Abstract
The nuclear lamina (NL) consists of a thin meshwork of lamins and associated proteins that lines the inner nuclear membrane (INM). In metazoan nuclei, a large proportion of the genome contacts the NL in broad lamina-associated domains (LADs). Contacts of the NL with the genome are believed to aid the spatial organization of chromosomes and contribute to the regulation of transcription. Here, we will focus on recent insights in the structural organization of the genome at the NL and the role of this organization in the regulation of gene expression.
Introduction
The NL is a thin meshwork of type V intermediate lamin filaments that coat the INM with the exception of sites of nuclear pore complexes (NPCs). The NL in mammalian cells consist of A-type and B-type lamins and many associated proteins including proteins that are integral components of the INM [1]. The protein constitution of the NL-meshwork can vary extensively between cell types and A-type lamin protein levels are generally strongly reduced in undifferentiated cell types. In accordance with observations of classical electron micrographs [2], it has long been recognized that the chromatin in proximity to the NL is in a condensed state. More recently, a novel method that combines DNA-labelling and three-dimensional electron microscopy (ChromEMT) revealed that chromatin is organized into 5-nm–24-nm nucleosomal chains with increased packaging densities at the NL [3•]. By employing the DamID technology, the identity of the genomic regions that contact the NL was first revealed in Drosophila melanogaster [4]. Since this first report, LADs have been further characterized in the fruit fly but also in Caenorhabditis elegans and multiple mammalian cell types [4–7]. LADs are of particular interest because, in addition to playing an important role in genome architecture, regions that contact the NL differ with respect to cell type-specific gene expression, suggesting a role for LADs in gene regulation [7,8]. This review will focus on the molecular mechanisms that may be involved in the organization of LADs and the possible contributions of genome–NL contacts to the regulation of transcription during cellular differentiation and development.
Genome organization at the NL
In mammalian cells, the genome contains approximately 1000–1500 LADs with a median domain size of ~0.5 Mb [5,7]. In addition to lamina association, chromatin has been shown to be structured in the three dimensional nuclear space in topologically associating domains (TADs), characterized by a high level of intra-domain contacts in contrast with few interactions occurring between TADs [9]. At a larger scale, TADs have been grouped into A and B compartments, corresponding to active and inactive chromatin regions, respectively. Likely because of their smaller size and threshold-dependent domain calling, only some TAD boundaries overlap with LAD boundaries. However, higher-order B compartments have been shown to generally coincide with LADs [9,10]. Although LADs are generally characterized as being gene-poor, they comprise about 30–40% of the genome and thus still encompass thousands of genes that are generally lowly transcribed. Interestingly, although Drosophila LADs are about fivefold smaller, the number of genes per LAD is comparable to that of mammalian LADs. This suggests that gene number is a potentially defining evolutionary constraint on LAD structure [11].
In addition to low gene density, LADs in mammalian cells are characterized by high A/T content and a high LINE (long interspersed nuclear element) density. These features are most prominent for LADs that invariably associate with the NL across cell types [8]. These constitutive LADs (cLADs) were, therefore, postulated to form a structural backbone of spatial genome positioning at the NL, as opposed to the cell type-specific positioning of facultative LADs (fLADs) [8]. Interestingly, LADs in totipotent zygotes and pluripotent mouse embryonic stem cells (mESCs) are particularly enriched in typical cLAD features, suggesting that genome–NL contacts in undifferentiated cells are preferentially established on a structural core backbone [our own unpublished data, 8]. This organization may represent a default state of LAD organization that is overruled by lineagespecific transcriptional programs. Indeed, in zygotes the genome–NL contacts are established before zygotic genome activation (ZGA) and enrichment for cLAD features is decreased with the commencement of transcription in early embryogenesis and upon differentiation in mESCs [our own unpublished data, 7]. Therefore, genome–NL contacts of backbone cLADs seem to occur independently of transcription.
In support of an anchoring role for cLADs, single-cell DamID has revealed that LADs that are present in most cells largely overlap with cLADs [10]. After considering over one hundred single-cell LAD profiles at a 100 kb resolution, this study showed that LADs associate with the NL in long stretches of continuous contacts, suggestive of multivalent interactions. A model in which multivalent interactions support genome–NL associations was further strengthened by the observation that longer LADs and LAD-dense chromosomes are more frequently positioned at the NL [10]. Interestingly, following cell division, LADs often do not return to the NL and, in some cases, localize to the vicinity of nucleoli instead [12]. This is consistent with the observation that nucleolus-associated domains (NADs) partially overlap with LADs [13,14]. LADs that maintain their localization at the nuclear periphery after mitosis are likely to be cLADs. To further elucidate NL versus nucleolar localization of chromatin regions, single-cell NAD profiles could be obtained from cells expressing Dam fused to nucleolar proteins.
Mechanisms of LAD formation
Multivalent interactions of A/T rich regions appear to support robust NL-associations, but what are the anchors that mediate these contacts? Lamins could directly mediate tethering as they have been shown to bind chromatin and DNA [15,16]. Indeed, in Drosophila, depletion of the B-type lamin causes detachment of certain gene loci from the nuclear periphery in S2 cells [17]. Similarly, loss of the sole lamin protein in C. elegans causes perinuclear release of large heterochromatic arrays [18]. Recent work in mESCs showed that overall LAD content is largely unaffected in cells depleted of all lamins, indicating that lamins are dispensable for tethering the genome to the NL. However, upon closer examination of these cells by DNA-FISH and Hi-C, local chromatin expansion of cLADs was observed (Figure 1, bottom left). The authors suggest a meshwork caging model in which lamins contribute to the structural organization of the genome at the NL as opposed to a role in anchoring [19••]. Previous work employing a microscopy version of the DamID technique revealed LAD-embedding into pockets formed in the lamin meshwork, which is consistent with this meshwork caging model [12]. Each lamin isoform forms similar but distinct filamentous networks, hence the meshwork properties could differ between cell types with different NL compositions [20–22]. Indeed, mouse embryonic fibroblast (MEF) nuclei depleted of Lamin A/C or Lamin B1, but not Lamin B2, resulted in a less dense lamin meshwork with different properties [21]. Similar studies conducted in cells with naturally different NL protein constitutions should reveal how different structural properties of the meshwork relate to cell type-specific LAD organization.
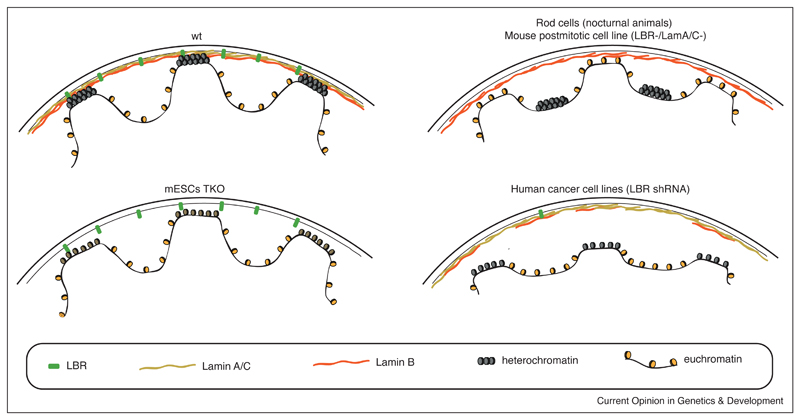
Figure 1. Schematic representation of lamina composition impact on chromatin conformation.
On the top left is a wild-type nucleus consisting of a nuclear lamina containing LBR and all lamins. The compacted heterochromatin is located at the nuclear periphery, while euchromatin is located more at the nuclear interior. In triple knockout mESCs that do not express any lamins (bottom left), genome–NL interactions are maintained but cLAD chromatin is decondensed [19••]. Rod cells of nocturnal animals that naturally lack LBR and Lamin A/C and mouse postmitotic cells in which LBR and Lamin A/C were mutated show inverted chromatin organization with heterochromatin localizing at the nuclear interior and euchromatin localizing at the periphery (top right) [23,25]. In human cancer lines in which LBR is downregulated, Lamin B1 expression is also affected and results in the detachment of heterochromatin from the NL (bottom right) [26••].
In combination with lamins, other integral proteins of the NL could potentially mediate genome–lamina contacts. Rod cells that naturally lack the Lamin B-receptor (LBR) and Lamin A/C display an inverse spatial organization of chromatin with interior heterochromatin and peripheral euchromatin positioning (Figure 1, top right) [23]. Similarly, in mouse olfactory neurons, the downregulation of LBR inherent to the final stages of the differentiation process results in the aggregation of silent olfactory receptor genes in heterochromatic foci at the nuclear interior [24]. A role in tethering heterochromatin toward the nuclear periphery was confirmed upon knockout of Lamin A/C and LBR in mouse postmitotic cells, which leads to the same inverse chromatin organization (Figure 1, top right) [25]. Also, depletion of LBR in two human cancer cell lines resulted in concomitant decrease in Lamin B1 expression and repositioning of pericentric heterochromatin toward the nuclear interior (Figure 1, bottom right) [26••]. Upon oncogene-induced senescence (OIS), LBR is downregulated and cLADs have been reported to detach from the nuclear lamina, likely contributing to the formation of heterochromatic foci in the nuclear interior. However, in this case, the absence of LBR alone was insufficient to cause such dramatic changes in chromatin architecture and other nuclear envelope transmembrane proteins (NETs) could be involved [27]. Indeed, certain NETs have been shown to play a role in chromatin tethering to the nuclear periphery of specific cell types [28,29].
LADs are typically covered by histone marks that contribute to a repressed chromatin state and may be involved in genome–lamina interactions. Indeed, it was found that interactions of LBR with chromatin can either occur directly through binding to H4K20me2 [30] or via LEM-domain mediated interaction with heterochromatin proteins such as HP1 [31,32]. Additionally, HP1 interactions with histone 3 lysine 9 methylated chromatin may help mediate LAD-positioning at the NL [33,34]. Indeed, H3K9me2 domains in mESCs show high concordance with LADs [35] and can thus serve to ‘hook’ LADs to the NL, even in the absence of lamins.
In human cells, H3K9me2-enriched chromatin was shown to project toward the nuclear periphery in a G9a (H3K9me2 methyltransferase)-dependent manner [36], and similarly, in human HT1080 cells, G9a depletion resulted in reduced NL-association of a number of LADs [12]. In addition, knock-out of G9a in mESCs caused selective upregulation of genes in LADs [37]. Targeting of LADs with a viral transcriptional activator peptide resulted in the loss of H3K9me2-enriched LADs from the NL-surface [12]. A similar strategy in mESCs resulted in the heritable repositioning of selected genes toward the nuclear interior [38]. Interestingly, in both studies, the employed viral peptide caused chromatin decondensation without transcriptional activation, indicating that nuclear organization is mediated by chromatin changes rather than by transcription. Taken together, these results clearly suggest an important role for H3K9me2 in LAD-positioning toward the NL.
In addition to H3K9me2, H3K9me3 has also been shown to be enriched in LADs [19••]. An elegant study dissecting the targeting properties of a bacterial artificial chromosome (BAC) encompassing the human beta-globin (HBB) locus showed that NL-positioning of regions within the ~200 kb locus involves NL-positioning via both H3K9me2 and H3K9me3 [39]. Thus, in this example, at least two independent mechanisms act abreast, and perturbation of both H3K9 methylation pathways are required to dislodge the endogenous HBB locus [39]. Similarly, in C. elegans only the removal of both SET-2 (necessary for the H3K9me1 and me2 marks) and MET-25 (necessary for H3K9me3) results in the repositioning of a heterochromatin array and a partial loss of the peripheral localization of endogenous loci [18]. In a follow-up study, the NL-associated CEC-4 protein was identified to anchor H3K9-methylated chromatin to the NL via its chromodomain [40]. A similar NL-anchor protein in mammals has not yet been identified, although the protein PRR14 may serve a similar function by anchoring H3K9-methylated chromatin via HP1α [41•].
A role for H3K27me3 in LAD organization is less clear. H3K27me3 was found to be enriched at LAD borders [5,42] and reduction of H3K27me3 was shown to affect the peripheral localization of ectopically integrated LADs [42]. However, H3K27me3 is not always found associated with LADs [10] and, therefore, a role in LAD positioning could be cell type-specific or LAD-specific. In flies, cell type-specific chromatin signatures associated with LADs were also observed. LADs in embryonic Kc167 cells are partially associated with Polycomb (Pc) [11,43] but not with HP1a/H3K9me2, while in differentiated cell types and particularly neurons, LAD-profiles display striking concurrence with HP1a/H3K9me2 [44•]. Collectively, a picture emerges of multiple non-exclusive chromatin-mediated processes that drive NL-association via LAD-specific and cell type-specific mechanisms.
LADs and the regulation of gene expression
Genes within LADs are generally lowly transcribed, which is suggestive of a role for LADs in gene silencing. The role of the NL in gene regulation may entail direct involvement in gene repression, for example, by exclusion of genes from the transcriptionally active nuclear interior, and/or indirect via the reinforcement or locking in of chromatin states. Random genomic integrations of thousands of reporters resulted in ~5–6-fold attenuation of gene activity when inserted in LADs relative to other genomic regions [45]. In three independent studies, artificial recruitment of reporter genes to the NL resulted in mixed outcomes of either partial repression of the reporter and some flanking genes [46,47] or no effect on transcription at all [48]. Thus, localization toward the NL is in general associated with gene repression even though exceptions occur. It still needs to be addressed whether the differential transcriptional sensitivities observed in these studies may be related to differences in flanking chromatin signatures or to the properties of adjacent LADs (cLADs or fLADs). A recent study, employing a new method to simultaneously measure LADs and mRNA from the same single cell, may provide a platform to systematically decipher the molecular interactions causing the differences in transcriptional dependencies for individual LADs in various cell types [49].
Active gene repression at the NL may be mediated via various non-mutually exclusive mechanisms. One straightforward mechanism would involve NL association of histone modifying enzymes with repressive signatures. LADs are generally depleted of active histone marks [5,43,50]. The absence of histone acetylation could be mediated by local interactions between the INM components Emerin and Lap2ß with histone deacetylases such as HDAC3 [51–55]. Suggestive of an active mechanistic interaction at the NL is the finding that HDAC3 activity is increased upon interaction with Emerin [54].
Another potential mechanism of gene repression through genome–NL interaction involves the sequestration of chromatin-bound transcription factors (TFs) [52,56,57]. These mechanisms are likely to occur only on a subset of fLADs and only in the specific cell types in which the TF is expressed. An emerging TF-mediated regulatory role for the NL is illustrated in a recent report, which describes the retention of chromatin at the NL via an interaction of the GLI1 TF with the NL protein Lap2ß. Transcriptional activation involves egress from the NL through ‘pulling’ by the competing nucleoplasmic isoform Lap2α [58••]. Nuclear shuttling of TFs between the nucleoplasm and the NL could provide a fast and tunable gene expression mechanism. Further studies are required to verify how general this type of gene-regulation is across cell types.
A more passive role for the NL in transcriptional regulation may be to preserve and reinforce gene inactivity and thereby locking in transcriptional states. Gene repression may not directly involve NL association, yet confinement in the NL territory may secure an irreversible repressive transcriptional state. During neuroblast differentiation in Drosophila, the hunchback gene translocates toward the NL after transcriptional silencing. Thus, the NL is not directly involved in hunchback silencing. However, prevention of hunchback association with the NL by lamin depletion results in a delay in cellular commitment and a prolonged neuroblast competence window [59]. Similar lineage specification defects have been observed in C. elegans upon disruption of the NL [40,60]. In mammalian cells, the role of the NL of locking in genes in a repressive state remains mostly unexplored. However, it was shown that the overall chromatin density at the nuclear periphery increases in more differentiated cell types [61] in combination with increased H3K9me3 levels [62]. Despite these changes, the proportion of the genome that contacts the NL appears constant between populations of mESCs and terminally differentiated cells [7]. This apparent discrepancy might be explained by differences in the proportion of LADs that associate with the NL in individual cells. Indeed, while cells in a more undifferentiated state show more heterogeneity between LAD content of individual cells [10,49], terminally differentiated single cells may have a higher proportion of LADs consistently contacting the NL, perhaps due to the increase in H3K9me3 levels. Interestingly, inhibition of H3K9 methylation results in a differentiation delay [62] and, removal of H3K9me3 alleviates binding restrictions of pluripotency factors to DNA and facilitates reprogramming [63]. Lamin A has also been reported to constitute a barrier to induced reprogramming [64]. Thus, the increased expression levels of Lamin A, perhaps in combination with increased H3K9me3, may help consolidate transcriptional states in more differentiated cell types [62,65].
Conclusions
Genome-nuclear lamina interactions play an important structural role in the three-dimensional organization of the genome and are likely to be involved in gene regulation. The integration of new insights into a preexisting framework of literature reveals a scenario whereby NL composition, chromatin state of LADs and presence of DNA-binding proteins cooperatively regulate gene expression at the nuclear periphery.
Acknowledgements
We would like to thank the members of the Kind group for critical reading of the manuscript. This work was supported by a Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) VIDI (016.161.339) and ERC-Stg EpiID (678423) grant to J.K. and an EMBO ALTF 1214-2016 fellowship to I.G. The Oncode Institute is supported by KWF Dutch Cancer Society.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.de Leeuw R, Gruenbaum Y, Medalia O. Nuclear lamins: thin filaments with major functions. Trends Cell Biol. 2018;28:34–45. doi: 10.1016/j.tcb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- 3.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [• The authors use a novel chromatin visualization technique to show that chromatin is organized into 5–24 nm chains with higher density at the nuclear lamina.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 5.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 6.Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 2010;11:R120. doi: 10.1186/gb-2010-11-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meuleman W, Peric-Hupkes D, Kind J, Beaudry J-B, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kind J, Pagie L, de Vries Sandra S, Nahidiazar L, Dey Siddharth S, Bienko M, Zhan Y, Lajoie B, de Graaf Carolyn A, Amendola M, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, van Steensel B. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One. 2010;5:e15013. doi: 10.1371/journal.pone.0015013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries Sandra S, Janssen H, Amendola M, Nolen Leisha D, Bickmore Wendy A, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 13.van Koningsbruggen S, Gierliński M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stierlé V, Couprie J, Östlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin J-C, Duband-Goulet I. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–4828. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- 16.Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L. The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO J. 1993;12:4413–4424. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Towbin Benjamin D, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser Susan M. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Hu J, Yue S, Kristiani L, Kim M, Sauria M, Taylor J, Kim Y, Zheng Y. Lamins organize the global three-dimensional genome from the nuclear periphery. Mol Cell. 2018;71:802–815.e807. doi: 10.1016/j.molcel.2018.05.017. [•• The authors analyze changes in chromatin spatial organization as a result of triple lamin knock-out in mESCs. Absence of all lamin isoforms from the NL is found to result in altered intra-TAD interactions and decondenzation of cLADs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, Medalia O. The molecular architecture of lamins in somatic cells. Nature. 2017;543:261. doi: 10.1038/nature21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell. 2015;26:4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie W, Chojnowski A, Boudier T, Lim John SY, Ahmed S, Ser Z, Stewart C, Burke B. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr Biol. 2016;26:2651–2658. doi: 10.1016/j.cub.2016.07.049. [DOI] [PubMed] [Google Scholar]
- 23.Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 24.Clowney EJ, LeGros Mark A, Mosley Colleen P, Clowney Fiona G, Markenskoff-Papadimitriou Eirene C, Myllys M, Barnea G, Larabell Carolyn A, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solovei I, Wang Audrey S, Thanisch K, Schmidt Christine S, Krebs S, Zwerger M, Cohen Tatiana V, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Lukášová E, Kovařík A, Bačíková A, Falk M, Kozubek S. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem J. 2017;474:281. doi: 10.1042/BCJ20160459. [•• In this paper, the silencing of LBR which caused decreased lamin B1 expression, resulted in heterochromatin relocation to the nucleoplasm and decondenzation.] [DOI] [PubMed] [Google Scholar]
- 27.Lenain C, de Graaf CA, Pagie L, Visser NL, de Haas M, de Vries SS, Peric-Hupkes D, van Steensel B, Peeper DS. Massive reshaping of genome–nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017;27:1634–1644. doi: 10.1101/gr.225763.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robson Michael I, de las Heras Jose I, Czapiewski R, Lê Thành P, Booth Daniel G, Kelly David A, Webb S, Kerr Alastair RW, Schirmer Eric C. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol Cell. 2016;62:834–847. doi: 10.1016/j.molcel.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuleger N, Boyle S, Kelly DA, de las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 2013;14:R14. doi: 10.1186/gb-2013-14-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J Biol Chem. 2012;287:42654–42663. doi: 10.1074/jbc.M112.397950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polioudaki H, Kourmouli N, Drosou V, Bakou A, Theodoropoulos PA, Singh PB, Giannakouros T, Georgatos SD. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001;2:920. doi: 10.1093/embo-reports/kve199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye Q, Callebaut I, Pezhman A, Courvalin J-C, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 33.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 34.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 35.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Yammine S, Shi C, Tark-Dame M, Göndör A, Ohlsson R. The visualization of large organized chromatin domains enriched in the H3K9me2 mark within a single chromosome in a single cell. Epigenetics. 2014;9:1439–1445. doi: 10.4161/15592294.2014.971633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci USA. 2009;106:19363. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bian Q, Khanna N, Alvikas J, Belmont AS. β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol. 2013;203:767. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez-Sandoval A, Towbin Benjamin D, Kalck V, Cabianca Daphne S, Gaidatzis D, Hauer Michael H, Geng L, Wang L, Yang T, Wang X, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- 41.Poleshko A, Mansfield Katelyn M, Burlingame Caroline C, Andrake Mark D, Shah Neil R, Katz Richard A. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013;5:292–301. doi: 10.1016/j.celrep.2013.09.024. [• In this study, the authors show that, during cardiac progenitor lineage restriction, Hdac3 plays a role in tethering lineage-specific genes to the nuclear lamina.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. 2015;208:33. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pindyurin AV, Ilyin AA, Ivankin AV, Tselebrovsky MV, Nenasheva VV, Mikhaleva EA, Pagie L, van Steensel B, Shevelyov YY. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenetics Chromatin. 2018;11:65. doi: 10.1186/s13072-018-0235-8. [• The authors obtain LAD profiles of Drosophila differentiated cell types and find that, contrary to embryonic cells, HP1a strongly overlaps with lamina associated genomic regions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akhtar W, de Jong J, Pindyurin Alexey V, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels Lodewyk FA, van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 47.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rooijers K, Markodimitraki C, Rang F, de Vries S, Chialastri A, de Luca K, Mooijman D, Dey S, Kind J. Simultaneous quantification of protein-DNA contacts and transcriptomes in single cells. bioRxiv. 2019 doi: 10.1101/529388. unpublished data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010;471:480. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2β interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 52.Zullo Joseph M, Demarco Ignacio A, Piqué-Regi R, Gaffney Daniel J, Epstein Charles B, Spooner Chauncey J, Luperchio Teresa R, Bernstein Bradley E, Pritchard Jonathan K, Reddy Karen L, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 53.Milon BC, Cheng H, Tselebrovsky MV, Lavrov SA, Nenasheva VV, Mikhaleva EA, Shevelyov YY, Nurminsky DI. Role of histone deacetylases in gene regulation at nuclear lamina. PLoS One. 2012;7:e49692. doi: 10.1371/journal.pone.0049692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demmerle J, Koch AJ, Holaska JM. Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome Res. 2013;21:765–779. doi: 10.1007/s10577-013-9381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poleshko A, Shah PP, Gupta M, Babu A, Morley MP, Manderfield LJ, Ifkovits JL, Calderon D, Aghajanian H, Sierra-Pagán JE, et al. Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell. 2017;171:573–587.e514. doi: 10.1016/j.cell.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Columbaro M, Mattioli E, Maraldi NM, Ortolani M, Gasparini L, D’Apice MR, Postorivo D, Nardone AM, Avnet S, Cortelli P, et al. Oct-1 recruitment to the nuclear envelope in adult-onset autosomal dominant leukodystrophy. Biochim Biophys Acta (BBA) 2013;1832:411–420. doi: 10.1016/j.bbadis.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Malhas AN, Lee CF, Vaux DJ. Lamin B1 controls oxidative stress responses via Oct-1. J Cell Biol. 2009;184:45. doi: 10.1083/jcb.200804155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirza AN, McKellar SA, Urman NM, Brown AS, Hollmig T, Aasi SZ, Oro AE. LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell. 2019;176:198–212.e115. doi: 10.1016/j.cell.2018.10.054. [•• In this study, the authors identify a LAP2 isoform-dependent mechanism to control nuclear localization of GLI1 in basal cell carcinomas.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohwi M, Lupton Joshua R, Lai S-L, Miller Michael R, Doe Chris Q. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattout A, Pike Brietta L, Towbin Benjamin D, Bank Erin M, Gonzalez-Sandoval A, Stadler Michael B, Meister P, Gruenbaum Y, Gasser Susan M. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol. 2011;21:1603–1614. doi: 10.1016/j.cub.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 61.Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, Inman M, Tsang H, Warr M, Passegué E, et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep. 2015;5:728–740. doi: 10.1016/j.stemcr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soufi A, Donahue G, Zaret Kenneth S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuo B, Yang J, Wang F, Wang L, Yin Y, Dan J, Liu N, Liu L. Influences of lamin A levels on induction of pluripotent stem cells. Biol Open. 2012;1:1118. doi: 10.1242/bio.20121586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J, Adli M, Zou James Y, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager Philip L, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]