Increasing evidence demonstrates that perturbation of excitation-contraction coupling in cardiomyocytes leads to cardiac dysfunction and heart failure 1. Critical to excitation-contraction coupling are cardiac dyads, microdomains at membrane contact sites between transverse-tubules (t-tubules) and junctional sarcoplasmic reticulum (SR) membranes 1, 2. Junctophilin 2 (JPH2), spanning the cleft from the T-tubule to the SR, plays a central role in formation and maintenance of T-tubule/SR junctions 3. Multiple mutations in JPH2 are associated with human cardiomyopathies. Consequently, there is growing interest in increasing our understanding of proteins that interact with JPH2 to comprise the cardiac dyad proteome.
Methods that rely on affinity pulldown coupled with mass spectrometry to characterize membrane protein-protein interactions (PPIs) are subject to limitations owing to the hydrophobic nature of membrane proteins that necessitates harsh extraction conditions, thus potentially disrupting relatively weak or transient PPIs. To overcome these limitations, a biotin ligase-based system, biotin identification (BioID), has been developed to assess PPIs 4. BioID has proved to be a useful strategy to screen for PPIs. However, this method has the limitation of artefacts that might arise owing to overexpression of the transfected or transduced target-biotin ligase fusion protein. To overcome these limitations, we developed a mouse knock-in strategy utilizing BioID2, a second generation of BioID 4, fusing BioID2 to the endogenous JPH2 coding sequence, and used this to investigate the cardiac dyad proteome in living cardiomyocytes.
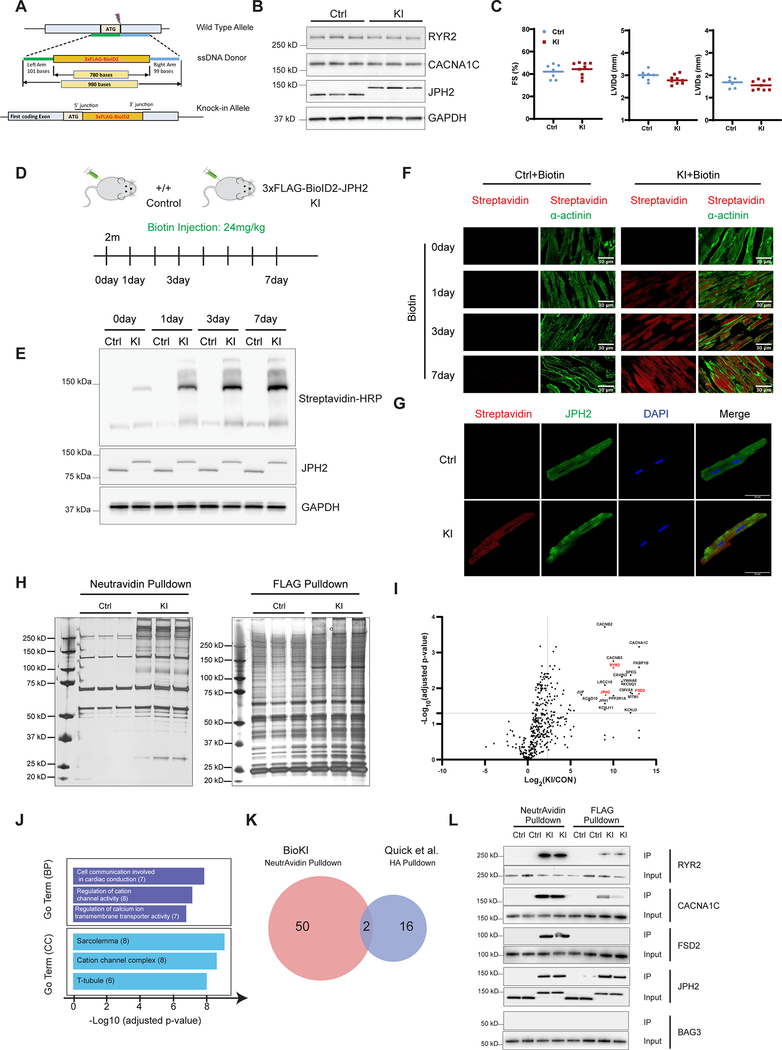
To knock-in the 3xFlag-BioID2 cassette into the JPH2 locus, we utilized the CRISPR-Cas9 genome-targeting system (Figure A). All mouse protocols were approved by Institutional Animal Care and Use Committee. The 3xFlag-BioID2-JPH2 fusion protein was expressed at levels equivalent to those of wild-type JPH2 protein (Figure B). Protein levels of two well-known JPH2 binding partners in junctional membrane complexes (RYR2 and CACNA1C) were not altered (Figure B). We also found no significant differences in cardiac function between homozygous KI and control mice (Figure C). Altogether, these data confirmed that expression of the BioID2-JPH2 fusion protein had no adverse effects on JPH2 expression and function. Immunoblot and immunofluorescence analyses with streptavidin antibody demonstrated that self-biotinylation and proximity labeling saturation in homozygous KI mouse hearts was achieved following 3 days of biotin administration (Figure [D–F]). Confocal imaging of isolated cardiomyocytes from BioID2-JPH2 KI mice demonstrated striking co-localization of JPH2 antibody signal with the streptavidin signal (Figure G). To identify biotinylated proteins that were in proximity to JPH2 within the cardiac dyad, we performed a NeutrAvidin pulldown and a FLAG affinity purification on whole heart lysates from KI mice and controls. As shown in Figure H, NeutrAvidin pulldowns were more specific and resulted in retrieving greater amounts of JPH2 proximal proteins than comparable FLAG affinity pulldowns. Characterization of biotinylated proteins utilizing label-free MS/MS enabled identification of 550 (> 2 peptides for each protein) JPH2-proximal proteins in homozygous KI mice. The distribution of biotinylated proteins between control and homozygous KI mice was illustrated using a Volcano plot (Figure I). Self-biotinylated JPH2 and known interacting partners within the cardiac dyad, such as RYR2 and CACN1AC were highly enriched in the fraction from KI mice. Gene Ontology (GO) enrichment analysis showed that the top cellular components were sarcolemma, cation channel complexes, and T-tubules (Figure J).
Figure. BioID1 knock-in mouse strategy identifies cardiac dyad proteome in vivo.
A, The JPH2 wild-type allele, the 3xFlag-BioID2 ssDNA donor designed for insertion and the final knock-in allele. The lengths of ssDNA and homology arms are shown. B, Western blot analysis of RYR2, CACNA1C and JPH2 in heart tissues from Ctrl and homozygous KI mice at 2 months of age. GAPDH served as a loading control. C, Echocardiographic measurements for Ctrl (n = 7) and homozygous KI (n = 9) mice at 2 months of age. Graphs indicates no significant changes between KI and Ctrl mice in: percentage fractional shortening (%FS), left ventricular internal diameter at end-diastole (LVIDd) and left ventricular internal diameter at end-systole (LVIDs). An unpaired t-test was performed with a confidence of 95% (significant p-value <0.05) D, Schematic overview of Biotin injection. Adult (2-month-old) Ctrl and homozygous KI mice were treated with biotin by intraperitoneal injection once daily at a dose of 24 mg/kg body weight for seven days. Mouse hearts were collected at indicated days after biotin injection. E, Western blot analysis of biotinylated proteins in mouse heart for indicated days. Biotinylated proteins were detected with HRP-conjugated streptavidin after SDS-PAGE separation. F, Immunofluorescence staining of biotinylated proteins (streptavidin, red) and α-actinin (green) in mouse heart sections by biotin injection for indicated days. G, Adult cardiac myocytes isolated from either Ctrl or homozygous KI mice after 3 days of biotin injection were fixed and co-stained of biotinylated proteins (streptavidin, red) and JPH2 (green) antibodies. H, Silver staining of eluted proteins of NeutrAvidin pulldown and FLAG pulldown from Ctrl and homozygous KI mice hearts. I, Volcano plot from Mass Spectrometry data of differentially biotinylated proteins between control (Ctrl) and homozygous KI mice (n = 3). We calculated the adjusted P values using the Benjamini-Hochberg algorithm; proteins were considered significantly enriched if they had an adjusted P value <0.05 and a fold change >5. Top 20 significant proteins are reported and several known cardiac dyadic proteins are highlighted in red. J, GO enrichment analysis of differentially biotinylated proteins by proteomics. Number of genes belonging to each category is indicated. BP: biological process, CC: cell component. K, Overlapping- proteins identified as likely specific interactors of JPH2 in our BioKI method and in Quick et al HA-immunoprecipitated method. L, Western blot analysis of eluted proteins from NeutrAvidin pulldown and FLAG pulldown with antibodies against RYR2, CACNA1C, FSD2, JPH2 and BAG3.
Quick and colleagues recently identified cardiac dyad binding partners/neighboring proteins of JPH2 using a transgenic mouse line overexpressing a JPH2-HA fusion protein specifically in cardiomyocytes 5. Out of 18 proteins suggested to be specific interactors of JPH2 by Quick et al, only two (RYR2 and SPEG) overlap with proteins found to be proximal to JPH2 by our approach (Figure K). Notably, important members of the cardiac dyad known to interact with JPH2, such as the L-type calcium channel proteins (e.g. CACNA1C, CACNB2, CACNB3) and newly discovered potential dyadic proteins (e.g. FSD2, LRRC10) were detected utilizing our BioID2 knock-in (BioKI) strategy, but were not detected by the JPH2-HA transgenic approach, attesting to the greater sensitivity and specificity of our BioKI strategy. We next performed pulldown analyses with NeutrAvidin or Flag antibodies, followed by Western blot analyses for known cardiac dyad proteins (RYR2, CACNA1C and FSD2) and non-dyad proteins Bag3 (negative control) (Figure L). Results again confirmed that levels of biotinylated proteins from NeutrAvidin pulldown were much higher than levels of the same proteins precipitated by FLAG pulldown.
In summary, our study represents proof of concept for the use of BioKI in the study of PPIs in vivo, where the target protein fused to the biotin ligase is present at levels comparable to those of the endogenous protein, obviating potential artefacts arising from overexpressed target-fusion proteins. Our data indicated that BioKI was more sensitive and specific than other methods and provided a robust method to identify membrane PPIs in living mice. In addition, our work also identified some potential novel cardiac dyad proteins which may play essential roles in cardiac dyad formation, maintenance, and function.
Acknowledgments
Sources of Funding
JC and SME are funded by grants from the National Institutes of Health, the National Heart, Lung, and Blood Institute, and JC holds an American Heart Association Endowed Chair in Cardiovascular Research.
Footnotes
Disclosures
None.
Data Sharing: The data, analytical methods, and study materials that support the findings of this study will be available to other researchers from the corresponding authors on reasonable request. The MS raw data have been deposited in MassIVE: MSV000084352.
References
- 1.Hong T and Shaw RM. Cardiac T-Tubule Microanatomy and Function. Physiol Rev. 2017;97:227–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu C, Spinozzi S, Chen JY, Fang X, Feng W, Perkins G, Cattaneo P, Guimaraes-Camboa N, Dalton ND, Peterson KL, Wu T, Ouyang K, Fu XD, Evans SM and Chen J. Nexilin Is a New Component of Junctional Membrane Complexes Required for Cardiac T-Tubule Formation. Circulation. 2019;140:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo A, Wang Y, Chen B, Wang Y, Yuan J, Zhang L, Hall D, Wu J, Shi Y, Zhu Q, Chen C, Thiel WH, Zhan X, Weiss RM, Zhan F, Musselman CA, Pufall M, Zhu W, Au KF, Hong J, Anderson ME, Grueter CE and Song LS. E-C coupling structural protein junctophilin-2 encodes a stress-adaptive transcription regulator. Science. 2018;362:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gingras AC, Abe KT and Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr Opin Chem Biol. 2019;48:44–54. [DOI] [PubMed] [Google Scholar]
- 5.Quick AP, Wang Q, Philippen LE, Barreto-Torres G, Chiang DY, Beavers D, Wang G, Khalid M, Reynolds JO, Campbell HM, Showell J, McCauley MD, Scholten A and Wehrens XH. SPEG (Striated Muscle Preferentially Expressed Protein Kinase) Is Essential for Cardiac Function by Regulating Junctional Membrane Complex Activity. Circ Res. 2017;120:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]