Abstract
Gestational diabetes (GDM), a common pregnancy complication associated with obesity and long-term health risks, is usually diagnosed at approximately 28 weeks of gestation. An understanding of lipid metabolism in women at risk of GDM could contribute to earlier diagnosis and treatment. We tested the hypothesis that altered lipid metabolism at the beginning of the second trimester in obese pregnant women is associated with a diagnosis of GDM. Plasma samples from 831 participants (16–45 years, 15–18 weeks gestation, BMI ≥ 30) from the UPBEAT study of obese pregnant women were used. The lipid, sterol and glyceride fraction was isolated and analysed in a semi-quantitative fashion using direct infusion mass spectrometry. A combination of uni-, multi-variate and multi-variable statistical analyses was used to identify candidate biomarkers in plasma associated with a diagnosis of GDM (early third trimester; IADPSG criteria). Multivariable adjusted analyses showed that participants who later developed GDM had a greater abundance of several triglycerides (48:0, 50:1, 50:2, 51:5, 53:4) and phosphatidylcholine (38:5). In contrast sphingomyelins (32:1, 41:2, 42:3), lyso-phosphatidylcholine (16:0, 18:1), phosphatidylcholines (35:2, 40:7, 40:10), two polyunsaturated triglycerides (46:5, 48:6) and several oxidised triglycerides (48:6, 54:4, 56:4, 58:6) were less abundant. We concluded that both lipid and triglyceride metabolism were altered at least 10 weeks before diagnosis of GDM. Further investigation is required to determine the functional consequences of these differences and the mechanisms by which they arise.
Introduction
Gestational diabetes (GDM) occurs in 8–24% of all pregnancies in the UK.1 GDM is more common in obese (BMI ≥ 30) women, amongst whom approximately one third are diagnosed with the condition.2,3 The increasing global prevalence of obesity4,5 has therefore led to parallel trends in the diagnosis of GDM. GDM is associated with adverse outcomes for mother and child including pre-eclampsia, complications in labour, stillbirth, fetal macrosomia, increased risk of later Type 2 diabetes (T2DM) for the mother,6–10 and greater adiposity in the offspring in childhood and adolescence.11–16 There is also evidence from animal models that the over-nutrition associated with obesity and insulin resistance during pregnancy influences the metabolism of the resulting offspring.17
Currently, GDM is diagnosed at 24–28 weeks gestation using an Oral Glucose Tolerance Test (OGTT). This test involves assessing the subject’s response to a glucose challenge, with fasting, +1 h and +2 h post-ingestion blood glucose measurements. However, the diagnostic thresholds of blood glucose concentration used in the diagnosis vary considerably.18–21 Furthermore, there is mounting evidence that complications in labour consistent with GDM occur below some of these thresholds,22–24 despite diagnosis and treatment.25 This, and the complications associated with GDM and obesity, have motivated a focus on earlier diagnosis and prediction. However, most models employ a range of mainly non-molecular variables that have low sensitivity.26 This led us to consider whether molecular biomarkers which are not routinely measured might be more sensitive discriminators than clinical factors such as previous diagnosis of GDM, age and BMI measured alone or in combination with routinely measured molecular markers.
Current evidence suggests that the concentration of glucose in the blood in the first and second trimesters is not an accurate indicator of GDM, and that OGTT thresholds do not perform equivalently in earlier trimesters of pregnancy.27 This has led to a consensus that diagnosis of GDM using the OGTT is not reliable in the first and early in the second trimesters. However, studies of a cohort not selected for BMI found evidence for shifts in lipid metabolism associated with GDM, at a molecular level.28,29 We have also characterised the early second trimester lipoprotein profile in obese pregnancy and GDM and found that the abundance of VLDLs, small LDLs and HDLs differs in women who develop GDM compared to those who do not.30 Collectively, this evidence suggests that both lipid biosynthesis and distribution are altered in advance of a standard diagnosis of GDM. Given the strong association between GDM and obesity, this raises the question of whether lipid dysregulation at a molecular level in GDM differs from that of obesity. This led us to the hypothesis that the lipid metabolism in obese women who develop GDM is altered before the hyperglycaemia becomes evident.
To test this hypothesis, we profiled the organically-soluble fraction (containing the lipids, glyceride and sterols) using state-of-the-art lipidomics methods on plasma samples collected at 15–18 weeks gestation from the UPBEAT30–32 cohort of obese pregnant women. Molecular profiling involved direct infusion mass spectrometry (DI-MS28,33–37) with chip-based nanospray, in both positive and negative ionisation modes. A supervised multi-variate analysis (sparse Partial Least Squares-Discriminant Analysis, sPLS-DA) followed by a student’s T-test was used to identify lipid molecular species that distinguished participants who were later diagnosed with GDM from those who did not develop GDM, and to identify the variables that drive this distinction. Candidate biomarkers were identified using multivariable adjustment for confounding factors including maternal BMI, age, ethnicity, parity and diagnosis of pre-eclampsia.
Experimental
Participants
Participants from the UK Pregnancies Better Eating and Activity Trial (UPBEAT; isrctn.org registration number 89971375)38 was used for this study, characteristics are shown in Table 1. UPBEAT was a multi-centre randomised controlled trial of a complex dietary and physical activity intervention designed to prevent GDM in obese women and reduce the number of large-for-gestational age infants. Women with underlying medical conditions were excluded. The UPBEAT study comprised 1555 women recruited between 2009 and 2014. The present analysis was performed using data from 831 women who were Caucasian, black or Asian (all), had a full set of data from the OGTT and a non-haemolysed plasma sample taken between 15 weeks 0 days’ and 18 weeks 6 days’ gestation (15 + 0 and 18 + 6, ~17 weeks gestation). All had a BMI ≥ 30 (kg m−2) and age of 16–45 years. Ethnicity, parity (0–7), sex of infant, and intervention arm were recorded and used for factor-based adjustment but not for stratification.
Table 1. Characteristics of the subset of the UPBEAT cohort use for this study. Diagnosis of GDM was made using IADPSG thresholds (18).
| Cohort |
No GDM |
GDM |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor | Cat. | No. | Mean | Std. dev. | Min. | Max. | No. | Mean | Std. dev. | Min. | Max. | No. | Mean | Std. dev. | Min. | Max. | p-Value |
| Maternal BMIa | 831 | 36.27 | 4.8 | 30 | 63.4 | 582 | 35.88 | 4.43 | 30 | 59.9 | 249 | 37.18 | 5.47 | 30 | 63.4 | <0.001 | |
| Maternal agea | 831 | 30.73 | 5.4 | 16 | 45 | 582 | 30.24 | 5.63 | 16 | 45 | 249 | 31.88 | 4.62 | 18 | 44 | <0.001 | |
| BGC (mM) | Fasting | 831 | 4.75 | 0.56 | 3.5 | 8.4 | 582 | 4.52 | 0.31 | 3.5 | 5 | 249 | 5.31 | 0.6 | 3.9 | 8.4 | <0.001 |
| BGC (mM) | 1 h | 831 | 8.05 | 2.13 | 2.6 | 17.6 | 582 | 7.21 | 1.42 | 2.6 | 9.9 | 249 | 10 | 2.22 | 4.1 | 17.6 | <0.001 |
| BGC (mM) | 2 h | 831 | 6 | 1.53 | 2.4 | 14.6 | 582 | 5.55 | 1.14 | 2.4 | 8.4 | 249 | 7.06 | 1.79 | 3 | 14.6 | <0.001 |
| HbA1C (%) | 782 | 4.85 | 0.35 | 3.89 | 6.61 | 546 | 4.79 | 0.33 | 3.89 | 5.78 | 238 | 4.97 | 0.37 | 4.14 | 6.61 | <0.001 | |
| HbA1C (mMol M–1) | 782 | 29.49 | 3.85 | 19 | 48.7 | 546 | 28.89 | 3.61 | 19 | 39.7 | 238 | 30.86 | 4.05 | 21.7 | 48.7 | <0.001 | |
| No. | % | Cum. (%) | |||||||||||||||
| Sex of infant | Male | 429 | 51.6 | — | — | — | 308 | — | — | — | — | 121 | — | — | — | — | 0.254 |
| Ethnicity | Black | 169 | 20.34 | 20.34 | — | — | |||||||||||
| White | 563 | 67.75 | 88.09 | — | — | ||||||||||||
| Asian | 54 | 6.5 | 94.59 | — | — | ||||||||||||
| Other | 45 | 5.42 | 100 | — | — | ||||||||||||
| Parity | 0 | 379 | 45.61 | 45.61 | — | — | |||||||||||
| 1 | 265 | 31.89 | 77.5 | — | — | ||||||||||||
| 2 | 121 | 14.56 | 92.06 | — | — | ||||||||||||
| 3 | 43 | 5.17 | 97.23 | — | — | ||||||||||||
| 4 | 15 | 1.81 | 99.04 | — | — | ||||||||||||
| 5 | 6 | 0.72 | 99.76 | — | — | ||||||||||||
| 7 | 2 | 0.24 | 100 | — | — | ||||||||||||
| Pre-eclampsia | 772 | 59 | |||||||||||||||
| Study Arm | Control | 415 | 49.94 | ||||||||||||||
| Intervention | 416 | 50.06 | |||||||||||||||
At 17 weeks gestation. BGC, blood glucose concentration. Ethnicity is maternal.
Diagnosis of GDM
The diagnoses of GDM were made according to IADPSG (International Association of Diabetes and Pregnancy Study Groups) criteria, with diagnosis based on one or more GDM-positive plasma glucose values following an oral glucose load of 75 g.18 OGTTs were performed between 24+2 and 30+0 weeks’ (mean 27+5) gestation.
Ethics
The UPBEAT trial was granted ethical approval by the National Health Service Research Ethics Committee (UK Integrated Research Application System; reference 09/H0802/5) and all participants, including women aged 16 and 17 using Fraser guidelines, provided informed written consent38 in compliance with the Declaration of Helsinki principles.
Reagents and standards
Solvents were purchased from Sigma-Aldrich Ltd (Gillingham, Dorset, UK) of at least HPLC grade and were not purified further. Lipid standards were purchased from Avanti Polar lipids (Alabaster, AL; through Instruchemie, Delfzijl, NL) and used without purification. Consumables were purchased from Sarstedt AG & Co (Leicester, UK).
Isolation of the organically-soluble fraction
Lithium heparin plasma samples, stored at −80 °C and blinded to participant data and in random order were used. Samples had been freeze–thawed once before the extraction in the present study. The lipid, triglyceride and sterol fractions were isolated together using a high throughput technique developed from existing procedures.39,40 Briefly, aliquots of plasma (25 μL) were placed along with blank and QC samples in the wells of a glass-coated 2.4 mL per well 96w plate (Plate+™, Esslab, Hadleigh, UK). Water (100 μL, MilliQ) was added to each of the wells, followed by methanol (150 μL, HPLC grade, spiked with Internal Standards, see Table S1, ESI†), followed by tert-butyl methyl ether (TMBE, 750 μL). The plates were then sealed (aluminium microplate sealing tape), agitated (10 min, 600 rpm) and centrifuged (2 min, 3·2k × g). A multi-channel pipette was used to transfer 25 μL of the organic solution to a glass-coated 240 μL per week 384w plate (Plate+™, Esslab, Hadleigh, UK). The samples were reconstituted (TBME, 25 μL and MS-mix [7.5 mM ammonium acetate in IPA : CH3OH (2:1)], 90 μL), and the plate heat-sealed and stored at −20 °C.
Mass spectrometry (DI-MS)
All samples were infused into an Exactive Orbitrap (Thermo, Hemel Hampstead, UK), using a Triversa Nanomate (Advion, Ithaca US), for direct infusion mass spectrometry (DI-MS41). Samples (10 μL ea.) were ionised at 1·2 kV in the positive ion mode. The exactive started acquiring data 20 s after sample aspiration began. The exactive acquired data with a scan rate of 1 Hz (resulting in a mass resolution of 65 000 full width at half-maximum [fwhm] at 400 m/z). The automatic gain control was set to 3 000 000 and the maximum ion injection time to 10 ms. After 72 s of acquisition in positive mode the Nanomate and the exactive switched over to negative mode, decreasing the voltage to −1·5 kV and the maximum ion injection time to 250 ms. The spray was maintained for another 66 s, after which the analysis was stopped and the tip discarded, before the analysis of the next sample began. The sample plate was kept at 15 °C throughout the acquisition. Samples were run in row order. The instrument was operated in full scan mode from m/z 150–1200 Da.
Mass spectrometry (LC-MS)
LCMS was run in a similar manner to recent studies29,42,43
Chromatographic separation of lipid and triglycerides was achieved using a Waters Acquity UPLC CSH C18 (50 mm × 2.1 mm, 1.7 mm) LC-column with a Shimadzu UPLC system (Shimadzu UK Limited, Wolverton, Milton Keynes). The column was maintained at 55 °C with a flow rate of 0.5 mL min−1. A binary mobile phase system was used with mobile phase A; acetonitrile : water mix (3:2, respectively, with 10 mM ammonium formate), and mobile phase B; isopropanol : acetonitrile mix (9:1, respectively, with 10 mM ammonium formate). The gradient profile was as follows; at 0 minutes_40% mobile phase B, at 0.4 minutes_43% mobile phase B, at 0.45 minutes_50% mobile phase B, at 2.4 minutes_54% mobile phase B, at 2.45 minutes_70% mobile phase B, at 7 minutes_99% mobile phase B, at 8 minutes_99% mobile phase B, at 8.3 minutes_40% mobile phase B, at 10 minutes_40% mobile phase B. Mass spectrometry detection was performed on a Thermo Exactive orbitrap mass spectrometer (Thermo Scientific, Hemel Hempstead, UK) operating in positive ion and negative ion continuous switching mode. Heated electrospray source was used; the sheath gas was set to 40 (arbitrary units), the aux gas set to 15 (arbitrary units) and the capillary temperature set to 300 °C. The instrument was operated in full scan mode from m/z 150–1200 Da. Lipid species were identified by detecting a signal peak for the corresponding accurate mass at the correct retention time. Signals were normalized to the total lipid/glyceride signal for that sample and shown as per mille (%).
Data processing
The lipid signals obtained were relative abundance (‘semiquantitative’), with the signal intensity of each lipid expressed relative to the total lipid signal intensity, for each individual, per cent (%). The relative abundance of all species identified was calculated separately for positive and negative ionisation modes. Raw high-resolution mass spectrometry data were processed using XCMS (www.bioconductor.org) and Peakpicker v 2.0 (an in-house R script37). Lists of known species (by m/z) were used for both positive ion (n = 1740 incl. standards) and negative ion mode (n = 5075 including standards). Signals that deviated by more than 9 ppm were discarded, as were those with a signal/noise ratio of <2 and those pertaining to fewer than 75% of samples. The correlation of signal intensity to concentration of plasma in QCs (0.25, 0.5, 0.75, 1.0, 1.5×) was used to identify which lipid signals were linearly proportional to abundance in the sample type and volume used (threshold for acceptance was a correlation of >0.75). The variation across analytical plates was corrected by batch mean centring before the removal of outlier measurements (values > or < 4 s.d. from the average for that variable). Signals were then corrected (divided by the sum of signals for that sample), in order to be able to compare samples. Zero values were interpreted as not measured. All signals that passed the DI-MS quality control process were identified as their most likely molecular species and will be further called variables. Several of these were checked by LCMS (vide supra). Importantly, signals were identified by their m/z and several molecular species can contribute to one signal. All statistical calculations were done on these finalised values.
Statistical methods
The analysis was structured according to a prepared analysis plan. Univariate analyses were carried out using Excel 2013. Multivariate analyses (MVA) were carried out using MetaboAnalyst 4.0.44 Stata SE v. 13.0 was used for multivariable analyses.
Analyses of the variables collected in positive and negative modes were carried out separately. Principal component analyses (PCA) were used to identify sample outliers, which were excluded before further analysis. sparse Partial Least Squares-Discriminant Analyses (sPLS-DA, an unsupervised MVA) were used to identify individual variables that distinguished the two groups. The variables with the lowest probability of a false positive result (p-value, Student’s t-test) were regarded as the most important in driving the difference between groups. The p-values were corrected using a Benjamini-Hochberg FDR correction based on the p-values of 565 independent variables, with values below 0·05 after correction regarded as significant. (This gave the same twenty variables as for a corrected Bonferroni FDR threshold based on 565 variables, p = 0·0021). Finally, variables that were identified from multivariable analysis-adjusted data were classed as candidate biomarkers. Odds ratios and uncorrected p-values relating to association with GDM were adjusted for maternal age, maternal BMI, ethnicity, parity and the presence of pre-eclampsia. Once again, a Benjamini-Hochberg FDR correction based on the p-values of 565 independent variables was used, with values below 0·05 after correction being regarded as significant. Further adjustments for sex of infant and intervention arm did not materially affect the results.
Results
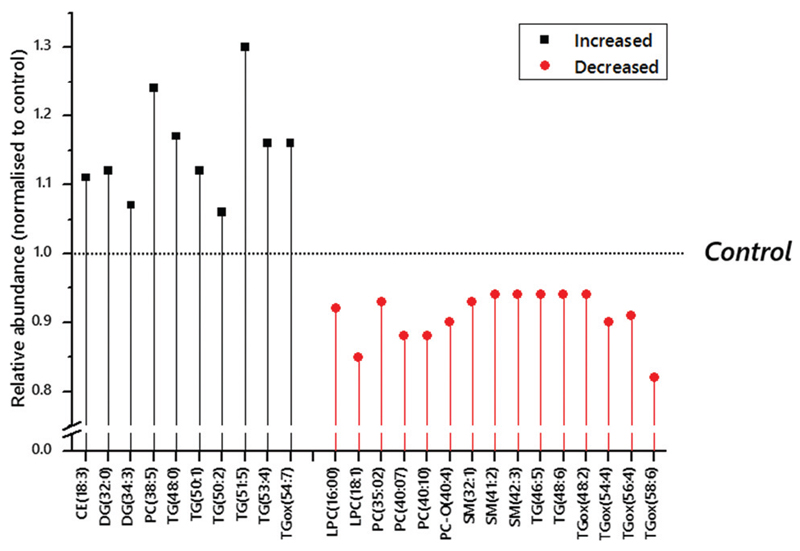
Direct infusion mass spectrometry (DI-MS) identified 215 variables in positive ion mode and 350 in negative ion mode. A combination of a sparse Partial Least Squares Discriminant Analysis (sPLS-DA) followed by a student’s t-test was used to identify which variables distinguished the groups and then the variables that drove the effect (Table S2 and Fig. S1, ESI†). This analysis suggested that the abundance of several triglyceride isoforms (50:1, 51:5; TG fragments DG–H2O(32:0) and (38:6)) was higher in obese women who went on to develop GDM than those who did not, whilst the abundance of oxidised forms of commonplace TGs, TGox(54:4) and TGox(56:4), were lower. This indicated that there was a significant shift in at least two aspects of triglyceride metabolism at least 10 weeks in advance of diagnosis of GDM. Several factors have been identified as additional risk factors for GDM that this statistical calculation using unadjusted data does not account for. These include adiposity, ethnicity and maternal age. Indeed, metadata for the participants indicated that the women who developed GDM were significantly older and had a higher BMI in the first trimester (Table 1). After adjustment for maternal age, BMI, ethnicity, infant sex and parity there was an increase in the number, and a small change in the variables that differed significantly between the two groups (Fig. 1 and Table S2, ESI†).
Fig. 1.
Normalised relative abundance of lipids identified as candidate biomarkers in the circulation of obese pregnant women at ~17 weeks gestation using p-values adjusted according to age, BMI, ethnicity, diagnosis of pre-eclampsia and parity. Candidate biomarkers that are more abundant than in controls are marked as Increased (shown in black). Eventual intervention arm (assigned randomly after the samples used in this study were collected) and sex of infant were not strongly associate with any of the variables tested. The Bonferroni corrected p-value threshold was 0·0021, based on 565 independent variables. The control group, against which the values shown were normalised, were an obese but otherwise typically healthy group (meta-data in Table 1).
Adjusted analyses (Fig. 1 and Table S3, ESI†) also showed that several TGs were more abundant in participants who went on to develop GDM. We used liquid chromatography mass spectrometry (LC-MS) to verify the assignments the signals. This also excluded certain possible isobaric species from identification (Table S4, ESI†). Species identified included TGs that comprise fatty acid residues with an odd number of carbon atoms and across a range of levels of unsaturation (0–5 olefin bonds). At least two lighter, more polyunsaturated TGs, TG(46:5) and (48:6), were less abundant in the GDM group and may both contain essential polyunsaturated fatty acids.
The TGox from four commonplace TGs were also less abundant in participants who went on to develop GDM (48:2, 54:4, 54:6, 58:6). None of the (more common) TG isoforms from which these TGox species originate was found to have a significantly different abundance. Sphingomyelins (32:1, 41:2, 42:3) were all found to be less abundant in participants who went on to be diagnosed with GDM. This was opposite to the shifts in abundance for cholesteryl esters (18:3 in the adjusted analysis, 20:5 in the unadjusted). The pattern for polyunsaturated PCs was less clear, with 38:5 higher in abundance but 35:2, 40:7 and 40:10 lower.
The rich but complicated pattern of shifts in phospholipid metabolism in the adjusted analysis raised questions about whether particular factors associated with GDM were associated with shifts in the abundance of particular species. We therefore carried out sensitivity analyses with respect to maternal age, BMI, ethnicity, parity, and intervention arm, in order to assess which of the candidate biomarkers identified in the analysis of adjusted data (Table 2) might be associated with classical risk factors (Table 3).
Table 2.
Unadjusted and adjusted odds ratios, confidence intervals and p-values for all variables that pass the Benjamini Hochberg (BH) FDR test for significance after adjustment for participants who go on to be diagnosed with GDM and those who do not. Variables are ordered alphabetically
| Not adjusted |
Adjusted |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | OR | SE | z | P > |z| | [95% Conf. interval] | OR | SE | z | P > |z| | [95% Conf. interval] | BH FDR corr. p-value | ||
| CE(18:3) | 1.089 | 0.033 | 2.82 | 0.005 | 1.026 | 1.156 | 1.110 | 0.036 | 3.23 | 0.001 | 1.042 | 1.183 | 0.033 |
| DG-H2O(32:0)a | 1.194 | 0.056 | 3.78 | 0 | 1.089 | 1.309 | 1.180 | 0.057 | 3.4 | 0.001 | 1.073 | 1.298 | 0.033 |
| DG-H2O(34:3) | 1.136 | 0.059 | 2.46 | 0.014 | 1.026 | 1.258 | 1.209 | 0.067 | 3.42 | 0.001 | 1.084 | 1.348 | 0.033 |
| lyso-PC(16:0)a | 0.830 | 0.042 | –3.66 | 0 | 0.751 | 0.917 | 0.836 | 0.044 | –3.41 | 0.001 | 0.754 | 0.926 | 0.033 |
| lyso-PC(18:1)a | 0.819 | 0.032 | –5.14 | 0 | 0.759 | 0.884 | 0.832 | 0.033 | –4.6 | 0 | 0.769 | 0.900 | 0.000 |
| PC(35:2) | 0.876 | 0.041 | –2.82 | 0.005 | 0.799 | 0.960 | 0.852 | 0.043 | –3.21 | 0.001 | 0.773 | 0.940 | 0.033 |
| PC(38:5) | 1.090 | 0.032 | 2.98 | 0.003 | 1.030 | 1.153 | 1.095 | 0.031 | 3.18 | 0.001 | 1.035 | 1.159 | 0.033 |
| PC(40:10) | 0.843 | 0.045 | –3.18 | 0.001 | 0.758 | 0.936 | 0.845 | 0.047 | –3.05 | 0.002 | 0.759 | 0.942 | 0.045 |
| PC(40:7) | 0.897 | 0.032 | –3.08 | 0.002 | 0.836 | 0.961 | 0.893 | 0.033 | –3.11 | 0.002 | 0.831 | 0.959 | 0.045 |
| PC-O(40:4)a | 0.893 | 0.027 | –3.74 | 0 | 0.841 | 0.947 | 0.909 | 0.028 | –3.1 | 0.002 | 0.855 | 0.965 | 0.045 |
| SM(32:1) | 0.866 | 0.043 | –2.89 | 0.004 | 0.786 | 0.955 | 0.842 | 0.043 | –3.34 | 0.001 | 0.761 | 0.931 | 0.033 |
| SM(41:2) | 0.872 | 0.047 | –2.53 | 0.011 | 0.784 | 0.970 | 0.844 | 0.047 | –3.03 | 0.002 | 0.756 | 0.942 | 0.045 |
| SM(42:3) | 0.894 | 0.040 | –2.49 | 0.013 | 0.819 | 0.976 | 0.864 | 0.040 | –3.13 | 0.002 | 0.789 | 0.947 | 0.045 |
| TG(46:5) | 0.829 | 0.051 | –3.07 | 0.002 | 0.736 | 0.934 | 0.798 | 0.051 | –3.54 | 0 | 0.704 | 0.904 | 0.000 |
| TG(48:0) | 1.112 | 0.037 | 3.19 | 0.001 | 1.042 | 1.187 | 1.109 | 0.038 | 3.02 | 0.002 | 1.037 | 1.186 | 0.045 |
| TG(48:6) | 0.853 | 0.048 | –2.81 | 0.005 | 0.763 | 0.953 | 0.819 | 0.048 | –3.38 | 0.001 | 0.73 | 0.92 | 0.033 |
| TG(50:1)a | 1.190 | 0.054 | 3.83 | 0 | 1.089 | 1.301 | 1.193 | 0.057 | 3.71 | 0 | 1.087 | 1.309 | 0.000 |
| TG(50:2) | 1.163 | 0.063 | 2.8 | 0.005 | 1.046 | 1.292 | 1.209 | 0.070 | 3.29 | 0.001 | 1.08 | 1.353 | 0.033 |
| TG(51:5)a | 1.131 | 0.035 | 4 | 0 | 1.065 | 1.202 | 1.145 | 0.037 | 4.15 | 0 | 1.074 | 1.221 | 0.000 |
| TG(53:4) | 1.122 | 0.042 | 3.09 | 0.002 | 1.043 | 1.207 | 1.136 | 0.044 | 3.33 | 0.001 | 1.054 | 1.225 | 0.033 |
| TGox(48:2) | 0.747 | 0.070 | –3.1 | 0.002 | 0.621 | 0.898 | 0.722 | 0.070 | –3.36 | 0.001 | 0.597 | 0.873 | 0.033 |
| TGox(54:7) | 1.151 | 0.055 | 2.95 | 0.003 | 1.048 | 1.263 | 1.165 | 0.058 | 3.07 | 0.002 | 1.057 | 1.284 | 0.045 |
| TGox(54:4)a | 0.741 | 0.059 | –3.79 | 0 | 0.635 | 0.865 | 0.735 | 0.060 | –3.77 | 0 | 0.627 | 0.863 | 0.000 |
| TGox(56:4)a | 0.793 | 0.058 | –3.18 | 0.001 | 0.687 | 0.915 | 0.788 | 0.059 | –3.18 | 0.001 | 0.68 | 0.912 | 0.033 |
| TGox(58:6) | 0.870 | 0.039 | –3.1 | 0.002 | 0.797 | 0.95 | 0.863 | 0.040 | –3.17 | 0.002 | 0.788 | 0.945 | 0.045 |
Variables that passed Bonferroni corrected-p-value FDR, based on 565 independent variables (unadjusted).
Table 3.
Sensitivity analysis of the significance of candidate biomarker across each confounding factor (adjusted together). Only p-values that pass at the Bonferroni-corrected threshold (0.0021, red) or at 0.05 (green) are shown, except for sex of infant and intervention arm, where the lowest p-value is shown. HbA1C was included in the sensitivity analysis in order to identify which variables were associated with measured hyperglycaemia
| HbA1C | Maternal BMI | Maternal Age | parity (0-7) | parity code* | Ethnicity (all) | Ethnicity (Black) | Ethnicity (White) | Ethnicity (Asian) | Diagnosis of PET | Sex if infant | Int. arm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CE(18:3) | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| CE(20:5)a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| Iyso-PC(15:0) a | 0.04 | 0.00 | 0.04 | 0.00 | ||||||||
| Iyso-PC(16:0) | 0.00 | 0.01 | ||||||||||
| Iyso-PC(18:1) | 0.00 | 0.01 | 0.00 | 0.02 | 0.02 | |||||||
| PC(36:5) a | 0.01 | 0.00 | 0.00 | 0.00 | ||||||||
| PC(38:5) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| PC(40:10) | 0.03 | |||||||||||
| PC(40:07) | 0.01 | 0.03 | 0.01 | 0.02 | ||||||||
| PC-O(18:1) a | 0.03 | 0.04 | 0.00 | 0.03 | 0.00 | |||||||
| PC-O(34:1) a | 0.01 | 0.00 | 0.00 | |||||||||
| PC-O(40:4) | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.04 | |||||
| PE(38:2) & PC(35:2) | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| PI(36:1) a | 0.00 | 0.00 | ||||||||||
| SM(32:1) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | |||||
| SM(41:2) | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| SM(42:3) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| DG(32:0) | 0.00 | 0.00 | 0.05 | |||||||||
| DG(34:3) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| DG(38:5) | 0.00 | 0.01 | 0.00 | |||||||||
| DG(38:6) a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| TG(46:5) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.07 | |||||
| TG(48:0) | 0.00 | 0.04 | ||||||||||
| TG(48:6) | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | |||||
| TG(50:1) | 0.00 | 0.00 | ||||||||||
| TG(50:2) | 0.04 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| TG(51:5) | 0.03 | 0.14 | ||||||||||
| TG(53:4) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| TGox(48:2) | 0.05 | |||||||||||
| TGox(54:4) | 0.04 | 0.04 | ||||||||||
| TGox(54:7) | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||
| TGox(56:4) | ||||||||||||
| TGox(58:5) a | 0.04 | 0.03 | 0.00 | 0.00 | ||||||||
| TGox(58:6) | 0.01 | |||||||||||
| No. (p<0.05) | 13 | 11 | 10 | 25 | 21 | 14 | 14 | 15 | 15 | 3 | 0 | 0 |
| No. (p <0.0021) | 6 | 3 | 6 | 22 | 14 | 11 | 13 | 14 | 13 | 0 | 0 | 0 |
Parity was also grouped into nulli- and mono-parous participants, and multiparous ones. Int. arm, Intervention arm.
Unadjusted analysis only.
These analyses showed that the factors (maternal BMI, age, parity, ethnicity) were associated with changes in the relative abundance of several variables. Ethnicity and parity had a similar profile, associated with increased abundance of CEs, PC(38:5) and TGs, but decreased abundance of sphingomyelins and TGox. There was some overlap between ethnicity/parity and maternal BMI, suggesting that some individuals may have a pronounced phenotype due to a summative effect.
Discussion
This study used detailed molecular profiling to identify alterations in the abundance, and thus metabolism, of plasma sterols, lipids and triglycerides of obese women in advance of a standard clinical diagnosis of GDM. A number of variables were identified as having a different abundance at ~17 weeks in women who were diagnosed with GDM at approximately 28 weeks. This study therefore offers evidence that lipid metabolism was altered at least 10 weeks before a clinical diagnosis of GDM was made.
The panel of lipids identified in the present study comprised several phospholipids, triglycerides and oxidised triglycerides, and at least one cholesteryl ester, as being associated with later onset of the condition. Shifts in phospholipid metabolism were characterised by a remodelling of polyunsaturated PCs, and a lower abundance of SMs, lyso-PCs and PE(38:2). PC(38:5) was more abundant whereas PC(40:10) and PC(40:7) were less abundant, suggesting that the number of species that may contain essential fatty acids such as FA(20:5) (EPA) or perhaps FA(22:6) (DHA) is reduced. This is intriguing in the light of evidence that lipoprotein composition can influence the fluidity of cell membranes45 and also that cell membrane composition is reflected in that of lipoproteins.46,47 This suggests that the phospholipid profile in lipoproteins is closely related to that of plasma membranes in vivo. As modulation of the composition of lipids in hydrated systems has a profound effect on the physical behaviour of membranes and other assemblies that comprise them,48–51 this raises questions about how shifts in the phospholipid composition of the system may alter its internal structures. A lower abundance of polyunsaturated PCs comprising longer chains (40 carbons) is consistent with thinner membranes rather than less fluid ones, especially with a concomitant increase in the proportion of PC(38:5). Furthermore, a decrease in the amount of lyso-PCs disfavours curvature away from the water (bulging). SM typically have relatively high melting transition temperatures and thus reduce fluidity in membranes.52 This evidence suggests that membranes in affected systems may be thinner and with modulated fluidity. Alterations in these membrane properties is consistent with general evidence that membrane composition affects protein activity.53–55 There are recognised relationships between membrane composition and protein activity in hyperglycaemia.56 In addition certain fatty acids, DG and ceramide have been shown to activate certain serine kinases that weaken insulin signalling pathways and therefore cause insulin resistance.57 The studies in this area have found that a greater dietary intake of n-3 PUFAs is associated with prevention of insulin resistance and greater insulin sensitivity, with phosphatidylethanolamine (PE) and sphingomyelin (SM) being independent predictors of insulin resistance.56 The present study identified several species comprising PUFAs and three isoforms of SM that were less abundant in participants who were later diagnosed with GDM, providing intriguing possibilities for further work about the specific molecular mechanisms that modulate insulin signal transduction. This would offer a mechanistic basis for the influence these species on insulin action.
The evidence that both the lengths of fatty acid residues and the number of olefin bonds in them differs between GDM and non-GDM groups in the present study may be the result of changes in how fatty acids are transferred between lipids and triglycerides. Pregnancy represents a period in which there is considerable change in the expression of lipases in several tissues.58–60 However, this may be altered in GDM,61 with evidence that a high fat maternal diet (associated with obesity and thus GDM),62 methylation of DNA63 and even the abundance of lipase inhibitor(s)64 affecting the lipase expression measured. The change in expression of placental endothelial lipase (PEL) is unclear at present, with conflicting results as to whether this increases in GDM.65,66 As PEL hydrolyses fatty acids from triglycerides that enter the placenta, changes in its expression may have a profound effect on the rate of transfer of FAs from the mother’s circulation to that of the fetus. This has been observed through an association between placental lipoprotein lipase activity being positively associated with adiposity of the infant.67 At present there are no studies that link particular lipid or triglyceride species to individual lipases or their expression, and thus a focused study is required to investigate that.
Similarly, fatty acid transporters may play a role in GDM, with several recent studies in this area.68–70 One recent report has shown that MFSD2a, a transporter for the essential fatty acid DHA (FA(22:6)), found in both placenta and the CNS,71,72 may be lower in GDM pregnancies and thus offers a possible mechanism for the lower availability of DHA in GDM.73 However, here too, focused study incorporating both labelled species and appropriate enzymology are not yet available.
Triglyceride metabolism was also modulated in advance of a diagnosis of GDM, with the most polyunsaturated species (46:5, 48:6) less abundant and others of a similar length or longer, with fewer double bonds being more abundant (48:0, 50:1, 50:2). Like the remodelling of PCs, the abundance of polyunsaturated species is reduced, however the average length of the fatty acid residues appears to be longer in TGs in affected systems. A shift towards less polyunsaturated TGs may explain why the oxidised derivatives of TGs (TGox) are less abundant in affected individuals; a lower abundance of polyunsaturated TGs reduces the concentration of species prone to non-specific oxidation.
The significant increase in the abundance of TG(51:5) and (53:4) in individuals who later developed GDM is consistent with a study of lipid metabolism in pregnant women with GDM in the Cambridge Baby Growth Study (CBGS) who were of heterogeneous BMI. The latter study identified a higher abundance of TG(51:1), a species that must comprise a fatty acid with an odd number of carbons, as being associated with GDM.29 The results of the CBGS and present study contrast with evidence that a higher dietary intake of odd-chain-containing species is associated with a lower risk of T2DM.74–76 However, the integration of a signal that represents both PC(35:2) and PE(38:2) is significantly lower than in controls, suggesting that at least one odd-chain containing phospholipid is lower in participants who go on to be diagnosed with GDM. Several or different lipid pathways may therefore be involved in the relationship between hyperglycaemia and lipid metabolism in GDM. However a further study would require information on dietary intake during pregnancy to characterise this interaction in GDM.
We also found an increase in mono-unsaturated species in association with GDM, e.g. TG(50:1), and a decrease in the abundance of species that may result from the release of mono-unsaturated fatty acids (lyso-PC(16:0) and lyso-PC(18:1)). A similar increase in abundance of PC(32:1) and TG(51:1) was previously found to be associated with the emergence of GDM in the CBGS cohort (heterogeneous for BMI).29 These species may contain or may have arisen from others that contained FA(16:1). Increased FA(16:1) in circulating phospholipids has been linked to a decrease in insulin sensitivity and increased adiposity.77 However, it is not clear why a greater number or abundance of such species should be present in obese individuals. These complicated effects may be explained by the evidence that the metabolism and distribution of C16:1 differs; a higher abundance of FA(16:1) in the circulation predicts metabolic syndrome,78 but the abundance of C16:1 in erythrocytes is relatively low in obese individuals79 and may offer protection against sudden cardiac arrest.80 Furthermore, infusions of FA(16:1) into obese ovines have been shown to reduce the size of intramuscular adipocytes and restore sensitivity to insulin.79 This pattern of distribution of fatty acids clearly has a profound effect on cardio-metabolic disease risk, and may have also have implications in GDM.
The importance of how lipids and triglycerides are distributed between lipoproteins in obesity and diabetes is intriguing in the context of recent work from our group that shows that the structure of lipid assemblies in the circulation differ in obese women from the UPBEAT cohort before development of GDM.30 White et al. showed that the profile of both VLDLs and HDLs differs at the same time point as the shifts in lipid profile detected in this study.30 This distribution of lipids between lipoproteins may be important for the effect(s) those lipids have. For example, the distribution may influence which proteins they interact with and thus their down-stream effects. Further work, in which lipoproteins are separated by size and undergo the same detailed molecular profiling as the overall plasma, is required to answer this question formally.
Our data raise questions about the nature of GDM-related shifts in lipid abundance, the role of those lipids in vivo and invite comparison with shifts in lipid metabolism in the CBGS cohort (not selected for BMI). In the CBGS cohort five candidate biomarkers changed in abundance before a diagnosis of GDM,29 viz. TG(51:1), TG(48:1), PC(32:1), PC-O(40:3), PC-O(40:4). Only the change in PC-O(40:4) is common to both studies, with the same trend of a lower abundance in the GDM groups. However, the isoform of a triglyceride found in the present study (48:0) is similar to (48:1), and the (51:1) found in CBGS may contain similar fatty acid residues to those in the isoforms (50:1) and (51:5) found in the present report. This suggests that there is some similarity between the shifts in lipid metabolism in advance of GDM between obese-only and mixed-BMI groups, and thus that there may be changes in lipid metabolism distinct from those associated with obesity.
Molecular profiling in two studies of T2DM has shown associations with the abundance of lipid and triglyceride species.81,82 Both demonstrate that isoforms of PC(38:6), very similar to the PC(38:5) identified in the present study, are more abundant in advance of T2DM.81,82 Other commonalities include the pattern of shifts in the abundance of triglycerides i.e. slightly larger, less unsaturated TGs are more abundant where slightly lighter, polyunsaturated TGs are less abundant in individuals who develop either condition.81 Since similar pathways appear dysregulated in T2DM and GDM, these relationships may underpin the association between GDM and the risk of T2DM post partum.7,83,84
The panels of lipids found to be associated with GDM in the present study, in the CBGS study,29 and in T2DM,81,82 are all notable for the considerable breadth of molecular species. This raises the question of why different lipids of contrasting molecular classes showed significant associations, where similar ones did not. One possible answer is that GDM has a range of aetiologies that emerge from one or more of several mechanisms. If such sub-groups are identified, risk stratification by sub-group according to risk may be desirable. For example, some participants had a high blood glucose concentration for about an hour after ingesting the sugar, others both a high 1 h and also high glucose 2 h after ingestion, and a third group had high blood glucose concentration in all three measurements and a fourth group with a high fasting blood glucose and a lower 1 h and 2 h. It seems unlikely that a short-term high glucose concentration in the circulation is the result of profound insulin insensitivity, where a high blood glucose concentration over a long period of time may indicate this. These responses to the OGTT may therefore reflect different pathophysiological pathways to hyperglycaemia in pregnancy. Recent work on possible genetic determinants of GDM found that both insulin sensitivity and secretion were associated with higher genetic risk scores,85 suggesting that such mechanisms also have a genetic basis.
The shifts in lipid metabolism observed in the present study and others may therefore be the result of the sum of up- and down-stream effects of changes to insulin secretion and sensitivity, such as altered abundance of fatty acids. A simple diagnosis of GDM using an OGTT may therefore describe a collection of aetiologies driven by one or more of related mechanisms. Grouping participants from a mechanistic perspective may therefore offer a different insight into the relationship with lipid metabolism. A finer understanding of the affected species in individuals who later develop GDM will indicate more clearly which pathways (e.g. biosynthesis of PC) are affected and which mechanisms (fatty acid distribution, oxidation of TGs) are altered. This approach may also be useful in the development of personalised treatment of GDM.
Changes in abundance of several lipids in the second trimester found to be related to later GDM development have also been identified as candidate biomarkers of higher birth weight in healthy pregnancies.86 This includes SM(32:1) and PC(35:2) (isobaric with PE(38:2)), both of which are less abundant in the circulation of women who deliver macrosomic babies from non-GDM pregnancies.86 Several isoforms of PC that are similar to PC(40:7) and (40:10) are similarly less abundant. However, it remains to be determined what, if any, are the functional roles of any of the predictive candidate biomarkers. Further work might focus on establishing the roles and distribution of the individual components identified here, but also how they emerge and are degraded.
Conclusions
This study tested the hypothesis that phospholipid and triglyceride metabolism were altered by the beginning of the second trimester in obese pregnant women who later developed GDM. Differences previously unknown in the abundances of lipid, sterol and triglyceride species were identified at least 10 weeks before diagnosis. This produced evidence that several aspects of triglyceride metabolism were modulated, including oxidation, and was consistent with previous work on the lipoprotein profile.30 The modulations observed invite studies into the emergence of changes in lipid and triglyceride metabolism as either a driver of hyperglycaemia or as an associated but independent metabolic effect. The early identification of altered lipid metabolism, before standard diagnosis through hyperglycaemia associated with GDM, implies that lipid species are involved in the aetiology of GDM, or in the same pathophysiological pathway.
Supplementary Material
† Electronic supplementary information (ESI) available. See DOI: 10.1039/c9mo00117d
Acknowledgements
The authors would like to thank Dr C. Gill for helpful conversations, laboratory assistance and sample transport and Dr S. G. Snowden for assistance with data processing and batch correction. The authors also gratefully acknowledge funding from the BBSRC (BB/M027252/1 for SF, BB/M027252/2 for SF and AK), NIHR (NIHR146281 for AK and BJJ), Diabetes UK (14/0004849 for SLW and 17/0005712 for CLM) and CAPES (BEX 9571/13-2) for MCV and MRC (MC_UU_12012/4 for SEO). The UPBEAT study received funding from the National Institute of Health Research (RP-PG-0407-10452), Medical Research Council UK (MR/L002477/1), Chief Scientist Office, Scottish Government Health Directorates (Edinburgh) (CZB/A/680), Biomedical Research Centre at Guys & St Thomas NHS Foundation Trust & King’s College London and the NIHR Bristol Biomedical Research Centre, Tommy’s Charity, UK (SC039280).
Footnotes
Samuel Furse: 0000-0003-4267-2051
Sara L. White: 0000-0001-7979-0508
Claire L. Meek: 0000-0002-4176-8329
Benjamin Jenkins: 0000-0003-0038-9709
Clive J. Petry: 0000-0002-6642-9825
Matias C. Vieira: 0000-0002-8076-4275
Susan E. Ozanne: 0000-0001-8753-5144
David B. Dunger: 0000-0002-2566-9304
Albert Koulman: 0000-0001-9998-051X
Author contributions
L. P. was PI of the UPBEAT RCT. S. F., S. L. W., L. P., D. B. D., C. L. M., M. C. V., S. E. O. and A. K. conceived and designed the study. S. F., S. L. W., C. J. P., L. P. and A. K. wrote the original analysis plan. S. F. extracted all samples, collected and processed all direct infusion mass spectrometry data, produced the figures and wrote the first draft of the manuscript. S. F. and C. L. M. conducted all statistical calculations. B. J. J. collected liquid chromatography-mass spectrometry data on a subset of samples, processed and prepared it for publication. S. F., S. L. W., C. L. M., C. J. P., M. C. V., S. E. O., L. P. and A. K. were involved in interpreting data. S. F., S. L. W., C. L. M., C. J. P., M. C. V., S. E. O., D. B. D., L. P. and A. K. read, commented on and provided feedback for correcting the manuscript. All authors approved the final version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Farrar D, Simmonds M, Griffin S, Duarte A, Lawlor DA, Sculpher MJ, Fairley L, Golder S, Tuffnell DJ, Bland M, Dunne FP, et al. Health technology assessment. 2016;20(86):1–348. doi: 10.3310/hta20860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu SY, Callaghan WM, Kim SY, Schmid CH, Lau J, England LJ, Dietz PM. Diabetes Care. 2007;30:2070–2076. doi: 10.2337/dc06-2559a. [DOI] [PubMed] [Google Scholar]
- 3.Kim SY, England L, Wilson HG, Bish C, Satten GA, Dietz P. Am J Public Health. 2010;100:1047–1052. doi: 10.2105/AJPH.2009.172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Obesity and overweight. http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
- 5.ONS. Statistics on Obesity, Physical Activity and Diet. https://files.digital.nhs.uk/publication/0/0/obes-phys-acti-diet-eng-2018-rep.pdf.
- 6.Kim C, Newton KM, Knopp RH. A Systematic Review. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 7.Noctor E, Dunne FP. World J Diabetes. 2015;6:234–244. doi: 10.4239/wjd.v6.i2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellamy L, Casas J-P, Hingorani AD, Williams D. Lancet. 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 9.Robitaille J, Grant AM. Genet Med. 2008;10:240. doi: 10.1097/GIM.0b013e31816b8710. [DOI] [PubMed] [Google Scholar]
- 10.Damm P. Int J Gynecol Obstet. 2009;104:S25–S26. doi: 10.1016/j.ijgo.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 11.Vohr BR, McGarvey ST, Tucker R. Diabetes Care. 1999;22:1284–1291. doi: 10.2337/diacare.22.8.1284. [DOI] [PubMed] [Google Scholar]
- 12.Catalano PM, Thomas A, Huston-Presley L, Amini SB. Am J Obstet Gynecol. 2003;189:1698–1704. doi: 10.1016/s0002-9378(03)00828-7. [DOI] [PubMed] [Google Scholar]
- 13.Plagemann A, Harder T, Kohlhoff R, Rohde W, Dörner G. Int J Obes. 1997;21:451. doi: 10.1038/sj.ijo.0800429. [DOI] [PubMed] [Google Scholar]
- 14.Martorell R, Stein AD, Schroeder DG. Journal of Nutrition. 2001;131:874S–880S. doi: 10.1093/jn/131.3.874S. [DOI] [PubMed] [Google Scholar]
- 15.Gillman MW, Rifas-Shiman S, Berkey CS, Field AE, Colditz GA. Pediatrics. 2003;111:e221–e226. doi: 10.1542/peds.111.3.e221. [DOI] [PubMed] [Google Scholar]
- 16.Vohr BR, Boney CM. J Matern-Fetal Neonat Med. 2008;21:149–157. doi: 10.1080/14767050801929430. [DOI] [PubMed] [Google Scholar]
- 17.Duque-Guimarães DE, Ozanne SE. Trends Endocrinol Metab. 2013;24:525–535. doi: 10.1016/j.tem.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 18.IADPSG. Metzger BE. Diabetes Care. 2010;33:676–682. doi: 10.2337/dc09-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NICE. Diabetes in pregnancy: management of diabetes and its complications from preconception to the postnatal period. 2015 [PubMed] [Google Scholar]
- 20.Zondag L. KNOV Factsheet Diabetes Gravidarum. 2018 [Google Scholar]
- 21.Deutsche Diabetes Gesellschaft. Deutsche Gesellschaft für Gynäkologie und Geburtshilfe: S3-Leitlinie Gestationsdiabetes mellitus (GDM), Diagnostik, Therapie und Nachsorge. 2018 [Google Scholar]
- 22.Seabra G, Saunders C, de Carvalho Padilha P, Zajdenverg L, da Silva LBG, de Souza Santos MMA. Diabetol Metab Syndr. 2015;7:17. doi: 10.1186/s13098-015-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riskin-Mashiah S, Younes G, Damti A, Auslender R. Diabetes Care. 2009;32:1639–1643. doi: 10.2337/dc09-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Group HSCR. N Engl J Med. 2008;358:1991–2002. doi: 10.1056/NEJMoa0707943. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigo N, Glastras SJ. J Clin Med. 2018;7:120. doi: 10.3390/jcm7060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamain-de Ruiter M, Kwee A, Naaktgeboren CA, Franx A, Moons KGM, Koster MPH. Diagnostic and Prognostic Research. 2017;1:3. doi: 10.1186/s41512-016-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosson E, Carbillon L, Valensi P. J Diabetes Res. 2017;2017:12. doi: 10.1155/2017/8921712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furse S, Snowden SG, Laurentya O, Prentice P, Ong K, Hughes IA, Acerini CL, Dunger DB, Koulman A. Sci Rep. 2019 doi: 10.1038/s41598-019-50693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu L, Koulman A, Petry CJ, Jenkins B, Matthews L, Hughes IA, Acerini CL, Ong KK, Dunger DB. Diabetes Care. 2016;39:2232. doi: 10.2337/dc16-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White SL, Pasupathy D, Sattar N, Nelson SM, Lawlor DA, Briley AL, Seed PT, Welsh P, Poston L. Diabetologia. 2017;60:1903–1912. doi: 10.1007/s00125-017-4380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira MC, Begum S, Seed PT, Badran D, Briley AL, Gill C, Godfrey KM, Lawlor DA, Nelson SM, Patel N, Sattar N, et al. Pregnancy Hypertension. 2018;13:267–272. doi: 10.1016/j.preghy.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White SL, Lawlor DA, Briley AL, Godfrey KM, Nelson SM, Oteng-Ntim E, Robson SC, Sattar N, Seed PT, Vieira MC, Welsh P, et al. PLoS One. 2016;11:e0167846. doi: 10.1371/journal.pone.0167846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharjee A, Prentice P, Acerini C, Smith J, Hughes IA, Ong K, Griffin JL, Dunger D, Koulman A. Metabolomics. 2017;13:25. doi: 10.1007/s11306-017-1166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koulman A, Prentice P, Wong MCY, Matthews L, Bond NJ, Eiden M, Griffin JL, Dunger DB. Metabolomics. 2014;10:1018–1025. doi: 10.1007/s11306-014-0628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prentice P, Koulman A, Matthews L, Acerini CL, Ong KK, Dunger DB. J Pediatr. 2015;166:276–281. doi: 10.1016/j.jpeds.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furse S, Billing G, Snowden SG, Smith J, Goldberg G, Koulman A. Metabolomics. 2019 doi: 10.1007/s11306-019-1589-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harshfield EL, Koulman A, Ziemek D, Marney L, Fauman EB, Paul DS, Stacey D, Rasheed A, Lee J-J, Shah N, Jabeen S, et al. J Proteome Res. 2019;18:2397. doi: 10.1021/acs.jproteome.8b00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubois V, Viljoen A, Laencina L, Le Moigne V, Bernut A, Dubar F, Blaise M, Gaillard J-L, Guérardel Y, Kremer L, Herrmann J-L, et al. Proc Natl Acad Sci U S A. 2018;115:E10147–E10156. doi: 10.1073/pnas.1812984115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graessler J, Schwudke D, Schwarz PEH, Herzog R, Shevchenko A, Bornstein SR. PLoS One. 2009;4:e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harshfield EL, Koulman A, Ziemek D, Marney L, Fauman EB, Paul DS, Stacey D, Rasheed A, Lee J-J, Shah N, Jabeen S, et al. J Proteome Res. 2019;18:2397–2410. doi: 10.1021/acs.jproteome.8b00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virtue S, Petkevicius K, Moreno-Navarrete JM, Jenkins B, Hart D, Dale M, Koulman A, Fernández-Real JM, Vidal-Puig A. Cell Reports. 2018;24:2005–2012. e2007. doi: 10.1016/j.celrep.2018.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders F, Acharjee A, Walker C, Marney L, Roberts L, Imamura F, Jenkins B, Case J, Ray S, Virtue S, Vidal-Puig A, et al. Genome Biol. 2018;19 doi: 10.1186/s13059-018-1439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cassim AM, Gouguet P, Gronnier J, Laurent N, Germain V, Grison M, Boutté Y, Gerbeau-Pissot P, Simon-Plas F, Mongrand S. Prog Lipid Res. 2019;73:1. doi: 10.1016/j.plipres.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Pritchard JKA, Schwarz SM, Medow MS, Stemerman MB. Am J Physiol. 1991;260:C43–C49. doi: 10.1152/ajpcell.1991.260.1.C43. [DOI] [PubMed] [Google Scholar]
- 46.Owen JS, Bruckdorfer KR, Day RC, McIntyre N. J Lipid Res. 1982;23:124–132. [PubMed] [Google Scholar]
- 47.Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Seddon JM, Templer RH. Philos Trans R Soc, A. 1993;344:377–401. [Google Scholar]
- 49.Seddon JM, Templer RH. In: The Handbook of Biological Physics. Lipowsky R, Sackman E, editors. I Elsevier Science; 1995. [Google Scholar]
- 50.Cullis PR, De Kruijff B. Biochim Biophys Acta, Rev Biomembr. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 51.Seddon JM. Biochim Biophys Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-t. [DOI] [PubMed] [Google Scholar]
- 52.Shaw KP, Brooks NJ, Clarke JA, Ces O, Seddon JM, Law RV. Soft Matter. 2012;8:1070–1078. [Google Scholar]
- 53.Furse S, Mak L, Tate EW, Templer RH, Ces O, Woscholski R, Gaffney PRJ. Org Biomol Chem. 2015;13:2001–2011. doi: 10.1039/c4ob02258k. [DOI] [PubMed] [Google Scholar]
- 54.Lee AG. Biochim Biophys Acta, Biomembr. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Phillips R, Ursell T, Wiggins P, Sens P. Nature. 2009;459:379. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perona JS. Biochim Biophys Acta, Biomembr. 2017;1859:1690–1703. doi: 10.1016/j.bbamem.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 57.Turner N, Cooney GJ, Kraegen EW, Bruce CR. J Endocrinol. 2014;220:T61. doi: 10.1530/JOE-13-0397. [DOI] [PubMed] [Google Scholar]
- 58.Alvarez JJ, Montelongo A, Iglesias A, Lasunción MA, Herrera E. J Lipid Res. 1996;37:299–308. [PubMed] [Google Scholar]
- 59.Herrera E, Lasunción MA, Gomez-Coronado D, Aranda P, López-Luna P, Maier I. Am J Obstet Gynecol. 1988;158:1575–1583. doi: 10.1016/0002-9378(88)90193-7. [DOI] [PubMed] [Google Scholar]
- 60.Rebuffé-Scrive M, Enk L, Crona N, Lönnroth P, Abrahamsson L, Smith U, Björntorp P. J Clin Invest. 1985;75:1973–1976. doi: 10.1172/JCI111914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gallo LA, Barrett HL, Dekker Nitert M. Placenta. 2017;54:59–67. doi: 10.1016/j.placenta.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Qiao L, Guo Z, Bosco C, Guidotti S, Wang Y, Wang M, Parast M, Schaack J, Hay WW, Moore TR, Shao J. Diabetes. 2015;64:3111–3120. doi: 10.2337/db14-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houde AA, St-Pierre J, Hivert MF, Baillargeon JP, Perron P, Gaudet D, Brisson D, Bouchard L. J Dev Origins Health Dis. 2014;5:132–141. doi: 10.1017/S2040174414000038. [DOI] [PubMed] [Google Scholar]
- 64.Ortega-Senovilla H, Schaefer-Graf U, Meitzner K, Abou-Dakn M, Herrera E. J Clin Endocrinol Metab. 2013;98:3430–3437. doi: 10.1210/jc.2013-1614. [DOI] [PubMed] [Google Scholar]
- 65.Gauster M, Hiden U, van Poppel M, Frank S, Wadsack C, Hauguel-deMouzon S, Desoye G. Diabetes. 2011;60:2457–2464. doi: 10.2337/db10-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrett HL, Kubala MH, Scholz Romero K, Denny KJ, Woodruff TM, McIntyre HD, Callaway LK, Nitert MD. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104826. e104826–e104826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heerwagen MJR, Gumina DL, Hernandez TL, Van Pelt RE, Kramer AW, Janssen RC, Jensen DR, Powell TL, Friedman JE, Winn VD, Barbour LA. Placenta. 2018;64:53–60. doi: 10.1016/j.placenta.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Devarshi PP, Grant RW, Ikonte CJ, Hazels Mitmesser S. Nutrients. 2019;11:1107. doi: 10.3390/nu11051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segura MT, Demmelmair H, Krauss-Etschmann S, Nathan P, Dehmel S, Padilla MC, Rueda R, Koletzko B, Campoy C. Placenta. 2017;57:144–151. doi: 10.1016/j.placenta.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 70.Pagán A, Prieto-Sánchez JE, Blanco-Carnero A, Gil-Sánchez JJ, Parrilla H, Demmelmair H, Koletzko B, Larqué E. Am J Physiol. 2013;305:E826–E833. doi: 10.1152/ajpendo.00291.2013. [DOI] [PubMed] [Google Scholar]
- 71.Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, Gu C. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen LN, Ma D, Shui G, Wong P, Cazenave-Gassiot A, Zhang X, Wenk MR, Goh ELK, Silver DL. Nature. 2014;509:503–506. doi: 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- 73.Prieto-Sánchez MT, Ruiz-Palacios M, Blanco-Carnero JE, Pagan A, Hellmuth C, Uhl O, Peissner W, Ruiz-Alcaraz AJ, Parrilla JJ, Koletzko B, Larqué E. Clin Nutr. 2017;36:513–521. doi: 10.1016/j.clnu.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 74.Imamura F, Fretts A, Marklund M, Ardisson Korat AV, Yang W-S, Lankinen M, Qureshi W, Helmer C, Chen T-A, Wong K, Bassett JK et al. Mozaffarian and Fatty Acids Outcomes Research Consortium. PLoS Med. 2018;15:e1002670. doi: 10.1371/journal.pmed.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riserus U, Marklund M. Curr Opin Lipidol. 2017;28:46–51. doi: 10.1097/MOL.0000000000000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jenkins B, West J, Koulman A. Molecules. 2015;20:2425. doi: 10.3390/molecules20022425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mozaffarian D, Cao H, King IB, Lemaitre RN, Song X, Siscovick DS, Hotamisligil GS. Am J Clin Nutr. 2010;92:1350–1358. doi: 10.3945/ajcn.110.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni Y, Zhao L, Yu H, Ma X, Bao Y, Rajani C, Loo LWM, Shvetsov YB, Yu H, Chen T, Zhang Y, et al. EBioMedicine. 2015;2:1513–1522. doi: 10.1016/j.ebiom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gianmattia del G, Carla F, Raffaele M, Dimitri P, Roux CWL, Federica del G, Limongelli P, Salvatore T, Ludovico D, Annibale Alessandro P. J Diabetes Metab. 2015;6 1000582. [Google Scholar]
- 80.Lemaitre RN, King IB, Sotoodehnia N, Knopp RH, Mozaffarian D, McKnight B, Rea TD, Rice K, Friedlander Y, Lumley TS, Raghunathan TE, et al. Metabolism. 2010;59:1029–1034. doi: 10.1016/j.metabol.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O’Donnell CJ, Carr SA. J Clin Invest. 2011;121:1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meikle PJ, Wong G, Barlow CK, Weir JM, Greeve MA, MacIntosh GL, Almasy L, Comuzzie AG, Mahaney MC, Kowalczyk A, Haviv I, et al. PLoS One. 2013;8:e74341. doi: 10.1371/journal.pone.0074341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herath H, Herath R, Wickremasinghe R. PLoS One. 2017;12:e0179647. doi: 10.1371/journal.pone.0179647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim C, Newton KM, Knopp RH. Diabetes Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 85.Powe CE, Nodzenski M, Talbot O, Allard C, Briggs C, Leya MV, Perron P, Bouchard L, Florez JC, Scholtens DM, Lowe WL, et al. Diabetes. 2018;67:2703–2709. doi: 10.2337/db18-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ciborowski M, Zbucka-Kretowska M, Bomba-Opon D, Wielgos M, Brawura-Biskupski-Samaha R, Pierzynski P, Szmitkowski M, Wolczynski S, Lipinska D, Citko A, Bauer W, et al. Prenatal Diagn. 2014;34:870–877. doi: 10.1002/pd.4386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.