan EM, Kaur C, et al. Notch-1 signaling regulates microglia activation via NF-kappaB pathway after hypoxic exposure in vivo and in vitro. PloS One. 2013;8(11):e78439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cao Q, Li P, Lu J, et al. Nuclear factor-kappaB/p65 responds to changes in the Notch signaling pathway in murine BV-2 cells and in amoeboid microglia in postnatal rats treated with the gamma-secretase complex blocker DAPT. J Neurosci Res. 2010;88(12):2701–2714. [DOI] [PubMed] [Google Scholar]
- [46].Tsao PN, Wei SC, Huang MT, et al. Lipopolysaccharide-induced Notch signaling activation through JNK-dependent pathway regulates inflammatory response. J Biomed Sci. 2011;18:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Arumugam TV, Chan SL, Jo DG, et al. Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med. 2006;12(6):621–623. [DOI] [PubMed] [Google Scholar]
- [48].Wei Z, Chigurupati S, Arumugam TV, et al. Notch activation enhances the microglia-mediated inflammatory response associated with focal cerebral ischemia. Stroke. 2011;42(9):2589–2594. [DOI] [PubMed] [Google Scholar]
- [49].Tao H, Si L, Zhou X, et al. Role of glyoxalase I gene polymorphisms in late-onset epilepsy and drug-resistant epilepsy. J Neurol Sci. 2016;363:200–206. [DOI] [PubMed] [Google Scholar]
- [50].Donato L, Scimone C. GLO1 gene polymorphisms and their association with retinitis pigmentosa: a case-control study in a Sicilian population. Mol Biol Rep. 2018;45(5):1349–1355. [DOI] [PubMed] [Google Scholar]
- [51].Donato L, Scimone C, Nicocia G, et al. Role of oxidative stress in Retinitis pigmentosa: new involved pathways by an RNA-Seq analysis. Cell Cycle. 2019;18(1):84–104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


