Abstract
Cytokinesis must be regulated in time and space in order to preserve genome integrity during cell proliferation and to allow daughter cells to adopt distinct fates and geometries during differentiation. The fission yeast Schizosaccharomyces pombe (S. pombe) has been a popular model organism for understanding spatiotemporal regulation of cytokinesis in a symmetrically dividing cell. Recent work on another member of the same genus, S. japonicus, suggests that S. pombe may have evolved an unusual division site placement mechanism based on a recently duplicated anillin paralog. Here we discuss an extraordinary evolutionary plasticity of cytokinesis within the fission yeast clade and argue that the comparative cell biology approach may provide functional insights beyond those afforded by scrutinizing individual model species.
Introduction
Cell division, or cytokinesis, is the final event in the cell cycle leading to individuation of daughter cells. Cytokinetic machinery physically divides the cytoplasm after mitotic chromosome segregation. In fungi and animal cells, this is achieved by constriction of the actomyosin ring, a dynamic structure assembled at the cellular cortex from filamentous actin (F-actin), myosin II complex and a number of associated proteins. Actomyosin dynamics are driving plasma membrane remodeling and in fungi, deposition of a dividing cell wall known as the septum. Whereas the bare bones of the actomyosin division machinery appear well conserved in the animal-fungal clade, different organisms and even different cells within the same organism exhibit a staggering variety of approaches to regulating cytokinesis in time and space [1]. How spatiotemporal dynamics of cytokinesis are rewired to produce novel behaviors and what underlies the emergence of new modes of cytokinesis regulation in evolution remains largely unknown.
The fission yeast S. pombe is a rod-shaped cell that assembles medial actomyosin rings and divides orthogonally to the long cell axis into two equally sized daughters. Several recent reviews provide extensive discussion on structural organization of the division ring and cell cycle-dependent regulation of its assembly and constriction [2–8]. Here we focus on the aspects of cytokinesis that diverged between S. pombe and its close relative, Schisozaccharomyces japonicus (S. japonicus) and discuss possible origins and functional implications of this evolutionary plasticity.
Mitosis and cytokinesis regulation diverged in fission yeasts
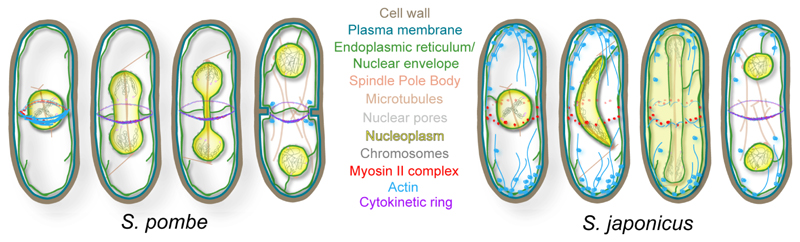
Both S. pombe and S. japonicus belong to the genus of fission yeasts, a small monophyletic clade at the base of the Ascomycota phylogenetic tree [9]. S. japonicus forms an early diverging branch with respect to the rest of Schizosaccharomyces species. Similar to S. pombe, the S. japonicus genome is encoded on three chromosomes, with relatively conserved gene order, gene content and gene structure [10]. Yet, the two species exhibit remarkable differences in physiology. In contrast to “closed” nuclear division in S. pombe (and, for that matter, the budding yeast) where the nuclear envelope (NE) remains intact throughout mitosis, S. japonicus breaks the NE during anaphase and reforms it around the segregated genomes at mitotic exit (Fig. 1) [11, 12]. This divergence has been linked to a simple scaling argument: since the nucleus maintains constant volume as it goes through closed division, cells need to expand the NE to form two spherical nuclei from one. S. japonicus does not expand the nuclear membrane during mitosis, unlike S. pombe and therefore, must break it to allow chromosome partitioning [11]. Mitotic NE breakdown in S. japonicus may have facilitated the emergence of a tethering mechanism that links the anaphase chromatin to the NE and promotes equal partitioning of the nuclear membrane and the nuclear pore complexes to the daughter nuclei [13]. Of note, animal cells break the NE during mitosis and use a variety of strategies to coordinate NE reformation with chromosome segregation.
Figure 1. Diagrams summarizing the modes of mitosis and cytokinesis in S. pombe (left) and S. japonicus (right).
Pictorial legend is included.
Importantly, the two species differ in the timing of actomyosin ring assembly and hence, have somewhat different requirements for regulating cytokinesis. S. pombe assembles a centrally positioned ring in metaphase, when chromosomes are not yet segregated (reviewed in [4]). Thus, the mitotic S. pombe cell must keep the ring from constricting until after anaphase to prevent physical damage to the genome. To this end, S. pombe controls ring constriction and septum deposition through a signaling system termed the septation initiation network (SIN). Full activation of SIN and proper localization of its downstream components coincide with the decline in mitotic CDK activity at the end of mitosis. Forced inactivation of CDK in metaphase cells is sufficient to induce cytokinesis, suggesting that this kinase inhibits SIN [14, 15]. On the other hand, CDK functions together with the positive regulator of SIN, the Polo kinase, to create permissive environment for SIN activation at the spindle pole bodies (SPBs) already during metaphase [16]. Thus, S. pombe uses an interlocked network of positive and negative cues to ensure that SIN activity peaks and cytokinesis occurs only after successful chromosome partitioning [14, 17–19]; for review see [3].
Yet, most cell types including animal cells, budding yeast and even S. japonicus postpone assembly of the division apparatus until chromosome segregation is complete [20–22]. Thus, the temporal mismatch between ring assembly and cytokinesis in S. pombe is rather uncommon and it may rely on species-specific mechanisms regulating ring formation and constriction. Most S. pombe cells can survive when the early metaphase pathway is disabled by mutations in the anillin-like protein Mid1, arguing that the species retained a form of ancestral post-mitotic assembly of the division apparatus [23]. Such cells exhibit grossly mispositioned and non-orthogonal rings. Curiously, inhibition of septum deposition allows S. pombe mid1 mutants sufficient time to correct ring orientation [24], suggesting that activation of the septum assembly machinery in S. pombe occurs before rings can be assembled via the post-mitotic pathway. Indeed, while mitotic events such as spindle assembly and chromosome segregation occur with comparable kinetics in S. pombe and S. japonicus cells, S. pombe deposits the division septum considerably earlier [21]. It would be of interest to assess the dynamics of SIN activation in S. japonicus – one of the predictions could be that SIN gets fully activated later in the cell cycle, as compared to S. pombe. A precocious, metaphase ring assembly may have arisen in S. pombe lineage to compensate for deregulation of the conventional pathway coordinating membrane and cell wall remodeling with actomyosin contractility driving ring compaction. Alternatively, early Mid1-dependent ring formation may have had an unknown contextual advantage at some point in S. pombe evolution.
Mechanisms of myosin II recruitment to the cortex in the two fission yeasts
Many ring components in both fission yeasts are assembled into large supramolecular complexes, or precursor “nodes”, at the equatorial cortex. Yet, the timing and the extent of cortical recruitment of individual node subunits may differ between the species. The list of proteins associated with these structures in S. pombe has been ever increasing. In fact, the S. pombe cortex appears occupied by distinct types of nodes that coexist at the plasma membrane during interphase before eventually coalescing into larger structures following mitotic commitment [6, 25].
Perhaps most relevant to this review are the cortical nodes centered on Mid1, a member of the anillin protein family. The S. pombe Mid1 was shown to harbor multiple structural determinants regulating its subcellular distribution and driving membrane binding and interactions with several ring components [26–30]; see review in [7]. Mid1 undergoes nucleocytoplasmic shuttling during interphase, showing predominantly nuclear steady-state distribution with only a minor fraction localizing to the cortical nodes overlying the nucleus where it co-localizes with the SAD-like kinase Cdr2 [31]. Polo kinase-dependent phosphorylation of Mid1 at the onset of mitosis stimulates its efflux from the nucleus and nearly quantitative relocalization to the equatorial cortex [32, 33].
Mid1 remains restricted at this position as a result of homodimerization stimulating high-affinity binding to specific phospholipids [30] and patterning of the plasma membrane by the underlying peripheral ER [34, 35]. Mid1 then recruits essential components of the cytokinesis machinery including the myosin II complex, the IQGAP protein Rng2, an actin nucleator Cdc12 and an F-BAR domain protein Cdc15 (reviewed in [4]). These proteins form a tight band of nodes that is rapidly reorganized into a single ring structure prior to anaphase [36]. Thus, S. pombe assembles actomyosin rings in an instructive, nucleus-guided manner based on a spatial cue provided by Mid1 [37–39]; reviewed in [7]. In addition to its function in specifying the division plane, the S. pombe Mid1 plays an essential structural role in cortical scaffolding of the actomyosin components during early mitosis.
A number of proteins including Blt1, the RhoGEF Gef2 and the kinesin Klp8 occupy cortical sites distinct from the Mid1/Cdr2 nodes during interphase. However, the overlap between the Blt1 nodes and the Mid1/Cdr2 nodes increases as cells approach the G2/M boundary [25, 40]. Expanding the network, Gef2 binds yet another protein, Nod1 [41, 42], which in turn interacts with Cdc15 [41]. Simultaneous disruption of Mid1 and Blt1 functions in S. pombe causes severe defects in myosin ring anchorage [40]. Thus it appears that S. pombe cells utilize multiple inter-connected protein components to ensure the fidelity of actomyosin ring assembly and constriction.
Given that S. japonicus does not assemble rings until the exit from mitosis, it is perhaps not surprising that mid1Δ or in fact, mid1Δblt1Δ S. japonicus cells divide normally ([21] and our unpublished data). It turns out that S. japonicus uses Mid1 for anchoring myosin II at the equatorial cortex during interphase rather than mitosis. The S. japonicus Mid1 has a 17-amino acid insertion that disrupts the positively charged region known to function as the nuclear localization signal in S. pombe, and thus, remains cortical throughout the cell cycle. The absence of Mid1 nucleocytoplasmic shuttling breaks the functional link between the nucleus and division site positioning and presumably allows the protein to form higher order complexes with a number of core ring components throughout most of the cell cycle. Interestingly, ring constriction rate in mid1Δ S. japonicus cells is considerably faster than in the wild type, suggesting that Mid1 could function as an actomyosin crosslinker [21].
In stark contrast to S. pombe where myosin remains cytosolic for the most of the cell cycle, S. japonicus cells recruit myosin, IQGAP and other associated proteins to the Mid1-based nodes at the equatorial cortex early in G2 [21]. Cortical association of the myosin II complex with Mid1 in S. pombe requires mitotic dephosphorylation of the myosin II heavy chain at the S1444 position and the myosin mutant refractory to phosphorylation accumulates at the medial cortex already in early G2 [43]. Suggesting that this level of regulation may be absent in S. japonicus, the Myo2 protein from this species is shorter than its orthologs from the other fission yeasts and lacks the C-terminal sequence comprising the putative phosphorylation site.
Interestingly, the budding yeast S. cerevisiae, evolutionarily distant from fission yeast [9], also localizes the myosin II complex to the future division site early in the cell cycle, shortly before bud emergence [44]. It remains to be seen if there are any interphase functions associated with the cortical band of myosin nodes – an interesting idea is that it may function in polarized growth by spooling actin cables or participating in vesicular trafficking [45]. It is not clear at this stage if the myosin II is active in interphase S. japonicus, but intriguingly, it appears to function as a bud neck-associated motor at this stage of the cell cycle in S. cerevisiae [46]. S. japonicus is a truly dimorphic species that readily undergoes a yeast-to-hyphal transition [47, 48]. Perhaps, such myosin relay mechanism could be advantageous in maintaining polarity in the context of a much larger hyphal cell. Importantly, whereas in S. japonicus the interphase cortical band of myosin nodes is normally dispensable for ring assembly and positioning, it appears to reinforce both of these processes upon insults to intracellular organization such as nuclear displacement [21].
Evolution of anillins in the fission yeast clade
Unlike fission yeasts that encode for two anillin-like proteins called Mid1 and Mid2, most fungi have a single anillin that is commonly termed Bud4, after the S. cerevisiae ortholog. At least in cases where it has been studied, Bud4 functions in scaffolding cytokinesis machinery including septins at the division site [49–51]. Interestingly, Mid2, a protein that functions in cell separation through its interactions with septins, clusters with the Bud4 clade, whereas Mid1 forms a clearly separate branch, unique for fission yeasts [21]. Thus, it appears that Mid1/Mid2 pair is a product of the fission yeast-specific duplication of an ancestral anillin. Notably, Mid1 proteins exhibit a signature of the rapidly evolving paralogs within the duplicated pair [52]. The two anillin genes in both S. pombe and S. japonicus show distinct patterns of gene expression, with Mid2 peaking at mitosis and Mid1 exhibiting constant levels of expression throughout the cell cycle [21, 53]. Thus, initial divergence in gene expression following the duplication event might have contributed to subfunctionalization of the two paralogs [54], with Mid1 specializing in scaffolding myosin II, IQGAP and other myosin-associated proteins, and Mid2 in organizing septins. Specific regulation of functions presumably diverged further following speciation. Interestingly, the S. pombe Mid1 has been linked to regulation of gene expression [55], a nuclear function that requires nucleocytoplasmic trafficking of the protein and thus may have facilitated emergence of a nucleus-guided, instructive division site positioning reliant on the myosin scaffold Mid1. On the contrary, in S. japonicus Mid1 plays a structural role in scaffolding myosin and the associated proteins at the cellular cortex, a function it shares with anillins in other fungi and metazoans [49, 56–63].
The myosin anchor Cdc15 ensures medial division in S. japonicus
The F-BAR protein Cdc15 is amongst the essential ring components recruited to the Mid1-centered nodes in S. japonicus. Some Cdc15 co-localizes with Mid1 already in interphase but the overlap between the two proteins increases at mitotic entry [21]. It is at some point late in mitosis when the myosin anchorage function is fully transferred to Cdc15. In cells where Cdc15 is inactivated by mutations, mitotic myosin recruitment is severely diminished and myosin II complex is not retained at the cortex after exit from mitosis. This is not dissimilar to S. pombe, where Cdc15 plays an essential role in anchoring the actomyosin ring during constriction [64, 65]. However, unlike its cousin, S. japonicus can recruit Cdc15 to the equatorial cortex without Mid1 and it is this recruitment that allows mid1Δ cells to form well-positioned rings and undergo seemingly normal cytokinesis. When both Mid1 and Cdc15 functions are impaired, actomyosin cables are formed with normal timing but are not anchored at the equatorial cortex [21].
Membrane association of Cdc15 is mediated through the N-terminally located F-BAR domain. F-BAR (also known as extended Fes/CIP4 homology) domains bind phospholipids and may promote membrane curvature through dimerization and subsequent oligomerization [66]. During interphase in both fission yeasts, Cdc15 is enriched at the cell tips, although in S. japonicus the protein also exhibits some localization to the medial cortex [21]. In cycling S. pombe, Cdc15 exists in multiple phosphoisoforms, yet it is largely dephosphorylated in mitosis [64, 66]. Dephosphorylation increases Cdc15 affinity to membranes and also promotes binding to its protein partners, such as Rng2 and the formin Cdc12. Mutants of Cdc15 that are refractory to phosphorylation localize to the medial cortex and recruit the myosin II complex, Cdc12 and Rng2 already in interphase [66]. Curiously, this localization does not depend on Mid1, suggesting that similarly to S. japonicus, hypo-phosphorylated Cdc15 in S. pombe can find the cell middle using a Mid1- and presumably, nucleus-independent mechanism [66].
How does it work? It appears that phosphoregulation of Cdc15 by the DYRK (dual specificity tyrosine regulated kinase) kinase Pom1 could be a part of the answer. Pom1 is enriched at the cell tips, forming cortical concentration gradients along the long axis of the cell. In S. pombe, these kinase gradients regulate mitotic entry through inhibition of Cdr2 [67, 68] and participate in division plane positioning by restricting the cortical Mid1/Cdr2 nodes at the cell center [28, 69, 70]. Fission yeast cells lacking Pom1 grow in a monopolar manner [21, 71]. In S. pombe, Pom1 controls cortical node assembly through phosphorylation of the basic region within Cdr2 that modulates membrane association in cooperation with the lipid-binding KA-1 domain. It also prevents formation of the cortical Mid1/Cdr2 nodes at the cell poles [69].
In interphase S. japonicus cells lacking Pom1, all tested Mid1 node components and Cdc15 mislocalize away from the equator, towards the non-growing tip. Interestingly, Pom1 also controls Cdc15 localization during mitosis in a Mid1-independent manner, and thus, defines the cortical area for actomyosin ring assembly [21]. Of note, Cdc15 was identified as a Pom1 substrate in phosphoproteomics studies in S. pombe [72] and given that Cdc15 localization is extensively regulated by Pom1 in S. japonicus, it is likely that this functional interaction is evolutionarily conserved. Interestingly, while the lack of Pom1 leads to Cdc15 relocalization to the growing tip in S. pombe, the opposite is true for S. japonicus where Pom1 deficiency drives Cdc15 accumulation at the non-growing cell tip [21]. What determines this disparate localization is unclear at the moment.
In addition to the positional effect of Pom1 on determining the cell division plane, actomyosin rings assembled in pom1Δ S. japonicus cells constrict at a slower rate than in the wild type [21]. This implies that Pom1 may continue to regulate Cdc15 throughout cytokinesis, modulating its affinity for membranes and interaction with other ring components.
Regulation of actin dynamics during actomyosin ring assembly
Nucleation, remodeling and bundling of actin are required for actomyosin ring assembly [73]. Cortical actin nucleation in S. pombe closely follows myosin recruitment to the medial nodes at the onset of mitosis. It is largely mediated by the essential formin Cdc12, which may also bundle actin filaments [74–77]. Cdc12 localizes to the cortex through two distinct interactions, with Cdc15 and Rng2/Myo2 [78, 79]. As mitosis progresses, Cdc12 acquires SIN-dependent phosphorylation that may regulate its F-actin nucleation and bundling activities [75]. Such phosphoregulation allows ring assembly in S. pombe cells lacking Mid1 and ring maintenance in the absence of septum deposition [75].
Unlike in S. pombe, actin filaments appear at the S. japonicus cortex late in mitosis, following NE breakdown [21, 22]. It is only at this point in the cell cycle that S. japonicus cells arrest tip growth, suggesting global remodeling of the actin cytoskeleton [21]. On the contrary, S. pombe stops elongating upon entry into mitosis, concurrently with ring assembly. Mechanistic studies of Cdc12 and other proteins driving actin remodeling will be crucial for understanding the temporal regulation of ring assembly in this organism. In particular, it will be important to assess Cdc12 interaction with Cdc15 and its potential regulation by SIN and possibly other mitotic kinases.
Interestingly, the anaphase nucleus in S. japonicus appears to spatially control actin nucleation. In cells where the nucleus is displaced by centrifugation and breaks away from the cell center, the site of F-actin nucleation is offset accordingly. This nuclear control of actin nucleation does not depend on the presence of Mid1-centered cortical nodes [21]. It is presently unknown if the signal that drives actin polymerization is released from the nuclear compartment or derives from a nuclear structure such as the SPB or the mitotic spindle. Formin activity is often controlled by small GTPases of the Rho family, including RhoA and Cdc42 [80–85]; see review in [86] and [87]. Although there is no evidence that Cdc42 controls Cdc12-dependent actin nucleation in S. pombe [88], it would be of interest to investigate the dynamics of Cdc42 activation at the equatorial cortex and possible links to Cdc12 activation in S. japonicus.
Concluding remarks
Work in S. pombe has elucidated many aspects of the actomyosin-dependent cytokinesis, ranging from ring assembly to its constriction to understanding how cytokinesis is entrained to the mitotic cell cycle [3]. Lately, S. pombe has been useful in illuminating mechanistic requirements for cytokinesis in vitro [89]. Whereas it will certainly continue producing important insights, it is worth keeping in mind that, probably just like any model system, S. pombe has its share of idiosyncrasies in the way it orchestrates cell division. Arguably the most unusual feature of S. pombe cytokinesis is its early, nucleus-instructed ring assembly relying on Mid1, one of the duplicated and highly diverged anillin paralogs [21, 52]. A pertinent, albeit fairly difficult to answer question is whether this nucleus-dependent ring formation was evolutionarily adaptive or if it had emerged as a spandrel that eventually became hard-wired into the logic of the cell cycle in this organism.
Importantly, comparative analyses of cytokinesis in S. pombe and S. japonicus show that different circuitries may be adapted to yield similar functional outcomes. Yeast cells of both species exhibit comparable geometry during growth and division yet rely on two different and distinctly regulated cortical myosin anchors to assemble the division machinery at the cellular equator (Fig. 2). It remains to be seen if nucleus-dependent division plane specification such as the one operating in S. pombe could be detrimental to S. japonicus upon yeast-to-hyphal transition.
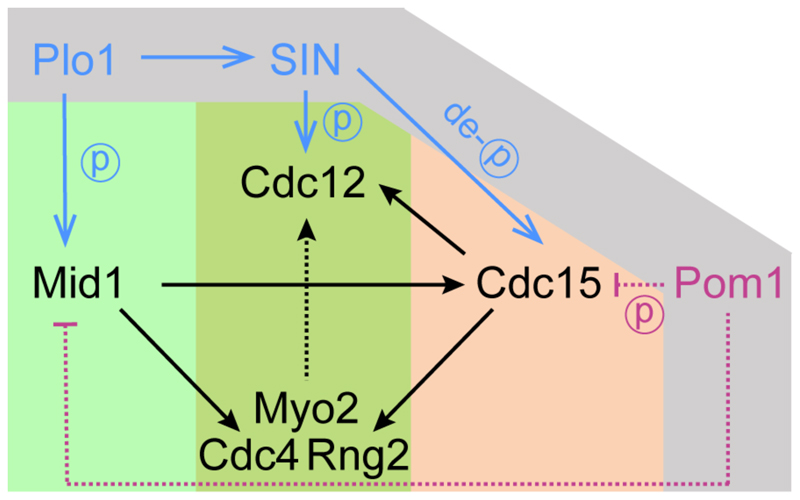
Figure 2. A diagram describing two modes of myosin II recruitment (Mid1- and Cdc15-dependent) to the equatorial cortex and ring assembly in fission yeasts.
Core cytokinetic ring components (in black) include the anillin-like protein Mid1, F-BAR domain protein Cdc15, IQGAP protein Rng2, myosin II heavy chain Myo2, myosin essential light chain Cdc4 and the cytokinetic formin Cdc12. Direction of black arrows indicates hierarchical organization of ring components according to protein localization dependency in cortical recruitment. Regulatory cascades controlling ring assembly and function are shown in grey outer area. The Polo kinase Plo1 and Septation Initiation Network coordinate actomyosin ring assembly and constriction through controlling phosphorylation status of core ring components. The tip-localized DYRK family protein kinase Pom1 prevents ring assembly at cell ends by inhibiting clustering of Mid1/Cdr2 nodes and likely keeping the mitotic myosin anchor Cdc15 in its inactive, “closed” conformation. Direct (solid lines) and genetic (dotted lines) interactions are mostly based on S. pombe data.
With a form of nuclear envelope breakdown and medial actomyosin ring assembly following exit from mitosis, S. japonicus is perfectly positioned to provide a much-needed evolutionary perspective to fission yeast studies and become a useful genetically tractable model for understanding the regulatory logic of cytokinesis prevalent in metazoans.
Acknowledgements
We are grateful to Oliferenko lab members for discussions and E. Makeyev for suggestions on the manuscript. Our work is supported by a Wellcome Trust Senior Investigator Award (103741/Z/14/Z) to S. O.
References
- 1.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes & development. 2009;23:660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 2.Bathe M, Chang F. Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding. Trends in microbiology. 2010;18:38–45. doi: 10.1016/j.tim.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson AE, McCollum D, Gould KL. Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton. 2012;69:686–699. doi: 10.1002/cm.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee IJ, Coffman VC, Wu JQ. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton. 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard TD. Progress towards understanding the mechanism of cytokinesis in fission yeast. Biochemical Society transactions. 2008;36:425–430. doi: 10.1042/BST0360425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nature reviews Molecular cell biology. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rincon SA, Paoletti A. Mid1/anillin and the spatial regulation of cytokinesis in fission yeast. Cytoskeleton. 2012;69:764–777. doi: 10.1002/cm.21056. [DOI] [PubMed] [Google Scholar]
- 8.Simanis V. Pombe's thirteen - control of fission yeast cell division by the septation initiation network. Journal of cell science. 2015 doi: 10.1242/jcs.094821. [DOI] [PubMed] [Google Scholar]
- 9.Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, et al. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic biology. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- 10.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yam C, He Y, Zhang D, Chiam KH, Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Current biology : CB. 2011;21:1314–1319. doi: 10.1016/j.cub.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 12.Aoki K, Hayashi H, Furuya K, Sato M, Takagi T, Osumi M, Kimura A, Niki H. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16:911–926. doi: 10.1111/j.1365-2443.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- 13.Yam C, Gu Y, Oliferenko S. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. Current biology : CB. 2013;23:2303–2310. doi: 10.1016/j.cub.2013.09.057. [DOI] [PubMed] [Google Scholar]
- 14.Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. The EMBO journal. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dischinger S, Krapp A, Xie L, Paulson JR, Simanis V. Chemical genetic analysis of the regulatory role of Cdc2p in the S. pombe septation initiation network. Journal of cell science. 2008;121:843–853. doi: 10.1242/jcs.021584. [DOI] [PubMed] [Google Scholar]
- 16.Rachfall N, Johnson AE, Mehta S, Chen JS, Gould KL. Cdk1 promotes cytokinesis in fission yeast through activation of the septation initiation network. Molecular biology of the cell. 2014;25:2250–2259. doi: 10.1091/mbc.E14-04-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Developmental cell. 2002;3:779–790. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 18.Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. The EMBO journal. 2011;30:341–354. doi: 10.1038/emboj.2010.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Current biology : CB. 2001;11:931–940. doi: 10.1016/s0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 20.Field C, Li R, Oegema K. Cytokinesis in eukaryotes: a mechanistic comparison. Current opinion in cell biology. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- 21.Gu Y, Yam C, Oliferenko S. Rewiring of cellular division site selection in evolution of fission yeasts. Current biology: CB. 2015;25:1187–1194. doi: 10.1016/j.cub.2015.02.056. [**This study describes a surprising divergence in cell division strategies between two fission yeast species. Unlike S. pombe that utilizes the nuclear-linked cue Mid1 for medial actomyosin ring positioning, S. japonicus cells rely on an F-BAR protein Cdc15 controlled by Pom1 kinase to anchor the myosin II complex to the medial cortex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfa CE, Hyams JS. Distribution of tubulin and actin through the cell division cycle of the fission yeast Schizosaccharomyces japonicus var. versatilis: a comparison with Schizosaccharomyces pombe. Journal of cell science. 1990;96(Pt 1):71–77. doi: 10.1242/jcs.96.1.71. [DOI] [PubMed] [Google Scholar]
- 23.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes & development. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. The Journal of cell biology. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akamatsu M, Berro J, Pu KM, Tebbs IR, Pollard TD. Cytokinetic nodes in fission yeast arise from two distinct types of nodes that merge during interphase. The Journal of cell biology. 2014;204:977–988. doi: 10.1083/jcb.201307174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Molecular biology of the cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Molecular and cellular biology. 2004;24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee IJ, Wu JQ. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. Journal of cell science. 2012;125:2973–2985. doi: 10.1242/jcs.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saha S, Pollard TD. Characterization of structural and functional domains of the anillin-related protein Mid1p that contribute to cytokinesis in fission yeast. Molecular biology of the cell. 2012;23:3993–4007. doi: 10.1091/mbc.E12-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L, Guan R, Lee IJ, Liu Y, Chen M, Wang J, Wu JQ, Chen Z. Mechanistic insights into the anchorage of the contractile ring by anillin and mid1. Developmental cell. 2015;33:413–426. doi: 10.1016/j.devcel.2015.03.003. [**Based on crystal structure and functional analyses of the human anillin and S. pombe Mid1, the authors suggest that synergistic interactions between different structural elements drive anillin association with membranes. They also explain differential requirements for Rho GTPase in driving anillin recruitment to the plasma membrane between human and S. pombe.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Current biology : CB. 2009;19:961–966. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 32.Almonacid M, Celton-Morizur S, Jakubowski JL, Dingli F, Loew D, Mayeux A, Chen JS, Gould KL, Clifford DM, Paoletti A. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Current biology : CB. 2011;21:473–479. doi: 10.1016/j.cub.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. The Journal of cell biology. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D, Vjestica A, Oliferenko S. The cortical ER network limits the permissive zone for actomyosin ring assembly. Current biology : CB. 2010;20:1029–1034. doi: 10.1016/j.cub.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Vjestica A, Oliferenko S. Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Current biology : CB. 2012;22:2048–2052. doi: 10.1016/j.cub.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 36.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Developmental cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 37.Daga RR, Chang F. Dynamic positioning of the fission yeast cell division plane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8228–8232. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes & development. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 39.Tolic-Norrelykke IM, Sacconi L, Stringari C, Raabe I, Pavone FS. Nuclear and division-plane positioning revealed by optical micromanipulation. Current biology : CB. 2005;15:1212–1216. doi: 10.1016/j.cub.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 40.Guzman-Vendrell M, Baldissard S, Almonacid M, Mayeux A, Paoletti A, Moseley JB. Blt1 and Mid1 provide overlapping membrane anchors to position the division plane in fission yeast. Molecular and cellular biology. 2013;33:418–428. doi: 10.1128/MCB.01286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu YH, Ye Y, Wu Z, Wu JQ. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Molecular biology of the cell. 2013;24:3187–3204. doi: 10.1091/mbc.E13-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jourdain I, Brzezinska EA, Toda T. Fission yeast Nod1 is a component of cortical nodes involved in cell size control and division site placement. PloS one. 2013;8:e54142. doi: 10.1371/journal.pone.0054142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. The Journal of cell biology. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. The Journal of cell biology. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moseley JB. Cytokinesis: does mid1 have an identity crisis? Current biology : CB. 2015;25:R364–366. doi: 10.1016/j.cub.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huckaba TM, Lipkin T, Pon LA. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. The Journal of cell biology. 2006;175:957–969. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuya K, Niki H. The DNA damage checkpoint regulates a transition between yeast and hyphal growth in Schizosaccharomyces japonicus. Molecular and cellular biology. 2010;30:2909–2917. doi: 10.1128/MCB.00049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto S, Furuya K, Nozaki S, Aoki K, Niki H. Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryotic cell. 2013;12:1235–1243. doi: 10.1128/EC.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eluere R, Varlet I, Bernadac A, Simon MN. Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell cycle. 2012;11:151–158. doi: 10.4161/cc.11.1.18542. [DOI] [PubMed] [Google Scholar]
- 50.Sanders SL, Herskowitz I. The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. The Journal of cell biology. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Si H, Rittenour WR, Xu K, Nicksarlian M, Calvo AM, Harris SD. Morphogenetic and developmental functions of the Aspergillus nidulans homologues of the yeast bud site selection proteins Bud4 and Axl2. Molecular microbiology. 2012;85:252–270. doi: 10.1111/j.1365-2958.2012.08108.x. [DOI] [PubMed] [Google Scholar]
- 52.Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic acids research. 2012;40:D695–699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nature genetics. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 54.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agarwal M, Papadopoulou K, Mayeux A, Vajrala V, Quintana DM, Paoletti A, McInerny CJ. Mid1p-dependent regulation of the M-G1 transcription wave in fission yeast. Journal of cell science. 2010;123:4366–4373. doi: 10.1242/jcs.073049. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Molecular biology of the cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. The Journal of cell biology. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Molecular biology of the cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Current biology : CB. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- 60.Frenette P, Haines E, Loloyan M, Kinal M, Pakarian P, Piekny A. An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PloS one. 2012;7:e34888. doi: 10.1371/journal.pone.0034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregory SL, Ebrahimi S, Milverton J, Jones WM, Bejsovec A, Saint R. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Current biology : CB. 2008;18:25–29. doi: 10.1016/j.cub.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 62.Reyes CC, Jin M, Breznau EB, Espino R, Delgado-Gonzalo R, Goryachev AB, Miller AL. Anillin regulates cell-cell junction integrity by organizing junctional accumulation of Rho-GTP and actomyosin. Current biology : CB. 2014;24:1263–1270. doi: 10.1016/j.cub.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Field CM, Coughlin M, Doberstein S, Marty T, Sullivan W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development. 2005;132:2849–2860. doi: 10.1242/dev.01843. [DOI] [PubMed] [Google Scholar]
- 64.Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Molecular biology of the cell. 2006;17:3254–3266. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. The Journal of cell biology. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Molecular cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 68.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 69.Rincon SA, Bhatia P, Bicho C, Guzman-Vendrell M, Fraisier V, Borek WE, Alves Fde L, Dingli F, Loew D, Rappsilber J, et al. Pom1 regulates the assembly of Cdr2-Mid1 cortical nodes for robust spatial control of cytokinesis. The Journal of cell biology. 2014;206:61–77. doi: 10.1083/jcb.201311097. [**This work explains mechanistically how Pom1 regulates positioning of Cdr2/Mid1 cytokinetic ring precursors at the cell center.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Celton-Morizur S, Racine V, Sibarita JB, Paoletti A. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. Journal of cell science. 2006;119:4710–4718. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- 71.Niccoli T, Arellano M, Nurse P. Role of Tea1p, Tea3p and Pom1p in the determination of cell ends in Schizosaccharomyces pombe. Yeast. 2003;20:1349–1358. doi: 10.1002/yea.1054. [DOI] [PubMed] [Google Scholar]
- 72.Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the fission yeast DYRK kinase Pom1. Molecular & cellular proteomics : MCP. 2015 doi: 10.1074/mcp.M114.045245. [*Using the SILAC-based phosphoproteomics approach this study identifies the Pom1 kinase targets in S. pombe and shows that they are enriched in proteins regulating cell polarity and cell cycle.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra M, Huang J, Balasubramanian MK. The yeast actin cytoskeleton. FEMS microbiology reviews. 2014;38:213–227. doi: 10.1111/1574-6976.12064. [DOI] [PubMed] [Google Scholar]
- 74.Johnson M, East DA, Mulvihill DP. Formins determine the functional properties of actin filaments in yeast. Current biology : CB. 2014;24:1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 75.Bohnert KA, Grzegorzewska AP, Willet AH, Vander Kooi CW, Kovar DR, Gould KL. SIN-dependent phosphoinhibition of formin multimerization controls fission yeast cytokinesis. Genes & development. 2013;27:2164–2177. doi: 10.1101/gad.224154.113. [*This interesting paper shows how the septation initiation network controls activity of formin Cdc12 in S. pombe.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coffman VC, Sees JA, Kovar DR, Wu JQ. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. The Journal of cell biology. 2013;203:101–114. doi: 10.1083/jcb.201305022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J, Huang Y, Yu H, Subramanian D, Padmanabhan A, Thadani R, Tao Y, Tang X, Wedlich-Soldner R, Balasubramanian MK. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. The Journal of cell biology. 2012;199:831–847. doi: 10.1083/jcb.201209044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. The Journal of cell biology. 2015;208:391–399. doi: 10.1083/jcb.201411097. [*This mechanistic study shows how an F-BAR protein Cdc15 interacts with Cdc12 formin, contributing to actin nucleation and ring assembly in S. pombe.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. The Journal of cell biology. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 81.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. The EMBO journal. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. The EMBO journal. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Current biology : CB. 2002;12:1864–1870. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- 84.Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Molecular biology of the cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Molecular cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annual review of biochemistry. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 87.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nature reviews Molecular cell biology. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 88.Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Molecular biology of the cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mishra M, Kashiwazaki J, Takagi T, Srinivasan R, Huang Y, Balasubramanian MK, Mabuchi I. In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nature cell biology. 2013;15:853–859. doi: 10.1038/ncb2781. [*The authors establish an in vitro actomyosin ring isolation platform in S. pombe and use it to investigate mechanistic requirements for ring constriction.] [DOI] [PubMed] [Google Scholar]