Abstract
The poor outcomes in esophageal adenocarcinoma (EAC) prompted us to interrogate the pattern and timing of metastatic spread. Whole genome sequencing and phylogenetic analysis of 388 samples across 18 EAC cases demonstrated in 90% of cases that multiple subclones from the primary tumor spread very rapidly from the primary site to form multiple metastases, including lymph nodes and distant tissues, a mode of dissemination that we term ‘clonal diaspora’. Metastatic subclones at autopsy were present in tissue and blood samples from earlier time-points. These findings have implications for our understanding and clinical evaluation of EAC.
Introduction
Metastatic spread to distant sites accounts for the majority of cancer deaths1. Understanding the anatomical extent of disease is essential to determine the optimum treatment strategy. This is challenging since cancer continually evolves at a microscopic scale, often beyond the resolution of clinical imaging techniques. Furthermore, the patterns of metastatic spread are often unpredictable in terms of time-course and anatomical location. Treatments may therefore be unnecessarily toxic (e.g. radical lymphadenectomy and chemotherapy) or insufficiently aggressive, leading to high recurrence rates2–4.
Esophageal cancer is the sixth most common cause of cancer-related death worldwide and the current median survival time is still <1 year5. Incidence rates for esophageal adenocarcinoma (EAC) have risen sharply and it is now the predominant subtype in developed countries. Prognosis is highly variable for EAC patients as shown by the wide range of 5-year survival (18-47% with lymph node involvement), making it difficult to advise patients when embarking on a long course of grueling treatment2,6.
Theoretical and experimental studies attempt to understand how tumor cell populations respond to selective pressures over time7. A number of models of tumor evolution have been proposed, including linear, branching, neutral and punctuated evolution, but the extent to which these are specific to a given cancer type or co-occur is controversial8,9. Genome sequencing studies have attempted to delineate different models of evolution10. However, many of these studies have focused solely on evolution within the primary site, and knowledge of how genetic diversity emerges during metastasis remains limited. The lack of understanding is in part due to the practical challenge of collecting multiple samples over space and time from advanced stage cancer patients.
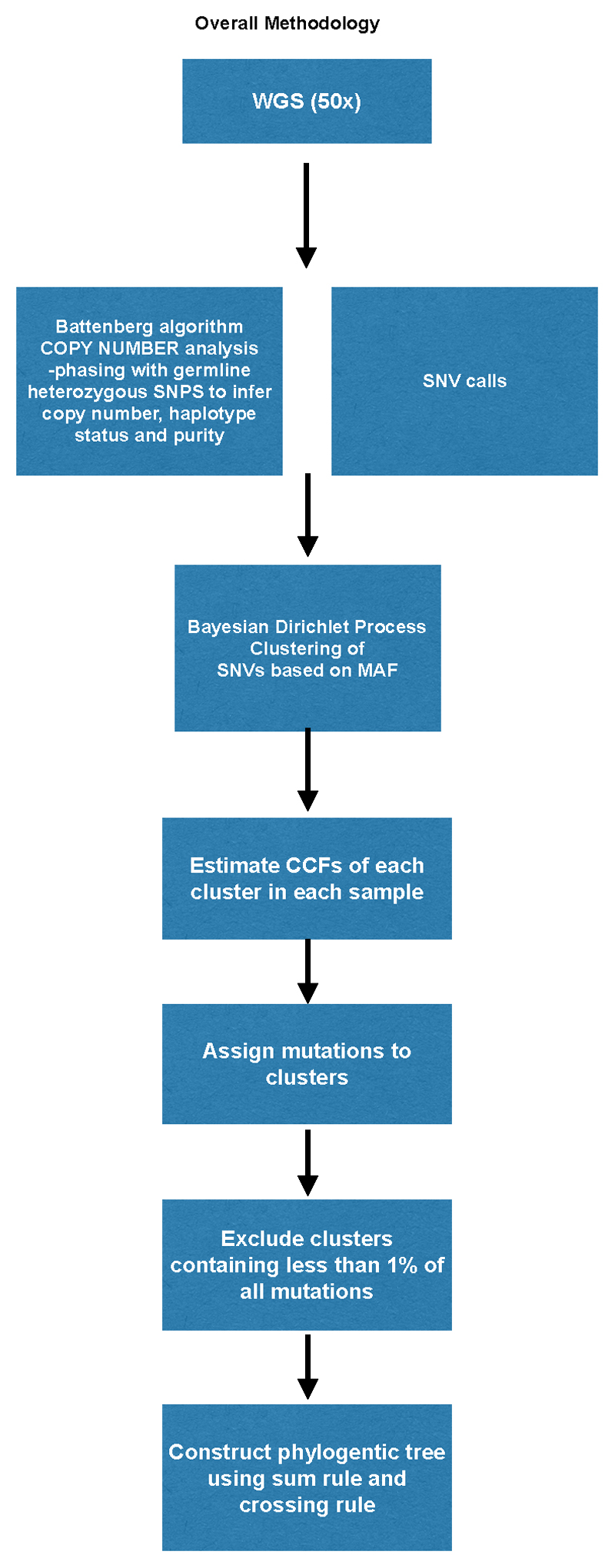
To better understand the evolution of EAC, we designed a prospective study with extensive sampling over time including samples from diagnosis, surgery and at warm autopsy (Figure 1). We used whole genome sequencing (WGS) at high depth (50x), to identify mutations, and at shallow (1x) coverage, to track known variants, to interrogate the clonal architecture across time and space.
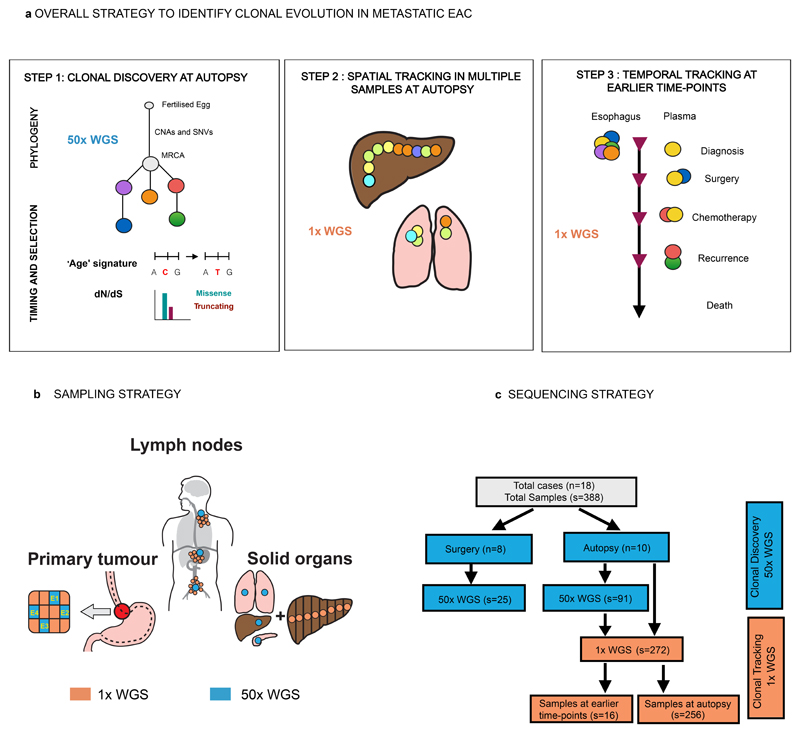
Figure 1. Overall project strategy and study design.
a. Overall Strategy to identify clonal evolution in metastatic EAC. There were three main steps in this study which comprised: Clonal discovery at autopsy (see Supplementary Note High Depth Whole Genome Sequencing (50x WGS), Mutation clustering and phylogenetic tree construction, dN/dS analysis and Mutational Signature Analysis); Spatial tracking at autopsy (see Supplementary Note Shallow Whole Genome Sequencing (1x WGS) and Temporal tracking at earlier time-points (see Supplementary Note Shallow Whole Genome Sequencing (1x) for Subclone identification, Supplementary Table 12 for precise samples for plasma and Extended Data Fig. 9 for FFPE diagnostic samples). Colored circles depict clones and subclones respectively. b. Sampling Strategy at Rapid Autopsy. Areas sampled for the 50x WGS part of the study are shown in blue and for 1x WGS are shown in orange. c. Study Design and Sequencing Strategy. The flow chart demonstrates the study design and how this relates to sequencing. Clonal Discovery is in blue and Clonal Tracking in orange. The sample distribution for 50x WGS and 1x WGS are shown. 50x WGS = High depth WGS (50x), 1x WGS = Shallow WGS (1x). n = number of cases, s = number of samples. †=248 solid tissue samples, and 8 ctDNA at autopsy. CNA, copy number alteration; SNV, single nucleotide variant; MRCA, most recent common ancestor.
Results
Genomic architecture of 18 cases
Eighteen cases were included in the study and the clinical demographics of these cases are shown in Supplementary Table 1 and 2, with details of the individual samples given in Supplementary Table 3 and 4. In the first part of the study (Figure 1a, Extended Data Fig. 1,2) we used 50x WGS to construct a phylogenetic tree for each case, to understand the relationship between the primary and metastatic tumors (Figure 2, Extended Data Fig. 3, Supplementary Figure 1, Supplementary Table 3, 4). Mutation clustering was performed, and the fractions of tumor cells carrying each set of mutations (Cancer Cell Fraction, CCF) within each sample were used to determine: 1) the clonal and sub-clonal architecture of each tumor (subclonal CCF <95%, clonal CCF ≥ 95%); 2) the hierarchy of events; and 3) the distance of these sub-clonal or clonal clusters from the most recent common ancestor (MRCA) (Figure 1a, Extended Data Fig. 1,2). The CCF and number of single nucleotide variants (SNVs) associated with each clone and subclone are shown in Supplementary Table 5 and 6, as is the tumor purity of each sample using the Battenberg algorithm11, in Supplementary Table 7 and the confidence intervals of the clonal and subclonal CCFs in Supplementary Table 8. Detailed information on experimental design is provided in the Life Sciences Reporting Summary.
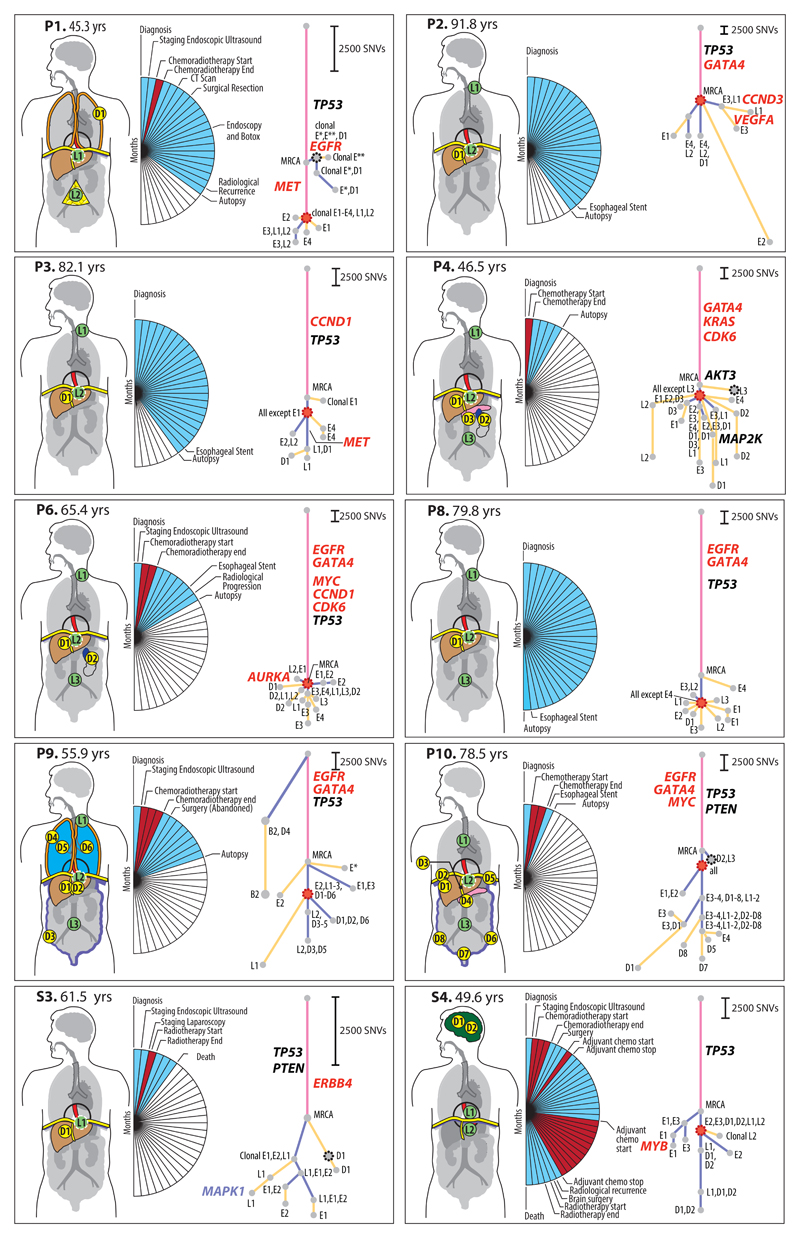
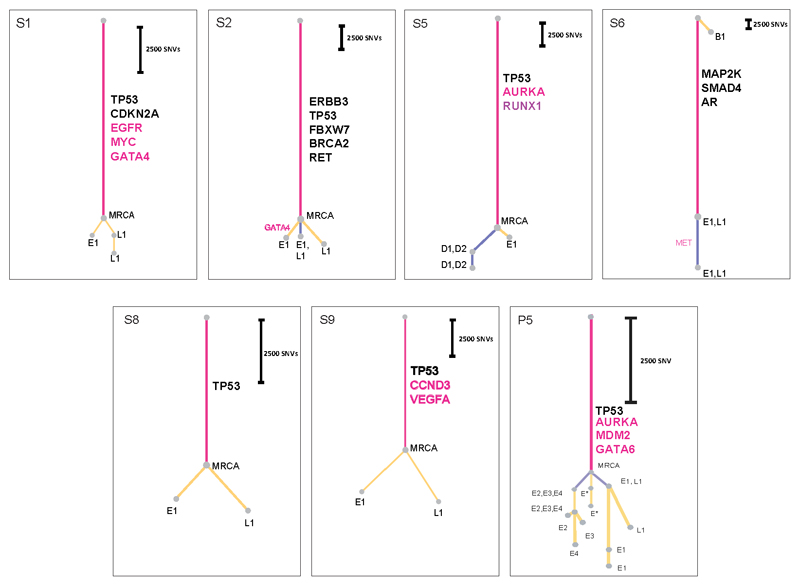
Figure 2. Phylogenetic Analysis of ten cases with nodal and distant metastases.
Patient body maps (S=surgical case, P=rapid autopsy) are shown. Green circles denote lymph node metastases and yellow circles distant metastases. The labels within each circle describe the specific location (see Supplementary Table 3, 4). An organ is shown in color if metastases were sequenced from that site. The adjacent wedged semi-circle depicts the clinical timelines for each patient. Each wedge corresponds to one month; blue wedges indicate the total lifetime of the patient and red wedges periods of therapy. Phylogenetic trees for each patient are shown and methodology is in Supplementary Note and Extended Data Fig. 1a-b; pink = truncal events shared by all samples, purple = branch events shared by more than one sample, yellow = leaves, events unique to a sample. The circle at the end of a trunk, branch or leaf represents a clone or subclone. Each clone or subclone is annotated to show which samples it is present in. E1-E4 = primary esophageal tumor, L1-L4= lymph nodes, D1-8 =distant metastases, B = Barrett’s Esophagus. A subclone annotated with E1, L2 for example indicates that this subclone is seen only in samples E1 and L2. The CCF of each subclone/clone (barring the MRCA) is in Supplementary Table 5 and 6. The length of the branches of the tree are reflective of the number of SNVs in the subclone/clone. The scales adjacent to each case are relative, given the variable number of SNVs per case. Trees are annotated with potential driver events, black: missense variants, red: amplifications. Gray dots outlined with a black dashed line denote the first subclone/clone to metastasize that would be classified as non-curative based on anatomical location. Red dots mark the stellate pattern on the phylogenetic tree.
These analyses enabled us to construct phylogenetic trees (Methods). In all cases we observed a long trunk compared to the rest of the tree (median 19,034 SNVs, IQR 11,299-63,908), consistent with previous studies in EAC12,13. The median size of clonal or subclonal clusters across all cases was 3,069 SNVs (IQR 1332-63908) and only 2/157 contained fewer than 200 SNVs (S1_3 and P5_11), Extended Data Fig. 3 and Supplementary Table 6.
The key driver events14,15 are depicted on each phylogenetic tree (Figure 2 and Extended Data Fig. 3). The events identified as most frequent in previous studies occurred in the trunks of the phylogenetic trees, consistent with their previous classification as drivers. TP53 was mutated in the trunk of 16 out of 18 cases, consistent with our knowledge of the disease14,16–19. Amplifications (gene names in red) were often truncal, but also observed on the branches of the phylogenetic tree, providing evidence of divergence during later evolutionary stages (Figure 2, Extended Data Fig. 3). The majority of events in driver genes were copy number alterations (CNAs) rather than SNVs or InDels (Figure 2, Extended Data Fig. 3)14,19,20. There was no significant difference in the overall number of structural variants between primary and metastatic samples (p=0.41, generalized linear model; Extended Data Fig. 4b). However, a larger proportion of structural variants in metastatic samples were retro-transpositions of mobile elements than in the primary samples (p=0.045, Extended Data Fig. 4c). This contrasts with pancreatic cancer, where deletions and fold-back inversions are more common in metastases, and breast cancer where tandem duplications dominate21. Interestingly, the high rate of L1 transposon activity in EAC has recently been associated with high activity in the germline22. Our results suggest a further increase in L1 activity in metastatic EAC. Furthermore, the proportion of structural variants found uniquely in metastases or in primary sites was higher than that of SNVs (Figure 2, Extended Data 4a), suggesting an increase in genomic instability in later stages of the disease. However, it cannot be ruled out that some structural variants have not been identified in every sample as a result of lower sensitivity in the detection of structural variants than SNVs.
Across the eighteen cases, 8 mutational signatures were observed, consistent with previous studies23–26 (Figure 3a), with varying prevalence across the cases. None of the signatures that we observe in patients in our cohort who had oncologic therapy have been associated with treatment with alkylating antineoplastic agents27, platinum therapy28 or radiation therapy29.
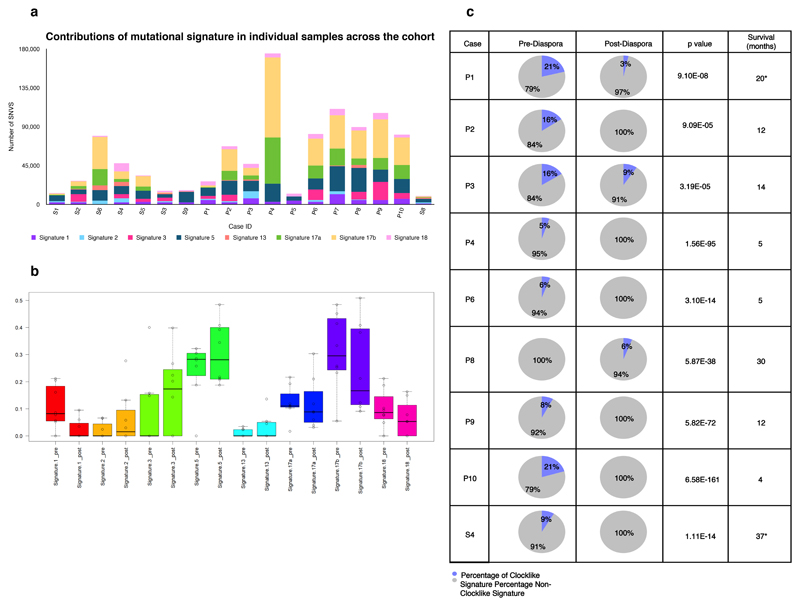
Figure 3. Mutational Signatures.
a. Contributions of mutational signature in 18 cases (n=122) across the cohort. The bar chart displays samples on a per case basis (X-axis) and depicts the number of SNVs contributing to each signature (Y-axis). b. Mutational signatures pre-and post-diaspora across all samples (n=122) in 18 cases.
Mutations were separately assigned to signatures and the proportion of mutations within each case assigned to each signature is shown. Dark lines = median, Boxes = 25th and 75th quartiles, whiskers extend to the most extreme point within 1.5× interquartile range of the box edge. Signatures 1 mutations have a significantly lower representation in post-diaspora mutations, while signature 3 mutations have significantly high. c. Mutational signature analysis of ageing signature (signature 1) pre-and post-diaspora in all cases (n=8) with local and distant spread (p<1.18 × 10-90 across all cases) Chi squared test was used to determine the p value. Survival is shown in months from the point of diagnosis *=cases which underwent surgery.
Early seeding of oligometastases
Ten of eighteen patients (S3, S4, P1-4, P6, P8-10) had both nodal and solid organ metastases, allowing a direct comparison of the genomic architecture between different metastatic sites (Figure 2).
In four of these ten cases, an isolated clone or subclone confined to 1 or 2 distant metastases, i.e. an oligometastasis, depicted as a dashed black node on the first branch of the phylogenetic tree, shared the highest congruence to the MRCA, (P1, P4, P10, S3 in Figure 2; Subclones P1_2, P4_3, P10_2, S3_2 in Supplementary Table 5). In P1, this clone (P1_2) was observed only in the primary tumor and a pleural metastasis. In S3 and P4, the clone involved in this isolated seeding was identified at a single distant site and not in the primary tumor (S3_2: liver metastasis (D1), P4_3: para-aortic lymph node (L3)). In P10, the early seeding clone (P10_2) was shared between a distant para-aortic node and a sub-clonal metastasis in the right hemi-diaphragm. The subclones associated with these isolated seeding events showed little divergence from the MRCA across these 4 cases (median 1,913 SNVs, range 832-8,591), suggesting early seeding to distant metastases. Notably, in P9 a subclone (P9_10, Supplementary Table 5) was found in a premalignant area of Barrett’s esophagus and a pleural metastasis but not in any of four areas of the primary tumor subject to 50x WGS. This subclone lineage shares no variants with the main lineage and appears to be an independent second cancer (Figure 2).
A single clone gives rise to multiple metastatic sites
A striking observation was that 9/10 cases had a clone (outlined in red on the phylogenetic tree in Figure 2) that was followed by dispersion of multiple subclones from the primary to discrete metastatic sites, resulting in a model of metastasis that we term ‘clonal diaspora’. In most cases, this dispersion was visually stellate in nature, this being defined as a feature of a phylogenetic tree involving 3 or more branches leading from a single founder clone (see details in Discussion). The subclones forming diasporas were located in both primary and metastatic tissue in eight cases (P1, P2, P3, S4, P4, P6, P8, P10) and in P9 were unique to metastases (Figure 2). The only two cases lacking a stellate pattern on the phylogenetic tree were P10 and S3. The latter is a non-autopsy case with limited tissue sampling and the early distant seeding in this case is consistent with a pattern of parallel evolution (Figure 2).
Subclonal spread is not constrained by location or tissue
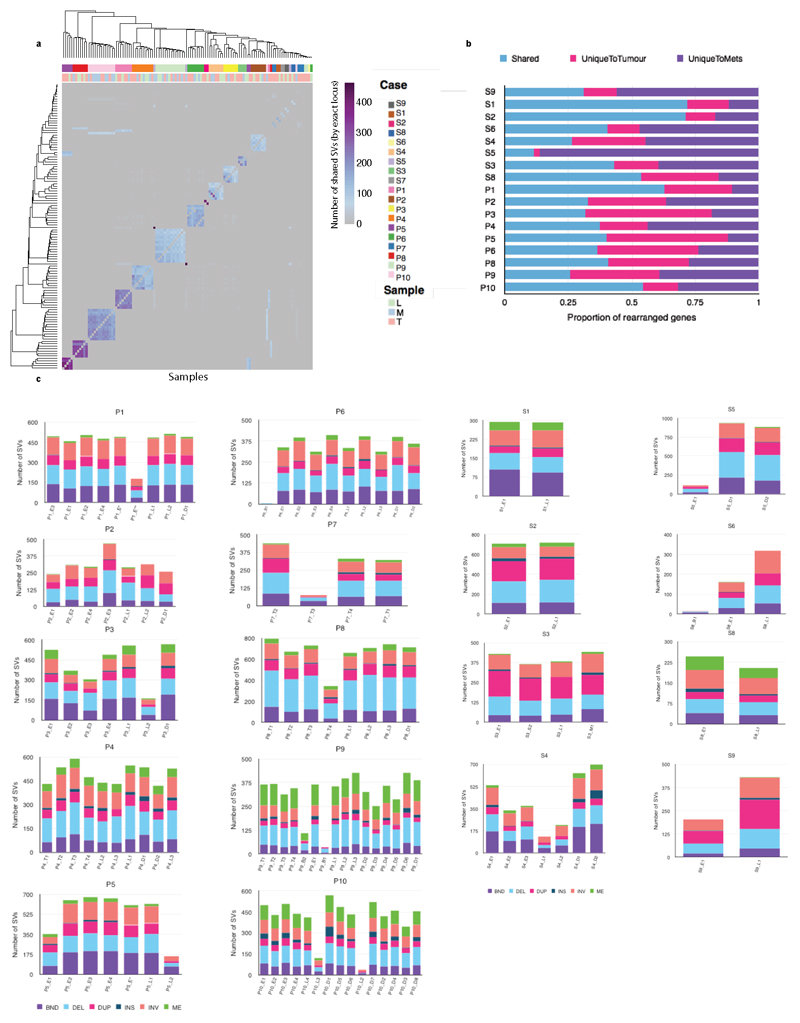
In the second step of the study we tracked the spread of metastases across a wider range of lymph node and distant tissue sites by performing 1x WGS in a further 248 tissue samples from 6 autopsy cases (Figure 1a,c). We did not call new mutations, as this would not be possible at 1x sequencing, but used this method to detect the spread of clones and subclones previously identified using 50x WGS (bioinformatic validation of methods in Extended Data Fig. 5 and 6, Supplementary Note; wet lab validation in Extended Data Fig. 7, Supplementary Table 9). The samples used in this part of the study are outlined in Supplementary Table 10. The median size of subclonal and clonal clusters (identified previously at 50x WGS) that we aimed to detect using 1x WGS was 3,784 (IQR 1,966-49,955). Sample sites were grouped according to their similarity based on the presence of subclones and clones previously detected with 50x WGS (Supplementary Note). The resulting groups of samples are color coded and numbered, and each sample site, colored by group, is shown on the adjacent body map (Figure 4, see also Supplementary Note). Notably, the samples that grouped together based on shared clonal origins were widely dispersed anatomically.
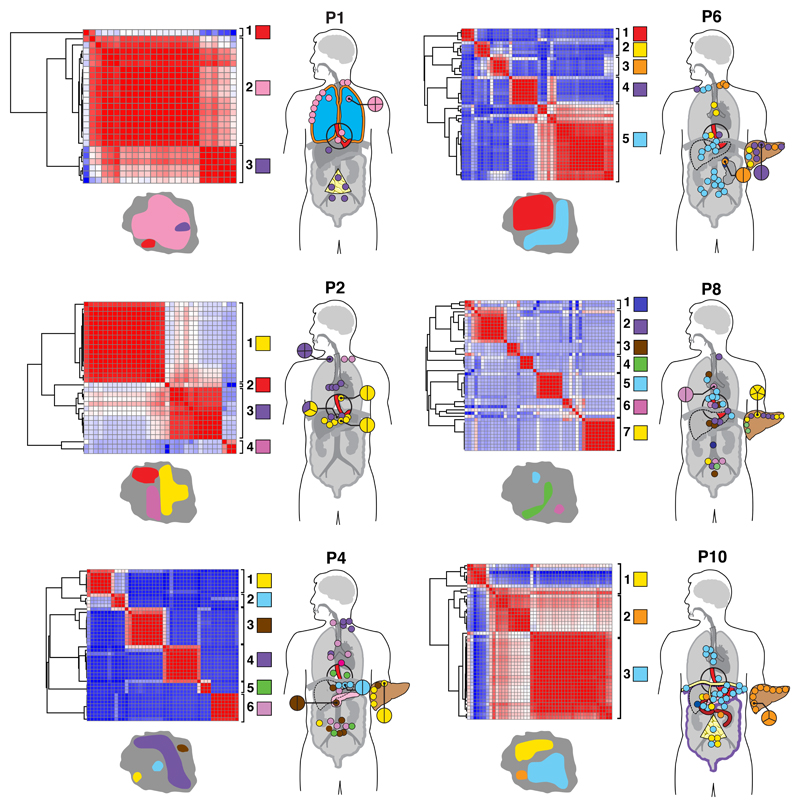
Figure 4. 1x WGS and similarity matrix clustering of 248 further tissue samples from six cases.
1x WGS was performed at an average depth of 1x to track subclones and clones previously discovered using 50x WGS for further tissue samples (n=248). Pearson correlation similarity matrix clustering was performed on all samples for each case (plotted against each other) with red indicating sample similarity (r=1) and blue indicating dissimilarity (r=-1). Sample sites used in this part of the study are shown in Supplementary Table 9 and the entire organ is highlighted if solid organ sites were sequenced. For example, liver metastases were only seen in P4, P6, P8, P10. Similarly, P2 had lymph nodes only (only colored dots are seen which represent lymph nodes, no solid organs are highlighted). Clustering was performed based on the presence of subclones and clones already detected using 50x WGS and distinct clusters were identified for each case as demonstrated by the adjacent key per case (each group is both colored and numbered). Samples are displayed on the adjoining body maps for which the color coding corresponds to the genomic clustering in the adjacent heatmap. Sites with multiple samples are magnified and the division of samples shown. Maps of the primary tumor with representation of metastatic subclones are shown with each case, with the colors of the subclones being the same as those in the matrix and body map. Areas shaded red in the primary tumor represent subclones that were not detected in the metastatic samples that underwent 1x WGS and were instead confined to areas of the primary tumor.
Four out of six cases with extensive spatial sampling (Figure 4) had liver metastases evaluated and three of these contained samples that were more similar to local lymph node metastases than neighboring liver metastases (P4, P6, P8 but not P10). The high number of groups within the liver (up to four) suggested seeding by multiple subclones (seen in P4, P6, P8), whereas the single group in the liver of P10 (orange, group 3) indicated seeding by a common progenitor or a set of closely related cells.
A comparison of lymph node location and genomic contiguity showed no evidence of tropism, i.e. genomically similar lymph nodes did not occupy nearby anatomical locations. Lymph nodes above and below the diaphragm were frequently seeded from common events (P2: groups 1, 3; P4: groups 5, 6; P6: group 5; P8: groups 2, 3,5, 6; P10: group 4), at odds with a progression from local to distant nodes. Similarly, a comparison of lymph node and solid organ metastases showed scant evidence for tropism, with the exception of P1 (Supplementary Note). This patient underwent surgical resection and subsequently had metastatic disease recurrence. In this cancer, separate subclones seeded lymph node and pleural metastases (Figure 2, 4). Notably, the distant metastasis (D1) was an early branching oligometastasis whereas the lymph nodes (L1, L2) constituted the later diaspora event (black and red circles, respectively, in Figure 2).
We further traced regions of the primary tumor at autopsy that had similar subclonal compositions to each of the metastases, shown as adjacent tumor maps (Figure 4, bottom left of each case). Subclones occupied spatially distinct areas in the primary tumor.
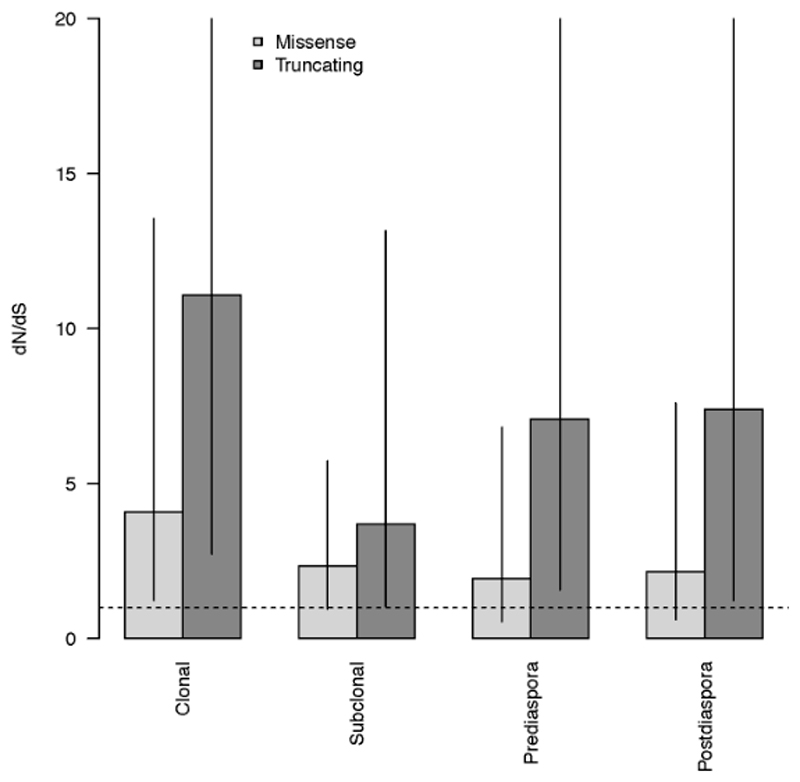
We also looked for driver amplifications post MRCA or post diaspora on a per case basis and identified selection in 6/10 cases. However, this is likely to be an under-estimate, since there may be non-copy number drivers present in additional cases. The ratio of non-synonymous to synonymous SNVs (dN/dS) was analyzed across all cases in order to assess the presence or absence of positive selection30. Results indicated positive selection in both clonal and subclonal genomes, albeit with lower levels of selection within subclones (Extended Data Fig. 8).
Metastatic spread is rapid in EAC
To examine the timing and speed of metastatic spread we analyzed base substitution mutational signatures, particularly the aging signature which features a predominance of C>T transition in the NpCpG trinucleotide context (Figure 1a, Figure 3).
Signature 1 arises from the spontaneous or enzymatic deamination of methylated cytosines, which is an endogenous process that occurs continuously in both healthy and cancerous cells. This has been shown to act as a molecular clock27,31–35, and was therefore used here as a method to examine the temporal relationship between metastases. Using a previously described method for deconvolving mutational signatures35, we observed that signature 1 was present in the trunk but absent in all subclones that constituted diaspora (following the red parental clone in Figure 2) for P2, P4, P6, P9, P10, S4 and it was significantly reduced for P1 (21% to 3%) and P3 (16% to 9%) (Wilcoxon signed rank test p=0.039, Figure 3c). To account for the possibility that the number of signature 1 mutations in branch subclones was below the resolution of our deconvolution methods, we also identified the number of mutations with the characteristic feature of signature 1, i.e. C>T mutations in a CpG context. To estimate the time of appearance of diaspora, we compared the number of these characteristic mutations that occurred along the trunk to the parental red clone marking the onset of diaspora with those that occurred on the longest branch leading from this point. The median proportion of such mutations occurring prior to the onset of diaspora was 0.911 (Figure 3b). Thus, in the majority of cases one might deduce that little time has elapsed between the appearance of the cell that is ancestral to disseminating cells and the individual cells that seeded each of the metastases. With the exception of P8, the proportion of mutations attributed to signature 1 was significantly lower after the parental (red) clone on the phylogenetic tree (p<9.1 × 10-5, Chi-squared test across all cases; Figure 3c) suggesting an increase in the activity of other processes in later evolutionary stages (Supplementary Table 11). Of note, there was an increase in the proportion of signature 3 in subclonal SNVs compared to clonal SNVs (Wilcoxon signed rank test p=0.019, Figure 3b), suggesting failure of DNA double strand break repair is predominantly a late-stage event in EAC.
Early detection from diagnostic samples
Next, we investigated eight cases (P1-4, P6, P8-10) for which the esophageal diagnostic FFPE biopsy or surgical sample (primary tumor at resection for P1 and lymph node from surgery for P9) were available, with a median time prior to autopsy of 12 months (range 5-30 months) (Figure 1). The diagnostic sample for P1 was snap frozen and sequenced to 50x (Figure 2; highlighted with * in Extended Data Fig. 9), while 1x WGS was performed on the remainder of the cases. Between 8% and 36% of the subclones and clones observed in samples taken from autopsy were also present in the diagnostic samples (Supplementary Note and Extended Data Fig. 9). In six cases, all subclones identified from the biopsy samples were also found in the primary samples from autopsy. Two diagnostic endoscopic samples from P4 also contained many of the mutations found in the lymph node L2 at autopsy, which had not been previously identified in the primary tumor at autopsy (Figure 2, subclone P4_17, Supplementary Table 5). Similarly, the biopsy sample from P10 contained a substantial number of mutations from both the oligometastasis that seeded D2 and L4 (Supplementary Table 5, P10_2), and the lineage that later metastasized to multiple sites (Figure 2). Notably, P4 and P10 had shorter survival times after diagnosis than the remaining patients (5 and 4 months, respectively).
Plasma sample analysis at autopsy and earlier time-points
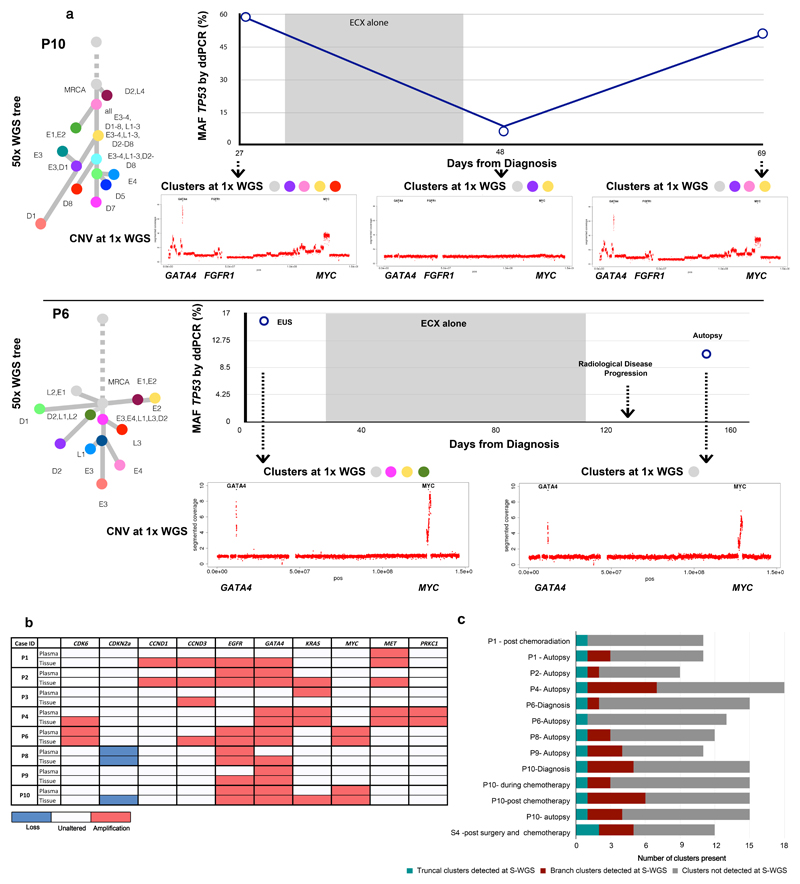
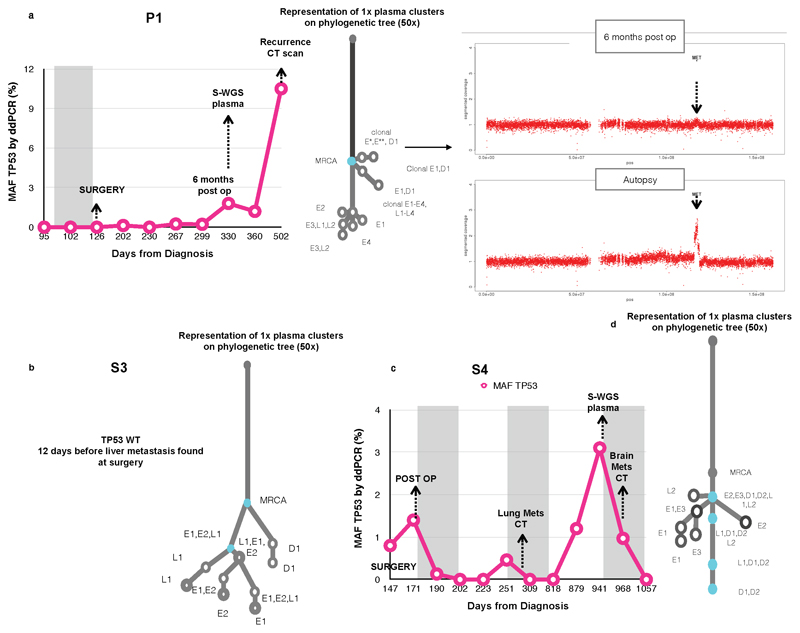
We assessed the clonal composition of circulating tumor DNA (ctDNA) at earlier time-points in seven blood samples from five cases (Figure 1, Figure 5a,c; Extended Data Fig. 10, Supplementary Table 12). Combined 1x WGS subclone/clone detection, copy number aberrations and TP53 fraction using digital PCR data are displayed for two of these cases (P6 and P10) in Figure 5a. Notably, P6 was a patient being treated with curative intent and had no radiological evidence of distant nodal or organ metastases at the time of clinical staging. However, at the time of diagnosis mutations from the truncal cluster and three subclonal clusters later found in the metastases were already present in the plasma (Figure 5a) along with amplifications in MYC and GATA4. Case S4 is noteworthy as the brain metastases (D1, D2 in Figure 2) appeared to have originated from a subclone shared between the primary and a local lymph node, both of which were removed at the time of surgery (Extended Data Fig. 10c). However, mutations from the truncal cluster and four subclonal clusters were already present in ctDNA prior to radiological recurrence.
Figure 5. Temporal and spatial tracing of metastatic subclones in plasma.
a. Plasma ctDNA 1x WGS and digital droplet PCR (ddPCR) analysis for TP53 mutant allele fraction (MAF) for P10 and P6. The MAF of TP53 (%) is shown on the Y-axis and days from diagnosis are shown on the X-axis. The shaded areas represent time periods of therapy. 1x WGS at select time-points was performed and the clonal composition of these samples are shown by the presence of colored clusters. The color of each corresponds to the color of the corresponding node on the adjacent 50x phylogenetic tree with the presence of colored clusters which correlate with the 50x tree. Moreover, copy number traces for each time point are shown for select chromosomes. b. The presence or absence of amplifications and deletions in plasma compared to tissue, detected from 1x WGS for 8 cases. Tissue refers to all samples collected at autopsy and at earlier time-points. c. Stacked bar charts to demonstrate the presence or absence of clusters across all plasma samples, including truncal and branch clusters using 1x WGS.
In eight cases, plasma was available from rapid autopsy. One case (P3) failed wet lab SNV validation and was hence removed from the SNV subclone analysis (Supplementary Note). Analysis of ctDNA demonstrated that in all cases the truncal cluster from autopsy was also represented in plasma (Figure 5c). In addition, mutations from between 0 and 7 subclonal clusters were identified from plasma (Figure 5c). The ratio of mutations detected from each subclone was very consistent between blood from earlier time points and autopsy (Pearson r range [0.851, 0.994], maximum P-value 8.9 × 10-4) and in 2 of 5 cases the proportion of mutations detected was higher in the earlier sample, suggesting an opportunity for earlier detection of heterogeneous cancer cell populations. Further, subclonal proportions estimated from exome sequencing of plasma samples were highly correlated with those from 1x WGS (Supplementary Table 9).
The majority of driver CNAs identified in the MRCA of each tumor from 50x WGS of tissue samples were also identified in plasma both at autopsy and at earlier time-points (Figure 5a,b). In addition, MET amplification, which was not present in the MRCA in P1 (Figure 2), was identified in plasma both at autopsy and an earlier time point (Extended Data Fig. 10a), suggesting opportunities for early detection of metastatic subclones. Notably, however, amplifications found only in oligometastases or in post-diaspora subclones from 50x sequencing were not identified in plasma, despite many of them being detected in 1x sequencing of tissue samples (Figure 5b). A plausible explanation for this observation is that each of the many metastasizing subclones contributed insufficient material to the sum of detected ctDNA to enable confident detection of CNAs.
Discussion
We have gathered multiple lines of evidence which suggest that, for the majority of EACs, a complex mode of spread is operative. These lines of evidence can be summarized as follows (Figure 6). We observe multiple subclones, each seeding multiple metastatic sites. These subclones are frequently derived from a single parental clone, generally resulting in a stellate pattern on the phylogenetic tree. Metastases in solid organs can bypass nodal involvement and samples within solid organ sites frequently resemble distant metastases more closely than neighboring metastases within the same organ, i.e. no tropism is observed. All metastases appear to have spread directly from the primary site, with little or no evidence of metastasis-to-metastasis seeding.
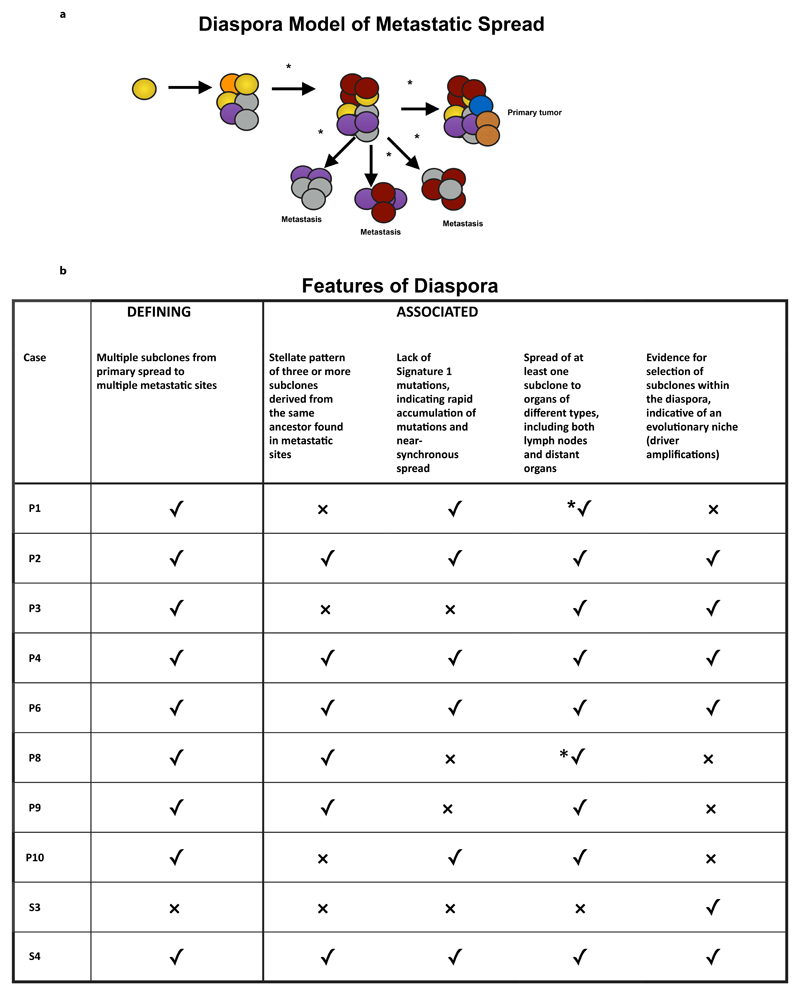
Figure 6. Diaspora model of metastatic spread and associated features.
Panel a depicts clonal diaspora with colored circles representing clones and subclones. *= evidence of selection. Panel b explains the five features seen in diaspora (one is defining, and the other are associated with diaspora) and whether these are present (✓) or absent (x) in each case. *✓ implies that the feature is present, and that the evidence was from 1x WGS.
These features differ in some important respects from previously described models of metastasis and we propose that they may constitute a distinct, additional model of evolution. We suggest that this pattern be referred to as a ‘diaspora’, by extension of the anthropological term to cancer36. Within this context, it is associated with the observation that multiple cell populations in metastatic sites are directly linked to the primary site of origin and that individual subclones seed multiple tissue types, analogous to a diaspora crossing multiple national boundaries.
A number of features were frequently associated with this phenomenon (Figure 6), with nine of the cases (all except S3) displaying at least two of the four following features: i) stellate pattern on the phylogenetic tree defined as three or more subclones emerging from the founder clones; ii) lack of signature 1 mutations post MRCA or post-diaspora; iii) spread of subclones to multiple organs of different type; iv) evidence for selection in post diaspora genotypes.
Until recently the genomic architectures of metastatic samples have not been defined with enough resolution to discern temporal or spatial patterns of metastatic spread. Several distinct patterns are now emerging which are not necessarily mutually exclusive or cancer-type specific. In pancreatic cancer, Yachida et al. demonstrated that distant organ seeding was a late event consistent with a linear progression model24. In prostate cancer, linear progression is often succeeded by multiple waves of seeding37. The same study further demonstrated widespread subclonal evolution in metastases and metastasis-to-metastasis spread, in keeping with the relatively long longevity of prostate cancer. Strikingly, a stellate pattern was not observed in any of the cases in that study, despite using a similar design to that used here.
In Supplementary Table 13 we compare the features of our proposed Diaspora model to the previously posited linear38 and parallel8 models. Whereas the linear model predicts that a single subclone seeding lymph node sites is followed by transmission to distant organs, the diaspora model posits simultaneous seeding of multiple sites directly from the primary. Unlike the parallel model, the diaspora model implies that metastasis formation occurs after the majority of evolution has occurred in the primary tumor, resulting in multiple subclones found in common between primary and metastatic tumors. Lymphatic and distant metastases in colon cancer have been shown to arise from independent subclones in the primary tumor with disparate evolutionary trajectories39. In contrast, in EAC we find that individual subclones frequently seed both lymph node and distant organs suggesting that disparate trajectories for nodal and solid organ metastases do not exist for this disease (Figure 2, 3). Of note we acknowledge that, despite the extensive and systematic sampling across all autopsy cases, further sampling may add further branches to our phylogenetic tree, although this is unlikely to affect the diaspora event itself.
In common with the Big Bang Model proposed for colorectal cancer40, our model predicts the occurrence of highly branching phylogenies. However, the Big Bang Model proposes neutral dynamics, whereas we observe strong evidence for selection in subclonal populations in the form of dN/dS ratios and the occurrence of subclonal driver amplifications (Figure 2, Extended Data Figure 8, Supplementary Figure 2). Moreover, the clonal maps of the primary tumor demonstrate subclones that occupy spatially discrete areas of the primary tumor (Figure 4), in contrast to the intermixed subclones predicted by the Big Bang Model40.
The sequence of events in metastatic progression may have clinical implications that require further study (Supplementary Table 13). Clonal architecture in EAC defies anatomical location of lymph node stations and distant sites, which is the current basis for the TNM staging and determines whether curative therapy is appropriate. It has been suggested that the high recurrence rate, 52% within one year, results from seeding of distant metastases that are not detected at the time of diagnosis26. This study provides molecular evidence for this observation and highlights the need for different systemic approaches to disease management, including consideration of more aggressive adjuvant therapy which is not currently the mainstay of treatment41–44. With advances in the sensitivity of ctDNA assays, metastatic subclones may be detectable in the blood, helping to determine when systemic therapy is required post-surgery and in detecting heterogeneity of acquired resistance45. Copy number variation in plasma may also be a future early detection strategy46.
The occurrence of metastasis is a pivotal event in the life history of a cancer. Understanding the drivers behind such an event would have potential relevance to patient stratification and predicting and preventing metastatic spread47. While we have identified many drivers on the trunks of the trees, prior to diaspora (Figure 2), we cannot be certain which event, if any, was the immediate trigger of diaspora in individual cases. In a number of cases, diaspora was coincident with an increase in the proportion of signature 3 mutations, associated with failure of DNA double-strand break-repair by homologous recombination (Figure 3b). Our findings are in keeping with the failure of DNA repair driving the appearance of genomic heterogeneity. Whether the heterogeneity observed is itself the driver of diaspora or merely a symptom is an important area for future study. Our investigations of the potential drivers of diaspora were limited to genomic factors, and further multi-platform studies looking at epigenetic and transcriptomic factors are other important avenues of future research. We anticipate that analyses of single cells or small clusters from primary sites, disseminated tumor cells and circulating tumor cells will also yield finer resolution of the processes of dissemination and metastasis.
In cancer there are currently very few in-depth studies examining the spatial and temporal evolution of metastases48. Further studies are required to ascertain the extent to which our diaspora theory pertains to other cancers.
Methods
Statistics
Unless otherwise stated, statistical analyses were performed using R, version 3.3.3. Clustering of mutations was carried out using a previously published Bayesian Dirichlet Process method, DPClust (https://github.com/Wedge-Oxford/dpclust), which calculates CCFs of each SNV, taking into account tumor purity and copy number aberrations as previously described49. Analysis of structural variants used generalized linear models, implemented with the R package MASS. Grouping of 1x WGS samples was performed with the GENE-E package (https://software.broadinstitute.org/GENE-E/download.html). Wilcoxon signed rank tests and Chi-squared tests were used as described in the main text. Simulations were used to ascertain the robustness of DPClust to violations of the infinite sites assumption and its sensitivity to detect small deviations from stellate patterns. Simulations were also used to confirm the correlation between the number of mutations detected from 1x WGS and CCF determined from 50x WGS, as described in Online Methods. dN/dS analysis was performed using the previously published package dndscv50 (https://github.com/im3sanger/dndscv).
Patient recruitment and Sample collection
EAC patients were recruited from Addenbrooke’s Hospital, Cambridge University Hospitals NHS Trust with the explicit aim to study the clonal evolution of metastases as a sub-study within OCCAMS (Oesophageal Clinical And Molecular Stratification). When it was clear that extensive sampling of metastases could not be achieved without multiple invasive procedures, the PHOENIX autopsy study was set up (Phylogenetic of Oesophageal Neoplasia – An Investigation of Clonal Expansion under REC 07/H0305/52, and REC EE/0043) with a prospective study design. Due diligence was undertaken to ensure compliance with ethical regulations at all times. Patients were eligible if they were at least 18 years of age and had received a confirmed diagnosis of EAC following central pathology review. Patients were only approached for the PHOENIX study following a palliative diagnosis of metastatic EAC, with the full involvement of the multidisciplinary team. Samples from the PHOENIX autopsy study were obtained within 6 hours of death and all post-mortems were carried out at Papworth Hospital NHS Trust, United Kingdom.
Samples from Cambridge OCCAMS patients were obtained during diagnostic oesophagogastroduodenoscopy (OGD), at endoscopic ultrasound (EUS) and/or from the surgical resection specimen. Where possible, multiple samples were taken from spatially distinct sites of the primary tumor or metastases. In two cases, brain metastases were sampled at a clinically indicated craniotomy. Blood or normal squamous esophageal samples, at least 5cm distant from the tumor, were used as a germline reference.
All tissue samples were snap-frozen in liquid nitrogen immediately after collection and stored at -80°C. Cancer samples were deemed suitable for DNA extraction only after consensus review of an H&E stained frozen section, from the same sample that would be sent for sequencing, by two expert pathologists who confirmed tumor cellularity at ≥70%.
Samples with overall ≥70% cellularity underwent dissection of the whole surface area with a scalpel, whereas marked areas of <70% underwent macrodissection or laser capture micro-dissection aided by methylene blue staining visualized on the PALM-Zeiss microscope (Zeiss, Oberkochen, Germany). An H&E stained slide was obtained before and after extraction to confirm tumor cellularity of the microdissected section.
DNA was extracted from frozen tissues using the All PrepDNA/RNA Mini Kit (Qiagen, Hilden, Germany) and from blood samples using the NucleonTM Genomic Extraction kit (Gen-Probe, San Diego, USA) according to the manufacturer’s instructions. Some samples were preserved in paraffin blocks after initially being stored in formalin. DNA from these samples was extracted using the QiAmp FFPE Kit (Qiagen). Plasma extraction (for ctDNA) was performed using the QiASymphony platform (Qiagen) as per the manufacturer’s instructions. All samples were eluted in 60μl of AE buffer and quantified using the High Sensitivity Qubit (Thermo Fisher Scientific, MA, USA).
We included 388 samples, predominantly from PHOENIX, and some additional samples from surgery and endoscopy (part of esophageal ICGC).
All samples were collected according to a strict SOP with quality control measures as already described. All demographic and clinical data was anonymized and stored on a central study database (OpenClinica and Labkey). The clinical characteristics of the patients are provided in Supplementary Table 1 and 2. In terms of specifics of sample collection at autopsy, the primary tumor was opened down the midline of the esophagus and the greater curve of the stomach to expose the lumen. The tumor was divided in 12 areas with sampling as shown. The size of tumors varied per case, but the division of sampling was always kept identical to preserve reproducibility. In terms of the strategy for genomic sequencing (as per Figure 1), up to 3 lymph nodes were chosen for 50x WGS in the areas shown (cervical, regional and para-aortic) and up to 24 lymph nodes in each case (8 further lymph nodes per cervical, regional and para-aortic areas (as per the Japanese Classification of nodal staging51) were chosen for the 1x WGS part of the study. At least one metastasis per solid organ was chosen for 50x WGS and for the 1x WGS part up to 8 samples were taken per organ for further analysis. In addition, 8 samples from metastatic sites which had previously been sequenced for 50x WGS were further sequenced for 1x WGS to assess the effects of metastatic heterogeneity.
Whole genome sequencing and data analysis strategy
We used the Illumina HiSeq platform to perform WGS on multiple regions collected from each primary tumor, lymph node and/or solid organ metastasis (Figure 1a,b, Supplementary Table 3, 4). All DNA extractions and WGS conformed with ICGC quality control standards and required ≥70% cellularity and a matched germline sample. WGS was performed at high depth (median coverage 66.3, IQR 56.1-87.2) to discover mutations in 122 samples from 18 patients (Supplementary Table 3, 4). In addition, low depth WGS (median coverage 1, IQR 1-5) was performed to track these mutations spatially in up to 48 solid tissue samples per case, (total=248) and 8 ctDNA samples at autopsy. Temporal tracking was performed in cases with archival biopsy material, and where historical bloods were available (Supplementary Table 12, Figure 5, Extended Data Fig. 6). For each patient the number of subclones and the cancer cell fraction within each subclone was inferred using an extension of a previously described Bayesian Dirichlet process11 and we applied a set of previously described rules to derive a phylogenetic tree (Additional Methods52). All sequencing data have been deposited in the European Genome-Phenome Archive under accession number EGAD00001005434. TP53 analysis in cell free tumor DNA (ctDNA) was performed using Digital PCR on the Bio-rad platform (Bio-rad, California) using validated TP53 assays (Supplementary Table 14).
Mutation clustering and phylogenetic tree construction
The workflow used to perform mutation clustering and phylogenetic tree construction is depicted in Extended Data Fig. 1a and illustrated with an example case, S3, in Extended Data Fig. 1b. For each patient, we inferred the number of subclones and the fraction of tumor cells within each subclone by using a previously described Bayesian Dirichlet process (BDP) to cluster mutations according to their mutation copy number49. We extended this process into n dimensions for patients with n related samples, where the number of mutant reads obtained from multiple related samples were modelled as independent binomial distributions. The BDP uses Markov chain Monte Carlo (MCMC) to sample the CCF values of the subclones in each sample. MCMC is run for 1000 iterations and outputs, for each iteration, the sampled position of each cluster, pih and the weight of each cluster, Vh, which is an estimate of the proportion of mutations assigned to that cluster. The first 200 iterations are considered as a ‘burn-in’ and are not used in subsequent steps. In order to obtain the set of subclones present within a tumor and their CCF values, the following procedure was followed:
Using the aforementioned MCMC sampling of CCF values from all n samples, for every possible triplet of samples, obtain posterior density estimates of CCF using the function kde in the R package ks, with input parameters x = pih, bandwidth = 0.1, w = Vh. Set gridsize such that density estimates are obtained to a resolution of 0.02. Identify local peaks in the posterior mutation density as locations higher than any other gridpoint within a range of 2 gridpoints. For each local peak, define a region representing a ‘basin of attraction’, defined by a set of planes running through the point of minimum density between each pair of cluster positions. Assign each mutation to the cluster in whose basin of attraction they are most likely to fall, using CCF values from MCMC sampling.
Across the set of all possible triplets, identify sets of mutations that are assigned to the same cluster in every triplet. Estimate the CCF of each cluster as the mean CCF of the mutations assigned to that cluster. Estimate the 95% confidence intervals as the [0.025,0.975] quantiles of the mean pih values of the mutations assigned to each cluster within MCMC sampling.
Finally, again using the aforementioned MCMC sampling of CCF values from all n samples, for every pair of samples, plot the mutation density, estimated using the function kde in the R package ks, with input parameters x = pih, bandwidth = 0.1, w = Vh.
Taking a conservative approach, clusters were identified as subclonal only if the 95% confidence intervals of the posterior estimate of the proportion of cells excluded the value 1. Clusters containing less than 1% of all mutations identified in a tumor were not included in phylogenetic reconstruction.
Occasionally, copy number states are incorrectly called in small regions of some cancer genomes. As a consequence, mutations falling in these regions have inaccurate estimates of CCF and can cause artefact clusters. Such clusters may be identified after mutation clustering since they contain a small percentage of mutations (less than 2.5%), the mutations within them are located in localized regions of the genome, and, often, they cannot be placed on the phylogenetic tree because they have discordant CCF values. We excluded these clusters from phylogenetic tree construction. The number of clusters excluded in total was seven (5 in P2, 1 in P3, 1 in P10). Two samples had low tumor content (36% in P3_E1, 14% in S5_T1). As a result, CCF estimates for subclones found in these samples are imprecise and led to violations of the sum rule (see below). The CCF values of the relevant clusters were manually corrected to enable them to be placed on the phylogenetic tree, as follows: P3_E1 only cluster adjusted from 1 to 0.85; S5_E1 truncal cluster adjusted from 0.85 to 1.
To determine the most likely phylogenetic tree, we applied two rules, previously described52. Briefly, the ‘sum rule’ (which is an extension of the pigeonhole principle described in Ref 11), asserts that if a subclone A is ancestral to both subclones B and C and if the summed CCFs of B and C exceed the CCF of A in any sample, the relationship between the subclones must be linear. The ‘crossing rule’ is applied to tree construction from multiple samples. It asserts that if the CCF of B is higher than the CCF of C in sample X and the CCF of B is lower than the CCF of sample C in sample Y then B and C must be in separate branches of the phylogenetic tree, i.e. they are not collinear. For all clonally related samples, the same underlying phylogenetic tree must exist. This exerts much greater stringency to the inferred ordering of subclonal clusters present in more than one sample and defines their position on the phylogenetic tree unequivocally. Note that P9 contains two independent cancers derived from Barrett’s esophagus and adenocarcinoma regions. CCF values are reported relative to the dominant cancer, so in P9_D4, which contains both cancers, the two cancers are reported with CCFs of 100% and 69%. This apparent violation of the sum rule results from the mathematical convenience of normalizing to the dominant cancer.
It should be noted that the sum rule and crossing rule only strictly apply when the infinite sites assumption (ISA) is obeyed. The ISA states that each mutation only occurs once during the lifetime of a tumor and that mutations never revert to normal. A recent study53 has shown, through analysis of targeted sequencing of single cells, that the ISA is not always followed in real data, for two reasons:
Copy number alterations (CNAs), specifically losses and loss of heterozygosity, have the effect of removing mutations in the deleted region, resulting in the apparent ‘reversion’ of a mutation.
The same mutation may occur on more than one occasion, particularly if the mutation is a driver mutation.
In our study, we take account of CNAs when calculating the CCF of each mutation. In regions that have undergone gain of one or both alleles, a mutation may be present on more than one chromosome copy, up to the number of copies of the most amplified chromosome copy. Conversely, if one or both chromosome copies have undergone loss in a particular sample, a mutation may be lost in that sample. In the situation where a mutation is unobserved in a sample and that sample has a copy number state lower than that observed in another sample in which the mutation is observed, we do not call the mutation as absent. Rather, we cluster it based on its CCF in the remaining samples, treating its CCF in the target sample as unknown.
Identification of cancer cell fraction
For each mutation we calculated the mutation copy number as previously described, using the mutant allele burden, tumor cellularity and locus specific copy number in the tumor and matched normal49. The mutation copy number reflects the percentage of tumor cells within a sample carrying that mutation, and permits the cross-comparison of the mutation in related samples despite differences in tumor purity and/ or copy number profiles. Mutations present on multiple copies of a chromosomal segment will have a mutation copy number greater than 1. To group mutations according to the percentage of cells containing it, or cancer cell fraction (CCF), the number of chromosomes carrying the mutation must be determined. For all mutations within amplified regions with a major allele copy number, the observed fraction of mutated reads was compared to the expected fraction of mutated reads resulting from a mutation present assuming a binomial distribution37.
Annotation of the trees with mutations
We annotated each tree with oncogenic or putative oncogenic alterations including substitutions and copy number changes. For substitutions, cluster assignment information from a multidimensional Dirichlet process was used.
For rearrangements and copy number changes, branch assignment was achieved by considering the set of samples containing the variant and the subclonal fraction of the associated copy number segment where applicable. All potential driver alterations were annotated. For substitutions, structural variants and copy number events, these included a set of genes compiled from the TARGET database from the Broad Institute and multiple sequencing datasets for OAC14–16,18,19.
Shallow Whole Genome Sequencing for Subclone Identification
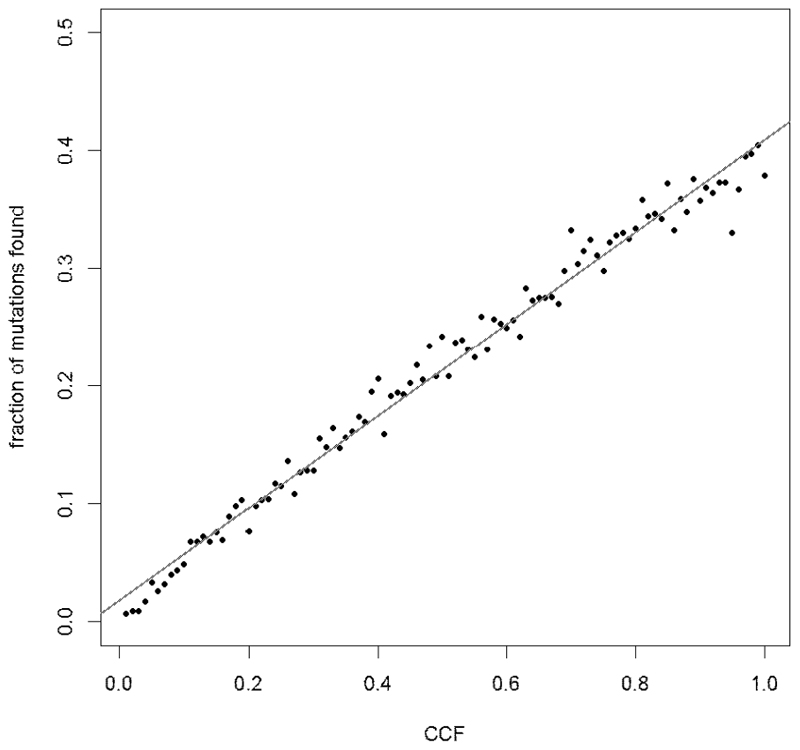
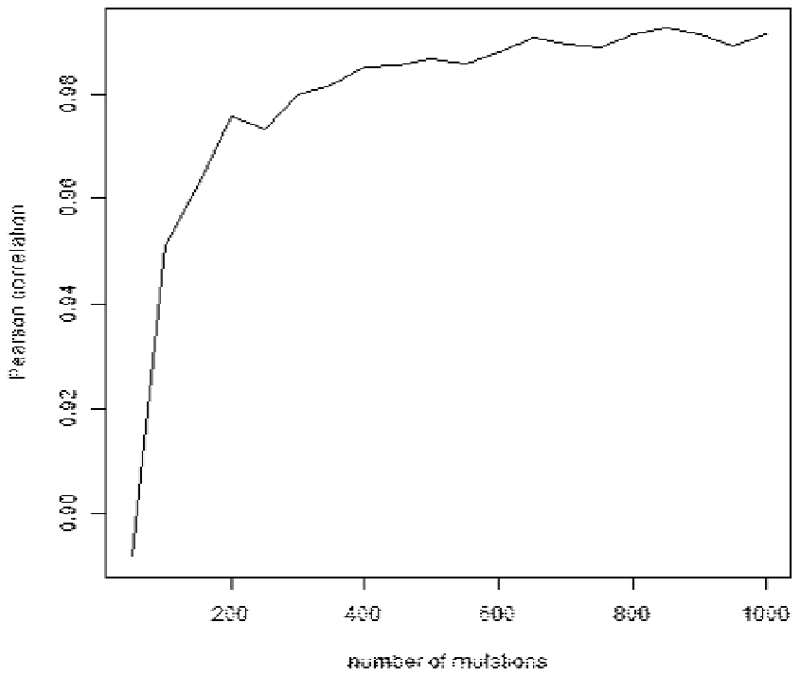
For shallow whole genome sequencing, samples were sequenced to a median depth of ~1x. It was not therefore feasible to call mutations de novo for these samples, but we were able to count the number of mutations from each subclone that reported a mutant read in 1x WGS sequencing. We performed simulations of 1x WGS data in order to ascertain the correlation between the number of mutations identified and the CCF of each subclone. First, we simulated subclones with CCF values between 0.01 and 1.00, assuming 1000 mutations per subclone, sequencing depth drawn from a Poisson distribution with expected value 1, and binomial sampling of WT and mutant reads. The correlation between the number of mutations detected and the CCF of the subclone was very high (Pearson r = 0.992, Extended Data Fig. 4). In order to test whether subclones containing fewer mutations also had good correlations between CCF and number of detected mutations, we performed further simulations of subclones containing between 50 and 1,000 mutations and ascertained that the correlation remained very high (> 0.997) for cluster sizes as small as 200 (Extended Data Fig. 5). Of the 169 subclones identified in our study, only two contained fewer than 200 mutations, indicating that the number of mutations detected is a good proxy for the CCF of a subclone.
SNVs from libraries sequenced to a minimum of 1x following filtering, were allocated to subclones previously identified at 50x WGS. Mapping quality and base quality of 10 were used. This resulted in tabulated counts for SNVs being allocated to subclones identified at 50x WGS for each sample. Normalization was performed according to the number of SNVs assigned to each subclone from 50x WGS, and to the total number of SNVs in that sample in order to account for potential differences in coverage, using the following equation:
in which ncluster and ntruncal are the numbers of loci in the target cluster and the truncal cluster that have mutant reads in the target sample and Hcluster and Htruncal are the number of mutations identified from 50x WGS in the target and truncal clusters. For each 1x WGS sample, this provides an estimate of the CCF of each subclone within that sample.
In all cases, near equal coverage was obtained and in cases of low cellularity further sequencing was performed in order to achieve this. After normalization, the GENE-E package (https://software.broadinstitute.org/GENE-E/download.html) was used to cluster the 1x WGS samples according to the similarity of their CCF profiles using Pearson correlation.
Extended Data
Extended Data Fig. 1. Flowchart describing key steps taken to construct phylogenetic trees.
A. The details of phylogenetic tree reconstruction is further elaborated in Supplementary methods, Mutation clustering and phylogenetic tree construction (p.25).
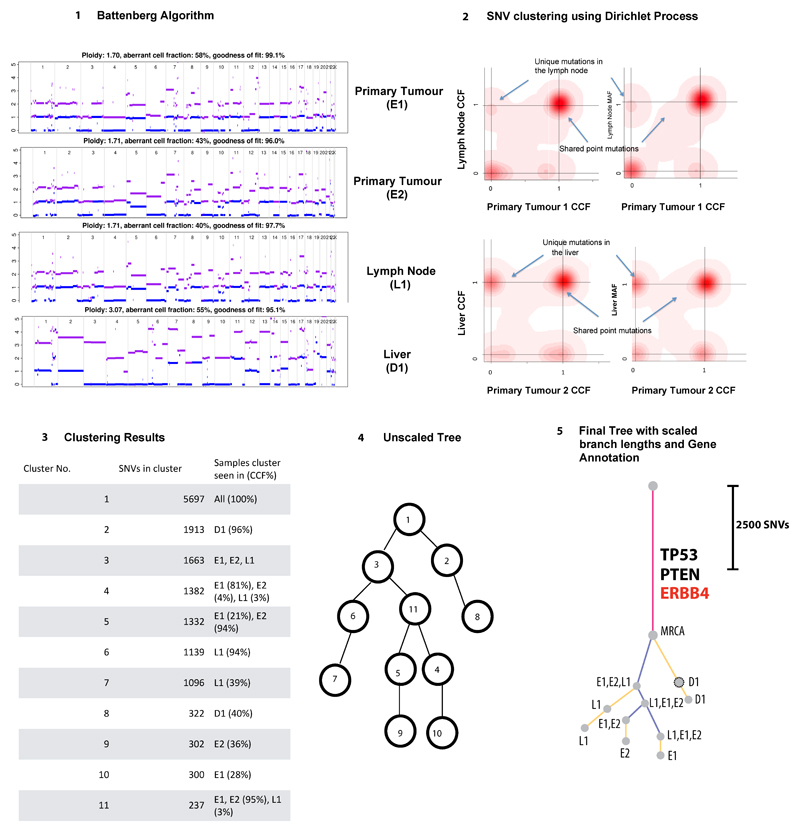
Extended Data Fig. 2. Phylogenetic tree construction for example case S3.
1) Battenberg algorithm to determine total copy number (purple line) and minor allele (blue line). Y-axis =number of chromosome copies, X-axis= chromosome and position. The average ploidy, aberrant cell fraction (cellularity) and goodness of fit to the model are shown for each sample, Primary E1, E2, Lymph node L1 and Distant metastasis D1. The goodness of fit is a measure of the amount of the genome with clonal, rather than subclonal copy number states. D1 has a subclonal mix of different copy number states resulting in noninteger total copy number, for example on chromosome 2, resulting in a goodness of fit below 100%. 2) Bayesian Dirichlet Process to cluster SNVs based on CCF in each sample. The density plots show the posterior probability of a mutational cluster, these are produced for every pair of samples and selected plots are shown High density at CCF of (0,0) indicates subclones that are not present in the pair of samples shown in a particular plot. 3) Clustering of results – Clusters are identified as local maxima in the posterior density. The table shows the number of SNVs assigned to each cluster, and their associated CCFs. 4) Unscaled Tree construction using the sum rule and crossing rule as detailed in Supplementary Methods p25. 5) Final Tree -The tree is drawn as seen in Figures 2 and Extended Data2, branch lengths are proportional to the number of SNVs assigned to each subclone. Scales vary on a per case basis depending on the total number of SNVs, in order to fit cases on one figure. Trees are annotated with the gene names of known drivers, and the colour of each branch represents a trunk (pink), branch (purple) or leaf (yellow). The grey circles represent clones and subclones and their CCFs are shown in Supplementary Table5 and 6.
Extended Data Fig. 3. Phylogenetic trees of cases in cohort with only nodal or distant organ disease, as derived from H-WGS.
E=esophagus, D=distant organ, L=lymph node, B= Barrett’s. For precise anatomical locations, refer to Supplementary Table3 and 4. MRCA=most recent common ancestor. Pink=trunk (shared events), Purple=branch (shared by more than one sample), Yellow=leaf (unique to one sample). Grey dots at the end of the lines (truncal, branches or leaves) represent subclones or clones, whose CCFs are shown in Supplementary Tables5 and 6. Trees are annotated with key driver events as identified from the literature14,16,19. Black=point mutations, Red=copy number alterations, purple= structural variants. The adjacent scales are relative to the number of SNVs in that particular case and hence constructed on a case by case basis.
Extended Data Fig. 4. Structural variation of 18 metastatic esophageal adenocarcinoma cases.
a. Similarity matrix based clustering for all SVs 122 genomes across 19 cases. SVs were deemed to refer to the same rearrangement event across cases if their corresponding breakpoint locations fell within a window of maximum 50 bp. The individual sample types are shown as a separate row on the x axis with the color key depicting the sample type. The purple scale indicates the number of shared SVs. (L=lymph nodes; M=metastasis; T=tumor). b. Histogram showing the percentage of rearranged genes that are concordant, unique to tumors and unique to metastases. Two-tailed Welch test P=0.2674 demonstrating no overall difference between total number of SVs in primary, local lymph nodes and distant metastases c. Stacked bar charts showing the composition of various SVs in each sample on a per patient basis INV= inversion, ME= mobile element, BND= translocation DEL=deletion, DUP=duplication, INS= insertion.
Extended Data Fig. 5. Random simulation model for S-WGS cluster detection.
The number of mutations detected correlates strongly with the CCF of the cluster (Pearson r=0.992, n=100). Number of mutations in each cluster =1000.
Extended Data Fig. 6. Correlation of fraction of mutations detected with CCF as a function of cluster size using simulated S-WGS data.
Pearson correlation coefficient is above 0.97 for clusters with 200 or more mutations.
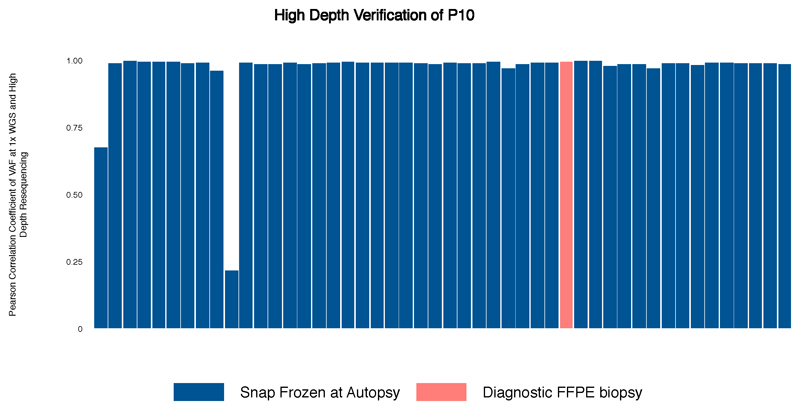
Extended Data Fig. 7. Bar chart demonstrating the Pearson correlation coefficient of VAF at 1xWGS and High Depth Resequencing (n=33).
Extended Data Fig. 8. Detection of Selection in subsets of mutations.
SNVs and indels from all cases (n=18) were aggregated into 4 different subsets: clonal = variants found in the MRCA (n=378453); subclonal = variants not found in the MRCA (n=516136); pre-diaspora = variants found above the diaspora founder clone in the phylogenetic tree (n=313545); post-diaspora = variants found in the diaspora founder or in clones below the founder in the phylogenetic tree (n=295316). Within each subset, dN/dS analysis was performed separately on: missense variants; truncating variants. Bars show maximum likelihood estimates of dN/dS values, with values greater than 1 (dashed line) indicating positive selection. Vertical lines = 95% confidence intervals, estimated using Wald test.
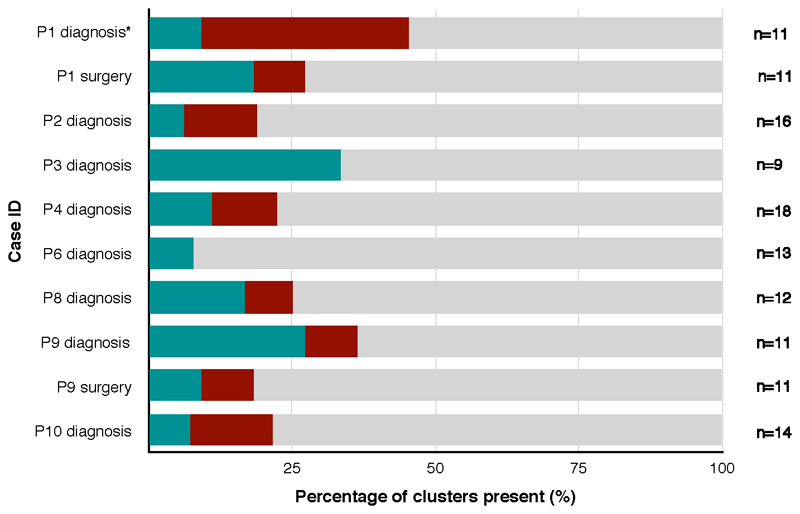
Extended Data Fig. 9. Percentage of truncal and branch clusters in tissue from earlier time-points.
Stacked horizontal bar chart representing the percentage of truncal and branch clusters present in tissue from earlier time-points on the x-axis and the Case ID on the y-axis. P1 diagnosis* is a frozen sample, while the rest are FFPE. Blue = truncal, maroon = branch, grey = not present. The number of clusters (n) is demonstrated for each case.
Extended Data Fig. 10. ctDNA analysis from historical plasma samples.
Digital PCR traces of mutant allele fraction for TP53 on the Y-axis and days from diagnosis on the X-axis, and grey areas indicate periods of therapy. Where subclones and clones are seen at 1xWGS on plasma, they are highlighted on the 50x phylogenetic tree (coloured blue). The samples in which these subcloens and clones are present in are shown in Supplementary Table3. There was no TP53 data for S3 as it was wild type for TP53 mutations. Copy number traces for P1 are shown, with the arrow demonstrating an area of MET amplification.
Supplementary Material
Acknowledgements
Above all, we are indebted to the patients who donated tissue samples to this project and thank them and their families who supported them through it. We would also like to thank the following individuals for their help with study set-up, patient liaison and tissue collection, Ben Smith, Nyrai Chinyama, Vijay Sujendran, Peter Safranek, Athanosios Xanthos, Tara Nuckcheddy-Grant, Rachel de la Rue, Sebastian Zeki, Rachael Fels Elliott, Peter Collins, Kitty Puttock, Sophie Rabey and staff at Arthur Rank Hospice and Luke A Wylie for scientific discussion and contribution. We would like to thank the Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium for providing the vehicle through which funding for the International Cancer Genome Consortium (ICGC) was obtained. We are grateful to Professor Simon Tavaré, FRS for his guidance and support for the esophageal whole genome sequencing project as a part of the International Cancer Genome Consortium (ICGC). We would like to thank Jo Westmoreland, LMB visual aids for her graphic art expertise. Thanks also go to the Cancer Research UK Cambridge Institute Genomics Core for their technical expertise. We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, from Addenbrooke’s Hospital. Additional infrastructure support was provided from the CRUK funded Experimental Cancer Medicine Centre in Cambridge. Computation by DCW used the Oxford Biomedical Research Computing (BMRC) facility, a joint development between the Wellcome Centre for Human Genetics and the Big Data Institute supported by Health Data Research UK and the NIHR Oxford Biomedical Research Centre.
Ayesha Noorani was funded through an MRC Clinical Research Fellowship. The work was funded through the above and an MRC core grant (RG84369) and an NIHR Research Professorship (RG67258) to Rebecca Fitzgerald. Funding for sample sequencing (50x WGS) was through the International Cancer Genome Consortium and was funded by a programme grant from Cancer Research UK (RG66287). All OCCAMS samples which were part of the surgical/endoscopy cohort were obtained from Cambridge patients. David Wedge is funded by the Li Ka Shing foundation and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre.
Footnotes
Data Availability
Sequencing data that support the findings of this paper have been deposited in the European Genome-phenome Archive with the accession code EGAD00001005434.
Code Availability
All code required to reproduce the analysis outline in this manuscript can be found in the main and supplementary methods. There are no restrictions to the accessibility of this code.
Author Contributions
AN designed and implemented the rapid autopsy study, collected the samples, performed the experiments, analyzed data and wrote the manuscript. MG and S.D.P contributed expertise in pathology and sample collection for the rapid autopsy study. ID-B and NG assisted in study implementation, and along with JC, assisted with sample collection at autopsy. M.S performed the structural variant analysis. M.D.E performed genomic data generation and QC. LB conducted data management. XL, PL-S and JW were involved with autopsy sample collection, advice on experiments and data analysis, and XL contributed to experiments, paper writing, and figure design. LA and IM assisted with data analysis. NG assisted with study Implementation. SMac coordinated the sequencing of samples from the OCCAMS project and contributed to paper writing. SM and AM provided pathology data. TT, SG, LP and DG assisted in implementation and ethical conduct of the autopsy study. R.H.H and AH were involved in surgical sample collection and providing surgical expertise. M.R.S contributed to critical evaluation of the study data and manuscript. D.C.W was responsible for data analysis, paper writing, and assuring integrity of data. The OCCAMS consortium was the vehicle through which the infrastructure and funding was obtained to support the study and the consortium contributed to discussions on the ICGC data and the clinical ramifications. R.C.F provided grant funding and was responsible for study design, supervision of the project, writing the paper and assuring integrity of the data.
The authors declare no competing interests.
References
- 1.Sporn MB. The war on cancer. Lancet. 1996;347:1377–81. doi: 10.1016/s0140-6736(96)91015-6. [DOI] [PubMed] [Google Scholar]
- 2.Waterman TA, et al. The prognostic importance of immunohistochemically detected node metastases in resected esophageal adenocarcinoma. Ann Thorac Surg. 2004;78:1161–9. doi: 10.1016/j.athoracsur.2004.04.045. discussion 1161-9. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda S, Takeuchi H, Kawakubo H, Kitagawa Y. Three-field lymph node dissection in esophageal cancer surgery. J Thorac Dis. 2017;9:S731–S740. doi: 10.21037/jtd.2017.03.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lou F, et al. Esophageal cancer recurrence patterns and implications for surveillance. J Thorac Oncol. 2013;8:1558–62. doi: 10.1097/01.JTO.0000437420.38972.fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smyth EC, et al. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham D, et al. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2008;358:36–46. doi: 10.1056/NEJMoa073149. [DOI] [PubMed] [Google Scholar]
- 7.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–12. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 9.Davis A, Gao R, Navin N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim Biophys Acta. 2017;1867:151–161. doi: 10.1016/j.bbcan.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murugaesu N, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–831. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstung M, et al. The evolutionary history of 2,658 cancers. bioRxiv. 2017 [Google Scholar]
- 14.Secrier M, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131–41. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulak AM, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–93. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver JM, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014;46:837–43. doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross-Innes CS, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–46. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nones K, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2014;5 doi: 10.1038/ncomms6224. 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frankell AM, et al. The landscape of selection in 551 Esophageal Adenocarcinomas defines genomic biomarkers for the clinic. bioRxiv. 2018 doi: 10.1038/s41588-018-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–9. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez-Martin B, et al. Pan-cancer analysis of whole genomes reveals driver rearrangements promoted by LINE-1 retrotransposition in human tumours. bioRxiv. 2018 179705. [Google Scholar]
- 23.Ajani JA, et al. Esophageal and esophagogastric junction cancers, version 1.2015. J Natl Compr Canc Netw. 2015;13:194–227. doi: 10.6004/jnccn.2015.0028. [DOI] [PubMed] [Google Scholar]
- 24.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–14. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariette C, et al. Pattern of recurrence following complete resection of esophageal carcinoma and factors predictive of recurrent disease. Cancer. 2003;97:1616–23. doi: 10.1002/cncr.11228. [DOI] [PubMed] [Google Scholar]
- 27.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02320-7. 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behjati S, et al. Mutational signatures of ionizing radiation in second malignancies. Nat Commun. 2016;7 doi: 10.1038/ncomms12605. 12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dentro SC, et al. Portraits of genetic intra-tumour heterogeneity and subclonal selection across cancer types. bioRxiv. 2018 [Google Scholar]
- 31.Lodato MA, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359:555–559. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Z, Wyman MJ, Sella G, Przeworski M. Interpreting the Dependence of Mutation Rates on Age and Time. PLoS Biol. 2016;14:e1002355. doi: 10.1371/journal.pbio.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letouze E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01358-x. 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–7. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pienta KJ, Robertson BA, Coffey DS, Taichman RS. The cancer diaspora: Metastasis beyond the seed and soil hypothesis. Clin Cancer Res. 2013;19:5849–55. doi: 10.1158/1078-0432.CCR-13-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foulds L. The experimental study of tumor progression: a review. Cancer Res. 1954;14:327–39. [PubMed] [Google Scholar]
- 39.Naxerova K, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357:55–60. doi: 10.1126/science.aai8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–16. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sjoquist KM, et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12:681–92. doi: 10.1016/S1470-2045(11)70142-5. [DOI] [PubMed] [Google Scholar]
- 42.Gabriel E, et al. Novel Calculator to Estimate Overall Survival Benefit from Neoadjuvant Chemoradiation in Patients with Esophageal Adenocarcinoma. J Am Coll Surg. 2017;224:884–894 e1. doi: 10.1016/j.jamcollsurg.2017.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burt BM, et al. Utility of Adjuvant Chemotherapy After Neoadjuvant Chemoradiation and Esophagectomy for Esophageal Cancer. Ann Surg. 2017;266:297–304. doi: 10.1097/SLA.0000000000001954. [DOI] [PubMed] [Google Scholar]
- 44.Pasquali S, et al. Survival After Neoadjuvant and Adjuvant Treatments Compared to Surgery Alone for Resectable Esophageal Carcinoma: A Network Meta-analysis. Ann Surg. 2017;265:481–491. doi: 10.1097/SLA.0000000000001905. [DOI] [PubMed] [Google Scholar]
- 45.Parikh AR, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–1421. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Roy N, et al. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clin Cancer Res. 2017;23:6305–6314. doi: 10.1158/1078-0432.CCR-17-0675. [DOI] [PubMed] [Google Scholar]
- 47.Hu Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 2019;51:1113–1122. doi: 10.1038/s41588-019-0423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5 doi: 10.1038/ncomms3997. 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martincorena Inigo, R KM, Gerstung Moritz, Dawson Kevin J, Haase Kerstin, Van Loo Peter, Davies Helen, Michael R, Stratton Michael R, Campbell Peter J. Universal Patterns Of Selection In Cancer And Somatic Tissues. Cell. 2017 doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Japanese Gastric Cancer, A. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011;14:101–12. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 52.Jiao W, Vembu S, Deshwar AG, Stein L, Morris Q. Inferring clonal evolution of tumors from single nucleotide somatic mutations. BMC Bioinformatics. 2014;15:35. doi: 10.1186/1471-2105-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuipers J, Jahn K, Raphael BJ, Beerenwinkel N. Single-cell sequencing data reveal widespread recurrence and loss of mutational hits in the life histories of tumors. Genome Res. 2017;27:1885–1894. doi: 10.1101/gr.220707.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.