Abstract
Listeria monocytogenes is a rapidly growing, Gram-positive, facultative intracellular pathogen that has been used for over 5 decades as a model to study basic aspects of infection and immunity. In a murine intravenous infection model, immunization with a sublethal infection of L. monocytogenes initially leads to rapid intracellular multiplication followed by clearance of the bacteria and ultimately culminates in the development of long-lived cell-mediated immunity (CMI) mediated by antigen-specific CD8+ cytotoxic T cells. Importantly, effective immunization requires live, replicating bacteria. In this review, we summarize the cell and immunobiology of L. monocytogenes infection and discuss aspects of its pathogenesis that we suspect lead to robust CMI. We suggest 5 specific features of L. monocytogenes infection that positively impact the development of CMI: (1) the bacteria have a predilection for professional antigen-presenting cells; (2) the bacteria escape from phagosomes, grow, and secrete antigens into the host cell cytosol; (3) bacterial secreted proteins enter the MHC class I pathway of antigen processing and presentation (4) the bacteria do not induce rapid host cell death; (5) cytosolic bacteria induce a cytokine response that favors CMI. Collectively, these features make L. monocytogenes an attractive vaccine vector for both infectious disease applications and cancer immunotherapy.
Keywords: bacteria, intracellular pathogen, macrophage, inflammasome, cancer, tumor, vaccine, dendritic cell
A brief primer on L. monocytogenes
L. monocytogenes is a rapidly growing, easily manipulated, Gram-positive bacterium that is taxonomically placed in the Firmicute phylum most closely related to bacterial members of the Bacilli, Lactobacilli and Enterococci (Radoshevich & Cossart, 2018). L. monocytogenes is a ubiquitous environmental inhabitant that lives a biphasic lifestyle as both a saprophyte and as a pathogen of many warm-blooded animals including livestock and humans (Freitag, Port, & Miner, 2009). L. monocytogenes is a common contaminant of a variety of fresh and processed foods. Humans often consume L. monocytogenes contaminated food, but the most identified illness occurs in pregnant women, neonates, the elderly, and individuals whose immune system is compromised where it often causes meningitis and CNS infection (Schlech, 2019). Although disease is relatively rare, it is often fatal, and in the developed world, represents a leading cause of death due to food-borne illnesses.
Although natural infection is clearly via the oral route, most basic research has been conducted in mice using either intravenous or intraperitoneal routes of administration. Indeed, beginning with the classic work of George Mackaness in the 1960s, L. monocytogenes emerged as a highly quantitative and reproducible murine model system to study basic aspects of innate and adaptive immunity (D’Orazio, 2019; Mackaness, 1962; McGregor, Koster, & Mackaness, 1970). To very briefly summarize decades of research, an effective innate immune response to L. monocytogenes is sufficient to contain the infection and relies on the orchestrated influx of neutrophils and macrophages to the sites of infection followed by the activation of macrophage bactericidal activity. Mice that lack B and T-cells do not succumb to infection, but are unable to clear the bacteria (Bancroft, Schreiber, Bosma, Bosma, & Unanue, 1987; Bhardwaj, Kanagawa, Swanson, & Unanue, 1998). In contrast, conventional mice that survive a primary challenge with a sub-lethal dose of L. monocytogenes clear the infection within 7–10 days and become highly resistant to a subsequent lethal challenge. Long-lived adaptive immunity is antibody-independent and depends on the expansion of antigen-specific CD8+ T cells and establishment of memory cells (cell-mediated immunity or CMI). Importantly, induction of adaptive immunity requires live, replicating bacteria; i.e., killed vaccines do not induce protective immunity (Berche, Gaillard, & Sansonetti, 1987; Von Koenig, Finger, & Hof, 1982). The observation that L. monocytogenes induces T-cell-mediated immunity suggested to numerous investigators that it might represent a highly amenable and potent recombinant vaccine vector for the induction of CMI (Goossens, Milon, Cossart, & Saron, 1995; Ikonomidis, Paterson, Kos, & Portnoy, 1994; Shen et al., 1995). Indeed, L. monocytogenes-based vaccines have been developed as therapeutic vaccines for cancer immunotherapy that have shown promising results in clinical trials (Flickinger, Rodeck, & Snook, 2018). Below, we describe the biological features of L. monocytogenes that make it such as potent inducer of CMI.
Cell biology of infection
Shortly after IV infection with L. monocytogenes, most of the bacteria are found within macrophages and dendritic cells (DCs) of the spleen and liver where the majority of bacteria are killed, but some of the bacteria escape from phagocytic vacuoles, replicate rapidly in the cytosol of infected cells and exploit host actin dynamics to spread to neighboring cells (Portnoy, Auerbuch, & Glomski, 2002; Serbina, Shi, & Pamer, 2012; Waite et al., 2011). This infectious process occurs in most, if not all adherent cells, including primary or immortalized cultures of bone marrow-derived macrophages (BMMs) which are ideal cells for studies on innate immunity (see below) and can be activated with IFN-γ to kill and degrade phagosomal bacteria (Herskovits, Auerbuch, & Portnoy, 2007).
Listeriolysin O (LLO), a cholesterol dependent cytolysin (CDC), is a secreted virulence factor and a primary determinant of L. monocytogenes pathogenesis (Nguyen, Peterson, & Portnoy, 2019). LLO is necessary to escape from both a primary phagosome and the secondary vacuole that forms upon cell-to-cell spread. LLO-damaged phagosomes and free bacteria in the cytosol are recognized by the host autophagy machinery, but the bacteria secrete two phospholipases C (PlcA/B), and utilize actin-based motility, that together allow them to bypass autophagy (Cheng, Chen, Engstrom, Portnoy, & Mitchell, 2018; Mitchell et al., 2018). Importantly, mutants lacking LLO are incapable of growth in BMMs and are 5-logs less virulent in mice (Portnoy, Jacks, & Hinrichs, 1988). Although LLO activity is essential, it is potentially cytotoxic and its activity must be compartmentalized to acidic cellular compartments. L. monocytogenes mutants that fail to properly restrict LLO activity to vacuoles kill the infected macrophage in vitro and are rendered avirulent in vivo. There are multiple mechanisms used by LLO to prevent cellular toxicity, which was recently reviewed (Nguyen et al., 2019).
Upon entering the host cell cytosol, L. monocytogenes dramatically up-regulates the expression of a cell surface transmembrane protein called ActA, which recruits and activates the host Arp2/3 complex to induce the polymerization of host actin filaments leading to intra- and intercellular spread (Gouin, Welch, & Cossart, 2005; Pillich, Puri, & Chakraborty, 2017). ActA mutants grow normally within the cytosol of infected cells, but are incapable of cell-to-cell spread and are approximately 1000-fold less virulent in mouse models (Brundage, Smith, Camilli, Theriot, & Portnoy, 1993). However, unlike LLO-minus mutants, which are poor inducers of CMI, ActA mutants are extremely potent inducers of CMI and are the primary basis of attenuation used in vaccine strains safely administered to humans (Flickinger et al., 2018).
Innate immune recognition of L. monocytogenes
MyD88-dependent responses:
Within minutes of IV infection, the majority of splenic L. monocytogenes can be found within several macrophages sub-types of the marginal zone, subsequently transitioning into dendritic cells (DCs) of the white pulp (Aoshi et al., 2009; Perez et al., 2017). Accordingly, most in vitro studies of innate immunity to L. monocytogenes utilize infection of BMMs from conventional and knockout mice. The initial innate immune response is somewhat generic and involves the detection of bacteria by Toll-like receptors (TLRs) on the cell surface and within phagosomes of macrophages, leading to the expression of chemokines and both inflammatory and regulatory cytokines (Bahjat et al., 2009; Serbina et al., 2003; Witte et al., 2012). This initial response can be studied using LLO-minus bacteria, which are trapped in phagosomes, and is entirely dependent on the MyD88 adapter molecule (Leber et al., 2008). As a Gram-positive bacterium, L. monocytogenes lacks LPS and consequently does not stimulate TLR4, but in non-activated BMMs is mostly detected by TLR2. In IFN-γ activated macrophages, the MyD88 response is mostly dependent on Unc93b1, which is a chaperone necessary for the trafficking of nucleic acid sensing TLRs 3, 7, and 9 to the phagosome (Tabeta et al., 2006). Since these TLRs detect nucleic acids, these results imply that lysis of the bacteria exposes their nucleic acids which are detected by Unc93b1-dependent TLRs. In addition to MyD88, there are numerous other innate immune factors that are essential for resistance to primary infection, notably IFN-γ, TNF, and CCR2, much of which is related to macrophage migration and activation (Pamer, 2004). Importantly, as few as 10 bacteria are lethal to MyD88 and IFN-γ KO mice, yet these same mice are not more sensitive to ActA-minus mutants which exhibit an abortive infection due to the defect in cell-to-cell spread and are likely killed by neutrophils subsequent to bacteriolysis (Harty & White, 1999).
STING-dependent responses in the cytosol.
Most of the BMM transcriptional response to L. monocytogenes is MyD88-dependent as discussed above. However, upon entering the cytosol, L. monocytogenes induces a unique set of IRF3-dependent genes, that leads to the expression of IFN-β and co-regulated genes (Leber et al., 2008). A hallmark of L. monocytogenes infection is the induction if type I interferons, that are detected by type I interferon receptor, IFNAR1. Clearly, monocyte recruitment is critical for innate resistance to L. monocytogenes (Shi & Pamer, 2011), and while cytosolic bacteria induce chemokine MCP-1 in a MyD88-independent manner, IFNAR MyD88 double mutant mice are defective for monocyte recruitment compared to single IFNAR mutant mice, and are more susceptible to infection than MyD88 KO mice alone (Jia, Leiner, Dorothee, Brandl, & Pamer, 2009), demonstrating that both phagosome and cytosolic pathways are redundant for innate resistance to infection.
The cytosol-specific response to L. monocytogenes is induced primarily by the molecule cyclic-di-AMP (c-di-AMP) (Woodward, Iavarone, & Portnoy, 2010). c-di-AMP is a small signaling molecule widespread among bacteria, especially Firmicutes, that controls aspects of bacterial metabolism and osmoregulation (Commichau, Heidemann, Ficner, & Stulke, 2019). L. monocytogenes secretes c-di-AMP through multidrug resistance transporters, although the role of its secretion, other than to stimulate the IFN-β response is not yet clarified (Crimmins et al., 2008; Huynh & Woodward, 2016). Bacterial cyclic-di nucleotides, c-di-AMP, c-di-GMP, and 3’−5’, 3’−5’cGAMP act by binding to and activating a host protein called STING (Stimulator of Interferon Genes) (Burdette et al., 2011). Importantly, the host response to DNA is also STING-dependent, but requires cGAS, which is activated by cytosolic dsDNA to synthesize a chemically distinct form of cGAMP 2’−5’, 3’−5’cGAMP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013). Thus, STING appears to be a central signaling hub for the recognition of many viral and bacterial pathogens by the recognition of either dsDNA and/or cyclic-di-nucleotides (Danilchanka & Mekalanos, 2013; Konno & Barber, 2014).
Inflammasome activation.
Inflammasomes are multiprotein complexes that assemble in response to microbial ligands, including bacterial flagellin and DNA, resulting in the activation of Caspase-1, and secretion of the inflammatory cytokines IL-1β and IL-18 and culminates in a form of inflammatory programed cell death called pyroptosis (Bergsbaken, Fink, & Cookson, 2009; Fink & Cookson, 2006; von Moltke, Ayres, Kofoed, Chavarria-Smith, & Vance, 2013). Many bacterial pathogens induce pyroptosis, although it typically plays only modest roles in host resistance to infection, probably because bona fide pathogens evolve to avoid it (Jorgensen & Miao, 2015). L. monocytogenes infection of BMMs results in relatively low levels of pyroptosis, but that which occurs is mostly due to the infrequent lysis of cytosolic bacteria and the activation of the DNA-dependent AIM2 inflammasome (McDougal & Sauer, 2018). In addition, LLO, like other pore-forming toxins, can activate the NLRP3 inflammasome, but this occurs from the outside of cells and probably is not directly relevant to L. monocytogenes pathogenesis (Sauer et al., 2010). Strains of L. monocytogenes engineered to secrete a fusion of ActA and the Legionella pneumophila flagellin activate the NLRC4-dependent inflammasome and are attenuated in WT mice but retain full virulence in NLRC4-minus mice (Sauer, Pereyre, et al., 2011; Warren et al., 2011). Therefore, it appears that avoidance of pyroptosis, and cell death in general, is an essential feature of L. monocytogenes pathogenesis. L. monocytogenes does not appear to actively inhibit cell death, but rather fails to stimulate it by growing without appreciable lysis, using multiple mechanisms to avoid LLO-mediated cytotoxicity, and not expressing its flagellum at 37°C (Shen & Higgins, 2006).
Generation of adaptive immunity
The hallmark of adaptive immunity to L. monocytogenes is that naïve mice that recover from a primary challenge develop long-term immunity to reinfection (Condotta, Richer, Badovinac, & Harty, 2012; D’Orazio, 2019). The most common assay used to measure adaptive immunity is to infect immunized mice with a dose of L. monocytogenes lethal to non-immunized mice, and determine the number of CFUs in the liver and spleen after 48 or 72h compared to the numbers observed in non-immunized mice. Typically, immune mice have approximately 5-logs fewer bacteria in their organs as compared to naïve mice. Although CD4+ T-cells play a role in the development of immunity, CD8+ T cells are the dominant effector T-cell, which is largely dependent on antigen-specific cytotoxicity (Harty, Schreiber, & Bevan, 1992; Sun, Williams, & Bevan, 2004). T-cell epitopes are derived largely from proteins secreted by L. monocytogenes into the host cell cytosol which are processed and presented in the MHC class I pathway of antigen presentation (Pamer, 2004; Pamer, Harty, & Bevan, 1991). Fusion proteins consisting of the N-terminus of ActA or LLO and a foreign protein or epitope lead to the generation of CD8+ T cells to the foreign epitope. Immunization with L. monocytogenes ActA mutants expressing an ActA-OVA fusion results in more than 10% of the primary CD8+ T cells being specific for OVA, and up to 20% upon secondary expansion (Bahjat et al., 2006).
What makes L. monocytogenes such a potent inducer of CD8+ T cells?
We hypothesize that there are five biological factors that contribute to L. monocytogenes as an effective inducer of CMI. Briefly, it requires the correct host cells, bacterial growth and secretion of antigens, the ideal cellular compartment for antigen presentation, lack of host cell death, and induction of a favorable cytokine milieu (Figure 1).
Figure 1: Biological factors that impact the development of cell-mediated-immunity (CMI) during L. monocytogenes infection and their role in tumor therapy:
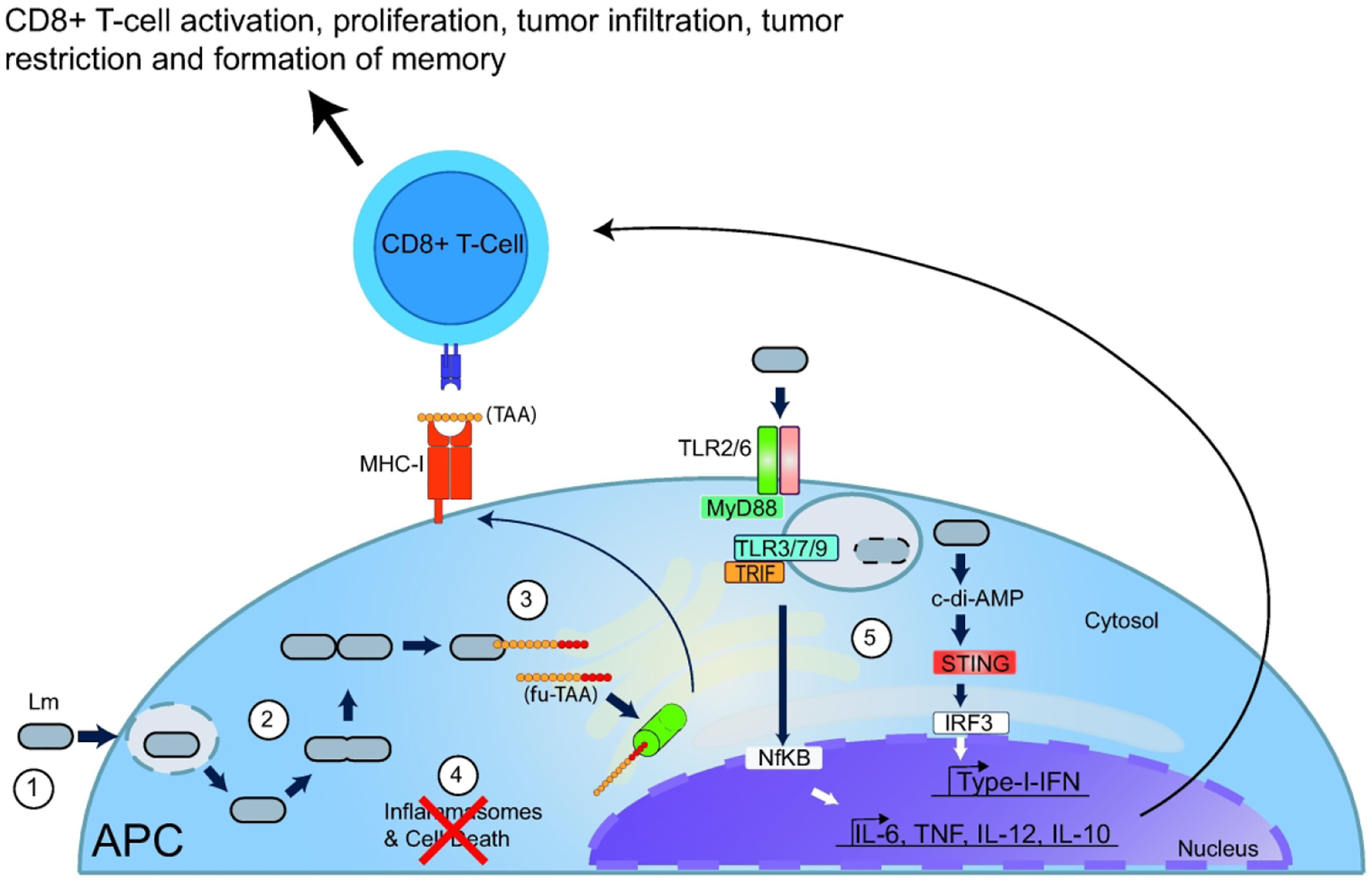
1) Upon intravenous inoculation, L. monocytogenes (Lm) is internalized by professional antigen presenting cells (APC), such as splenic macrophages or dendritic cells. 2) L. monocytogenes escapes the phagosome, enters the cytosol, rapidly replicates, and secretes proteins. 3) L. monocytogenes can be engineered to secrete recombinant tumor associated antigens fused to virulence factors, termed here as fusion-tumor-associated antigens (fu-TAA). Secreted proteins are degraded by proteasomes (green cylinder) and enter the MHC class I pathway of antigen presentation. 4) L. monocytogenes minimally activates inflammasomes and consequently, infection does not lead to rapid host cell death. 5) Unlike phagosome confined bacteria that can lyse within phagosomes and primarily activate the MyD88-dependent pathway through Toll Like Receptors (TLRs), cytosolic bacteria induce cytosolic innate immune pathways by secreting cyclic di-AMP (c-di-AMP) thereby inducing an overall balanced cytokine milieu that favors CMI and tumor restriction by antigen specific cytotoxic CD8+ T cells.
1. The bacteria have a predilection for professional antigen-presenting cells.
Within minutes of intravenous vaccination, the majority of splenic L. monocytogenes are found predominantly within macrophages of the marginal zone, where bacterial growth is restricted (Aoshi et al., 2009). As early as 1-hour post infection the majority of splenic L. monocytogenes can be found within CD169+ macrophages, such that depletion of CD169+ macrophages dramatically increases susceptibility to disease (Perez et al., 2017). In coordination with DCs, CD169+ macrophages support CMI by providing antigen and producing type I IFN. However, CD169+ macrophages can suppress CMI to self-antigens and phagosome-restricted content including apoptotic cells and exosomes (Grabowska, Lopez-Venegas, Affandi, & den Haan, 2018; McGaha, Chen, Ravishankar, van Rooijen, & Karlsson, 2011; Miyake et al., 2007; Saunderson, Dunn, Crocker, & McLellan, 2014). Indeed, several hours after infection, phagosome confined LLO-minus bacteria are still localized to CD169+ macrophages, whereas bacteria growing in the host cytosol are transferred to CD8-α+ DCs that serve as a protective niche for bacterial survival (Edelson et al., 2011; Neuenhahn et al., 2006; Perez et al., 2017). Batf3 knockout mice that specifically lack CD8-α+ DCs are resistant to infection, suggesting that this population of DCs represent a critical host cell for the pathogenesis of L. monocytogenes (Edelson et al., 2011). Clearly, professional antigen-presenting cells (APCs) like DCs, have a predominant role in presenting foreign antigens and orchestrating the development of antigen-specific cytotoxic T cells (Merad, Sathe, Helft, Miller, & Mortha, 2013). However, Batf3 knockout mice can still be immunized with high doses of L. monocytogenes, suggesting that APCs other than CD8-α+ DCs are sufficient for induction of CMI (Edelson et al., 2011).
2. The bacteria escape from phagosomes and grow in the host cell cytosol.
Once inside APCs, L. monocytogenes escapes from phagosomes and grows rapidly in the host cell cytosol. Entry into the host cytosol is necessary for bacterial replication and is a prerequisite for all subsequent steps in pathogenesis. Furthermore, cytosolic access is required for CMI, but in the absence of bacterial replication, cytosolic access is not sufficient to induce robust CMI, since killed but metabolically active L. monocytogenes fails to induce optimal CMI due to defective DC maturation (Bahjat et al., 2006). Indeed, cytosolic localization is required for recruitment of CD8-α+ DCs to the site of infected CD169+ macrophages (Perez et al., 2017). In addition, escape from a phagosome provides a mechanism to avoid the MyD88-dependent responses, such as IL-10 expression, which suppress immunity (Bahjat et al., 2009) as discussed below. Strikingly, whereas as few as 1000 CFUs of a cytosolic ActA-minus mutant can induce long-lived immunity, as many as 108 of a phagosome confined LLO-minus mutant fails to induce an appreciable level of immunity (Archer, Durack, & Portnoy, 2014; Bahjat et al., 2006; Berche et al., 1987; Orgun & Way, 2008).
3. Bacterial secreted proteins enter the MHC class I pathway of antigen processing and presentation.
Entry and growth of L. monocytogenes in the host cytosol promotes secretion of bacterial proteins directly into the cytosol where they are processed and presented into the MHC class I pathway without a requirement for cross presentation. In BALB/c mice, a dominant MHC I epitope is derived from LLO and inhibition of proteasomes blocks antigen presentation (Villanueva, Sijts, & Pamer, 1995). However, cytosolic access of secreted proteins, is strikingly more efficient at inducing CD8+ T cells compared to the cross priming of antigens derived from the bacterial cytosol (Shen et al., 1998).
4. The bacteria do not induce rapid cell death.
Whereas many pathogens cause cell death, L. monocytogenes grows in APCs without causing appreciable cell death until there are over 200 bacteria per cell (Brundage, Smith, Camilli, Theriot, & Portnoy, 1993). Bacteria that are engineered to kill infected cells prematurely by necrosis, apoptosis, or pyroptosis are defective for the induction of adaptive immunity (Theisen & Sauer, 2016). The simplest explanation is that premature killing of the APCs prevents all aspects of immunity including secretion of cytokines and presenting of antigen. However, infected host cell death does not preclude that killed cells are phagocytosed by DCs and processed by cross-presentation (Alloatti, Kotsias, Magalhaes, & Amigorena, 2016; Edelson, 2012), and indeed, although CMI is diminished, there remains substantial levels of immunity in mice immunized with cytotoxic strains (McDougal & Sauer, 2018; Sauer, Pereyre, et al., 2011).
5. Cytosolic bacteria induce a cytokine response that favors CMI.
Surprisingly, when mice are simultaneously co-infected with both phagosome confined LLO-minus bacteria, and ActA-minus mutants, immunity is decreased by over 100-fold (Bahjat et al., 2009). Analysis of serum cytokines revealed that the addition of LLO-minus bacteria suppressed the host inflammatory response, and resulted in acute elevation of IL-10, a MyD88-dependent cytokine known to suppress both innate and adaptive immunity to various microbial pathogens (Couper, Blount, & Riley, 2008; Moore, de Waal Malefyt, Coffman, & O’Garra, 2001; Saraiva & O’Garra, 2010; Slobedman, Barry, Spencer, Avdic, & Abendroth, 2009). Blockade of the IL-10 receptor at the time of immunization eliminated suppression caused by the LLO-minus bacteria and even enhanced immunity in mice immunized with LLO-minus alone (Bahjat et al., 2009). These data suggest that one of the reasons that LLO-minus bacteria fail to induce protective immunity is that they suppress immunity via the MyD88 pathway (Bahjat et al., 2009). Indeed, MyD88 KO mice vaccinated with ActA-minus bacteria show enhanced immunity compared to wild type mice, demonstrating the suppressive role MyD88 signaling can play during vaccination with live bacteria (Archer et al., 2014). In vitro, BMMs are potent inducers of MyD88-dependent cytokines in response to LLO-minus bacteria. However, the in vivo cellular source of IL-10 during immunosuppression by LLO-minus strains is not known, though it likely involves CD169+ macrophages, as they are the host cells that predominantly capture systemic bacteria and demonstrate prolonged retention of LLO-minus bacteria (Perez et al., 2017).
Phagosome-confined bacteria activate the MyD88-dependent pathway, which enhances the secretion of the immunosuppressive cytokine IL-10 and thus counteracts the proinflammatory signals needed for CMI (Bahjat et al., 2009). On the other hand, cytosolic L. monocytogenes not only induces expression of MyD88-dependent genes like IL-6, IL-12, and TNF, which are beneficial for CMI, but also secretes c-di-AMP which activates STING and leads to a strong type I IFN dependent pathway (Sauer, Sotelo-Troha, et al., 2011; Woodward et al., 2010). While under some experimental conditions, type I IFN promotes CMI (Diamond et al., 2011; Stetson & Medzhitov, 2006), in the case of L. monocytogenes, it paradoxically has the opposite effect, as mice lacking the type I IFN receptor or STING induce a stronger CD8+ T cell response (Archer et al., 2014). Interestingly, in sepsis mediated immune suppression, early type I IFN secretion by splenic macrophages decreases DC antigen presentation and inflammatory cytokine secretion (Schwandt et al., 2012) supporting the notion that during systemic bacterial infections overproduction of type I IFN negatively affects CMI. More surprisingly, a MyD88/STING double mutant, which produces almost no detectible cytokine response to L. monocytogenes is fully immunized with an ActA-minus mutant (Archer et al., 2014). Indeed, nearly all KO mice tested, other than those lacking T-cells or factors necessary for T-cell-mediated killing are still effectively vaccinated by the ActA-minus strain (Harty & White, 1999). One exception is mice defective for prostaglandin E2, that show a diminished level of immunity which correlates with fewer antigen specific CD8+ T cells (Theisen et al., 2018). Therefore, we suspect that multiple and redundant innate immune pathways lead to CMI.
L. monocytogenes vaccines
The observation that attenuated strains of L. monocytogenes induce robust CMI led to studies showing that L. monocytogenes-induced CMI can be directed towards elimination of cancer cells in vivo (Pan, Ikonomidis, Lazenby, Pardoll, & Paterson, 1995). L. monocytogenes engineered to secrete Tumor Associated Antigens (TAAs) can effectively treat established tumors in mice and the therapeutic response depends greatly on CD8+ T cells (Brockstedt et al., 2004). This approach has also proven safe in humans and has led to several clinical trials, some of which are ongoing (Flickinger et al., 2018; Maciag, Radulovic, & Rothman, 2009).
Although the L. monocytogenes strain used in all clinical trials, 10403S, is clearly potent at inducing CMI, it is difficult to imagine that the strain cannot be improved. There are several pros and cons to the therapeutic use of strain 10403S. 10403S has been studied extensively over the past several decades, is well characterized and genetically tractable. However, additional issues should be considered. For example, the streptomycin resistance mutation in 10403S reduces virulence by approximately 3-fold (Bishop & Hinrichs, 1987). Furthermore, unlike many L. monocytogenes isolates found in nature, which downregulate expression of flagellin at 37°C and during intracellular infection, 10403S contains a point mutation in the ribosome binding site for flagellar repressor gene mogR, such that 10403S still expresses some flagella, even at 37°C (Shen & Higgins, 2006). The effect of flagellar expression by 10403S may have unknown consequences such as affecting pyroptosis during therapeutic vaccinations.
On the other hand, some features conserved in 10403S may be well suited for CMI. Several clinical isolates contain mutations in tetR, a negative regulator of MDR transporters that efflux cdi-AMP (Schwartz et al., 2012). Indeed, TetR mutants express elevated levels of MdrT and secrete more c-di-AMP, leading to enhanced type I interferon expression (Crimmins et al., 2008). Overstimulation of the STING pathway may diminish CMI, as discussed above, whereas 10403S induces a balanced level of c-di-AMP secretion favoring CMI (Archer et al., 2014). Similarly, a balanced level of STING stimulation is important for tumor immunotherapy (Sivick et al., 2018).
Another consideration for vaccine development using live L. monocytogenes is providing the vector with optimal access to the cytosol. As mentioned above, the majority of injected bacteria are killed upon phagocytosis and likely induce unfavorable MyD88 signaling. Suppressive signaling may be reduced by using strains that contain a point mutation in the master virulence regulator PrfA that locks it in an active state, termed PrfA*. These strains escape more readily into the cytosol and are enhanced for CMI (Lauer et al., 2008; Qiu et al., 2011). Alternatively, pretreatment of L. monocytogenes with reducing agents, which mimic the reducing environment experienced by L. monocytogenes in the cytosol, can also pre-activate PrfA (Portman, Dubensky, Peterson, Whiteley, & Portnoy, 2017). Finally, we have recently completed a forward genetic screen for LLO-minus mutants that induce aberrant IL-10 induction in BMMs. We find that mutants that fail to stimulate TLR2, show diminished IL-10 induction, whereas mutants that are prone to lysis, induce enhanced Unc93b1 dependent IL-10 (Unpublished data, Nguyen & Chávez-Arroyo, Cheng, Krasilnikov, Louie, & Portnoy). This suggests that L. monocytogenes can be tailored for either enhanced or diminished induction of these pathways to optimize vaccination using live bacteria in cancer therapy.
Future Directions
An exciting future direction in the fields of CMI and microbial pathogenesis is to gain a better understanding of the relationship between virulence and the capacity to induce adaptive immunity. For example, loss of ActA-dependent cell-to-cell spread reduces L. monocytogenes virulence by approximately 1000-fold, yet CMI remains remarkably robust. On the other hand, LLO-minus bacteria remain in phagosomes, are up to 100,000-fold less virulent, and are poor inducers of adaptive immunity. Although not fully understood, it appears that immunosuppression occurs during vaccination with LLO-minus bacteria, and IL-10 is involved (Bahjat et al., 2009). Development of a strategy to bypass phagosomal escape while retaining the induction of CMI is an exciting prospect for the field of translational immunology, as it has the greatest potential for increasing the safety of live vaccines. A deeper understanding of the events that occur during the first few hours of infection is still needed. Many questions remain about the early responses that lead to CMI, including the host cell types involved and the optimal cytokines that lead to CMI.
The relevance of chronic versus acute infection is also an important area to consider, as chronic infections tend to induce T cell exhaustion (Wherry & Kurachi, 2015). We suggest that the acute nature of L. monocytogenes infection is an additional feature responsible for inducing a favorable environment for the induction of CMI because it does not lead to T cell exhaustion. A possible explanation for why acute infections with L. monocytogenes leads to robust CMI may involve the rapid and acute induction of type I IFN. In contrast, chronic infections with some viruses leads to immunosuppression due to prolonged type I IFN signaling (Dagenais-Lussier et al., 2017). This observation may explain why L. monocytogenes mutations that cause enhanced production of type I IFN show diminished CMI in mice, and may even explain why some clinical isolates harbor similar mutations (Archer et al., 2014; Schwartz et al., 2012).
Although many findings observed in mice are conserved in humans, key biological differences between humans and mice influence our ability to extrapolate findings. One example is in species-specific polymorphisms found in innate immune signaling proteins, as is the case with STING. While bacterial c-di-AMP is a potent inducer of mouse STING, it is less potent at activating certain human STING variants (Patel & Jin, 2019).
Finally, our understanding of how L. monocytogenes induces CMI, has motivated an exciting frontier for personalized L. monocytogenes vaccines for cancer immunotherapy (Flickinger et al., 2018). Advancements in sequencing and computational technology now allow for the identification of patient-specific tumor neoantigens. The identified neoantigen epitopes can be expressed by L. monocytogenes to direct CMI to those tumor cells in vivo (Figure 1). Indeed, combining the factors that make L. monocytogenes such a potent generator of CMI, along with newly developing therapies, like checkpoint blockade, is an exciting prospect for the future of cancer immunotherapy using live L. monocytogenes as vaccines (Deng et al., 2018; Rosenberg et al., 2016).
Clearly, decades of research have placed L. monocytogenes as an excellent model system for studying innate and adaptive immunobiology, in particular the development of potent CMI. We have reviewed five reasons why L. monocytogenes is such a robust inducer of CD8+ T cells. These, and other factors, yet to be discovered, make L. monocytogenes a powerful system to study CMI with implications for vaccine development and cancer immunotherapy.
Acknowledgments
This work was supported by National Institutes of Health grants 1P01 AI063302 (D.A.P.) and 1R01 AI027655 (D.A.P).
Footnotes
Conflicts of Interest
No conflicts of interest to report
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, … Hornung V (2013). cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature, 498(7454), 380–384. doi: 10.1038/nature12306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloatti A, Kotsias F, Magalhaes JG, & Amigorena S (2016). Dendritic cell maturation and cross-presentation: timing matters! Immunol Rev, 272(1), 97–108. doi: 10.1111/imr.12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshi T, Carrero JA, Konjufca V, Koide Y, Unanue ER, & Miller MJ (2009). The cellular niche of Listeria monocytogenes infection changes rapidly in the spleen. Eur J Immunol, 39(2), 417–425. doi: 10.1002/eji.200838718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer KA, Durack J, & Portnoy DA (2014). STING-dependent type I IFN production inhibits cell-mediated immunity to Listeria monocytogenes. PLoS Pathog, 10(1), e1003861. doi: 10.1371/journal.ppat.1003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahjat KS, Liu W, Lemmens EE, Schoenberger SP, Portnoy DA, Dubensky TW Jr., & Brockstedt DG (2006). Cytosolic entry controls CD8+-T-cell potency during bacterial infection. Infect Immun, 74(11), 6387–6397. doi: 10.1128/iai.01088-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahjat KS, Meyer-Morse N, Lemmens EE, Shugart JA, Dubensky TW, Brockstedt DG, & Portnoy DA (2009). Suppression of cell-mediated immunity following recognition of phagosome-confined bacteria. PLoS Pathog, 5(9), e1000568. doi: 10.1371/journal.ppat.1000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft GJ, Schreiber RD, Bosma GC, Bosma MJ, & Unanue ER (1987). A T cell-independent mechanism of macrophage activation by interferon-gamma. The Journal of Immunology, 139(4), 1104–1107. [PubMed] [Google Scholar]
- Berche P, Gaillard JL, & Sansonetti PJ (1987). Intracellular growth of Listeria monocytogenes as a prerequisite for in vivo induction of T cell-mediated immunity. The Journal of Immunology, 138(7), 2266–2271. [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, & Cookson BT (2009). Pyroptosis: host cell death and inflammation. Nat Rev Microbiol, 7(2), 99–109. doi: 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Kanagawa O, Swanson PE, & Unanue ER (1998). Chronic Listeria Infection in SCID Mice: Requirements for the Carrier State and the Dual Role of T Cells in Transferring Protection or Suppression. The Journal of Immunology, 160(1), 376–384. [PubMed] [Google Scholar]
- Bishop DK, & Hinrichs DJ (1987). Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol, 139(6), 2005–2009. [PubMed] [Google Scholar]
- Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, Luckett W, … Dubensky TW Jr. (2004). Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc Natl Acad Sci U S A, 101(38), 13832–13837. doi: 10.1073/pnas.0406035101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage RA, Smith GA, Camilli A, Theriot JA, & Portnoy DA (1993). Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc Natl Acad Sci U S A, 90(24), 11890–11894. doi: 10.1073/pnas.90.24.11890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, … Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature, 478(7370), 515–518. doi: 10.1038/nature10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MI, Chen C, Engstrom P, Portnoy DA, & Mitchell G (2018). Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell Microbiol, 20(9), e12854. doi: 10.1111/cmi.12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Heidemann JL, Ficner R, & Stulke J (2019). Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria. J Bacteriol, 201(1). doi: 10.1128/jb.00462-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condotta SA, Richer MJ, Badovinac VP, & Harty JT (2012). Probing CD8 T cell responses with Listeria monocytogenes infection. Adv Immunol, 113, 51–80. doi: 10.1016/b978-0-12-394590-7.00005-1 [DOI] [PubMed] [Google Scholar]
- Couper KN, Blount DG, & Riley EM (2008). IL-10: the master regulator of immunity to infection. J Immunol, 180(9), 5771–5777. doi: 10.4049/jimmunol.180.9.5771 [DOI] [PubMed] [Google Scholar]
- Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW Jr., & Portnoy DA (2008). Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci U S A, 105(29), 10191–10196. doi: 10.1073/pnas.0804170105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais-Lussier X, Loucif H, Murira A, Laulhe X, Stager S, Lamarre A, & van Grevenynghe J (2017). Sustained IFN-I Expression during Established Persistent Viral Infection: A “Bad Seed” for Protective Immunity. Viruses, 10(1). doi: 10.3390/v10010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, & Mekalanos JJ (2013). Cyclic dinucleotides and the innate immune response. Cell, 154(5), 962–970. doi: 10.1016/j.cell.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lira V, Hudson TE, Lemmens EE, Hanson WG, Flores R, … Dubensky TW Jr. (2018). Recombinant Listeria promotes tumor rejection by CD8(+) T cell-dependent remodeling of the tumor microenvironment. Proc Natl Acad Sci U S A, 115(32), 8179–8184. doi: 10.1073/pnas.1801910115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, … Schreiber RD (2011). Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med, 208(10), 1989–2003. doi: 10.1084/jem.20101158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, … Vance RE (2013). The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep, 3(5), 1355–1361. doi: 10.1016/j.celrep.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio SEF (2019). Innate and Adaptive Immune Responses during Listeria monocytogenes Infection. Microbiology Spectrum, 7(3). doi: 10.1128/microbiolspec.GPP3-0065-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson BT (2012). Dendritic cells in Listeria monocytogenes infection. Adv Immunol, 113, 33–49. doi: 10.1016/b978-0-12-394590-7.00006-3 [DOI] [PubMed] [Google Scholar]
- Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, … Murphy KM (2011). CD8alpha(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity, 35(2), 236–248. doi: 10.1016/j.immuni.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, & Cookson BT (2006). Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol, 8(11), 1812–1825. doi: 10.1111/j.1462-5822.2006.00751.x [DOI] [PubMed] [Google Scholar]
- Flickinger JC, Rodeck U, & Snook AE (2018). Listeria monocytogenes as a Vector for Cancer Immunotherapy: Current Understanding and Progress. Vaccines, 6(3), 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Port GC, & Miner MD (2009). Listeria monocytogenes — from saprophyte to intracellular pathogen. Nature Reviews Microbiology, 7(9), 623–628. doi: 10.1038/nrmicro2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, … Patel DJ (2013). Structure-function analysis of STING activation by c[G(2’,5’)pA(3’,5’)p] and targeting by antiviral DMXAA. Cell, 154(4), 748–762. doi: 10.1016/j.cell.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens PL, Milon G, Cossart P, & Saron M-F (1995). Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. International Immunology, 7(5), 797–805. doi: 10.1093/intimm/7.5.797 [DOI] [PubMed] [Google Scholar]
- Gouin E, Welch MD, & Cossart P (2005). Actin-based motility of intracellular pathogens. Current Opinion in Microbiology, 8(1), 35–45. doi: 10.1016/j.mib.2004.12.013 [DOI] [PubMed] [Google Scholar]
- Grabowska J, Lopez-Venegas MA, Affandi AJ, & den Haan JMM (2018). CD169(+) Macrophages Capture and Dendritic Cells Instruct: The Interplay of the Gatekeeper and the General of the Immune System. Front Immunol, 9, 2472. doi: 10.3389/fimmu.2018.02472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Schreiber RD, & Bevan MJ (1992). CD8 T cells can protect against an intracellular bacterium in an interferon gamma-independent fashion. Proc Natl Acad Sci U S A, 89(23), 11612–11616. doi: 10.1073/pnas.89.23.11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, & White D (1999). A knockout approach to understanding CD8+ cell effector mechanisms in adaptive immunity to Listeria monocytogenes. Immunobiology, 201(2), 196–204. doi: 10.1016/s0171-2985(99)80059-x [DOI] [PubMed] [Google Scholar]
- Herskovits AA, Auerbuch V, & Portnoy DA (2007). Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog, 3(3), e51. doi: 10.1371/journal.ppat.0030051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, & Woodward JJ (2016). Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol, 30, 22–29. doi: 10.1016/j.mib.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidis G, Paterson Y, Kos FJ, & Portnoy DA (1994). Delivery of a viral antigen to the class I processing and presentation pathway by Listeria monocytogenes. The Journal of Experimental Medicine, 180(6), 2209–2218. doi: 10.1084/jem.180.6.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Leiner I, Dorothee G, Brandl K, & Pamer EG (2009). MyD88 and Type I interferon receptor-mediated chemokine induction and monocyte recruitment during Listeria monocytogenes infection. J Immunol, 183(2), 1271–1278. doi: 10.4049/jimmunol.0900460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I, & Miao EA (2015). Pyroptotic cell death defends against intracellular pathogens. Immunol Rev, 265(1), 130–142. doi: 10.1111/imr.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, & Barber GN (2014). The STING controlled cytosolic-DNA activated innate immune pathway and microbial disease. Microbes Infect, 16(12), 998–1001. doi: 10.1016/j.micinf.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P, Hanson B, Lemmens EE, Liu W, Luckett WS, Leong ML, … Dubensky TW Jr. (2008). Constitutive Activation of the PrfA regulon enhances the potency of vaccines based on live-attenuated and killed but metabolically active Listeria monocytogenes strains. Infect Immun, 76(8), 3742–3753. doi: 10.1128/iai.00390-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, & Portnoy DA (2008). Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog, 4(1), e6. doi: 10.1371/journal.ppat.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag PC, Radulovic S, & Rothman J (2009). The first clinical use of a live-attenuated Listeria monocytogenes vaccine: a Phase I safety study of Lm-LLO-E7 in patients with advanced carcinoma of the cervix. Vaccine, 27(30), 3975–3983. doi: 10.1016/j.vaccine.2009.04.041 [DOI] [PubMed] [Google Scholar]
- Mackaness GB (1962). CELLULAR RESISTANCE TO INFECTION. 116(3), 381–406. doi: 10.1084/jem.116.3.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal CE, & Sauer JD (2018). Listeria monocytogenes: The Impact of Cell Death on Infection and Immunity. Pathogens, 7(1). doi: 10.3390/pathogens7010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaha TL, Chen Y, Ravishankar B, van Rooijen N, & Karlsson MC (2011). Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood, 117(20), 5403–5412. doi: 10.1182/blood-2010-11-320028 [DOI] [PubMed] [Google Scholar]
- McGregor DD, Koster FT, & Mackaness GB (1970). Biological Sciences: The Short Lived Small Lymphocyte as a Mediator of Cellular Immunity. 228(5274), 855–856. doi: 10.1038/228855a0 [DOI] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, & Mortha A (2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol, 31, 563–604. doi: 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G, Cheng MI, Chen C, Nguyen BN, Whiteley AT, Kianian S, … Portnoy DA (2018). Listeria monocytogenes triggers noncanonical autophagy upon phagocytosis, but avoids subsequent growth-restricting xenophagy. Proceedings of the National Academy of Sciences, 115(2), E210–E217. doi: 10.1073/pnas.1716055115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, & Tanaka M (2007). Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest, 117(8), 2268–2278. doi: 10.1172/jci31990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KW, de Waal Malefyt R, Coffman RL, & O’Garra A (2001). Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol, 19, 683–765. doi: 10.1146/annurev.immunol.19.1.683 [DOI] [PubMed] [Google Scholar]
- Neuenhahn M, Kerksiek KM, Nauerth M, Suhre MH, Schiemann M, Gebhardt FE, … Busch DH (2006). CD8alpha+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity, 25(4), 619–630. doi: 10.1016/j.immuni.2006.07.017 [DOI] [PubMed] [Google Scholar]
- Nguyen BN, Peterson BN, & Portnoy DA (2019). Listeriolysin O: A phagosome-specific cytolysin revisited. Cellular Microbiology, 21(3), e12988. doi: 10.1111/cmi.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgun NN, & Way SS (2008). A critical role for phospholipase C in protective immunity conferred by listeriolysin O-deficient Listeria monocytogenes. Microb Pathog, 44(2), 159–163. doi: 10.1016/j.micpath.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG (2004). Immune responses to Listeria monocytogenes. Nat Rev Immunol, 4(10), 812–823. doi: 10.1038/nri1461 [DOI] [PubMed] [Google Scholar]
- Pamer EG, Harty JT, & Bevan MJ (1991). Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature, 353(6347), 852–855. doi: 10.1038/353852a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan ZK, Ikonomidis G, Lazenby A, Pardoll D, & Paterson Y (1995). A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat Med, 1(5), 471–477. doi: 10.1038/nm0595-471 [DOI] [PubMed] [Google Scholar]
- Patel S, & Jin L (2019). TMEM173 variants and potential importance to human biology and disease. Genes Immun, 20(1), 82–89. doi: 10.1038/s41435-018-0029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez OA, Yeung ST, Vera-Licona P, Romagnoli PA, Samji T, Ural BB, … Khanna KM (2017). CD169(+) macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen. Sci Immunol, 2(16). doi: 10.1126/sciimmunol.aah5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillich H, Puri M, & Chakraborty T (2017). ActA of Listeria monocytogenes and Its Manifold Activities as an Important Listerial Virulence Factor In Mannherz HG (Ed.), The Actin Cytoskeleton and Bacterial Infection (pp. 113–132). Cham: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Portman JL, Dubensky SB, Peterson BN, Whiteley AT, & Portnoy DA (2017). Activation of the Listeria monocytogenes Virulence Program by a Reducing Environment. MBio, 8(5). doi: 10.1128/mBio.01595-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Auerbuch V, & Glomski IJ (2002). The cell biology ofListeria monocytogenesinfection. The Journal of Cell Biology, 158(3), 409–414. doi: 10.1083/jcb.200205009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Jacks PS, & Hinrichs DJ (1988). Role of hemolysin for the intracellular growth of Listeria monocytogenes. The Journal of Experimental Medicine, 167(4), 1459–1471. doi: 10.1084/jem.167.4.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yan L, Chen J, Chen CY, Shen L, Letvin NL, … Chen ZW (2011). Intranasal vaccination with the recombinant Listeria monocytogenes DeltaactA prfA* mutant elicits robust systemic and pulmonary cellular responses and secretory mucosal IgA. Clin Vaccine Immunol, 18(4), 640–646. doi: 10.1128/cvi.00254-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshevich L, & Cossart P (2018). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nature Reviews Microbiology, 16(1), 32–46. doi: 10.1038/nrmicro.2017.126 [DOI] [PubMed] [Google Scholar]
- Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, … Dreicer R (2016). Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet, 387(10031), 1909–1920. doi: 10.1016/s0140-6736(16)00561-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M, & O’Garra A (2010). The regulation of IL-10 production by immune cells. Nat Rev Immunol, 10(3), 170–181. doi: 10.1038/nri2711 [DOI] [PubMed] [Google Scholar]
- Sauer JD, Pereyre S, Archer KA, Burke TP, Hanson B, Lauer P, & Portnoy DA (2011). Listeria monocytogenes engineered to activate the Nlrc4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc Natl Acad Sci U S A, 108(30), 12419–12424. doi: 10.1073/pnas.1019041108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, … Vance RE (2011). The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun, 79(2), 688–694. doi: 10.1128/iai.00999-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, & Portnoy DA (2010). Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe, 7(5), 412–419. doi: 10.1016/j.chom.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunderson SC, Dunn AC, Crocker PR, & McLellan AD (2014). CD169 mediates the capture of exosomes in spleen and lymph node. Blood, 123(2), 208–216. doi: 10.1182/blood-2013-03-489732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlech WF (2019). Epidemiology and Clinical Manifestations of Listeria monocytogenes Infection. Microbiology Spectrum, 7(3). doi: 10.1128/microbiolspec.GPP3-0014-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwandt T, Schumak B, Gielen GH, Jungerkes F, Schmidbauer P, Klocke K, … Limmer A (2012). Expression of type I interferon by splenic macrophages suppresses adaptive immunity during sepsis. Embo j, 31(1), 201–213. doi: 10.1038/emboj.2011.380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, & Leber JH (2012). Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun, 80(4), 1537–1545. doi: 10.1128/iai.06286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina NV, Kuziel W, Flavell R, Akira S, Rollins B, & Pamer EG (2003). Sequential MyD88-independent and -dependent activation of innate immune responses to intracellular bacterial infection. Immunity, 19(6), 891–901. doi: 10.1016/s1074-7613(03)00330-3 [DOI] [PubMed] [Google Scholar]
- Serbina NV, Shi C, & Pamer EG (2012). Monocyte-Mediated Immune Defense Against Murine Listeria monocytogenes Infection. In (pp. 119–134): Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, & Higgins DE (2006). The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog, 2(4), e30. doi: 10.1371/journal.ppat.0020030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Miller JF, Fan X, Kolwyck D, Ahmed R, & Harty JT (1998). Compartmentalization of Bacterial Antigens: Differential Effects on Priming of CD8 T Cells and Protective Immunity. Cell, 92(4), 535–545. doi: 10.1016/s0092-8674(00)80946-0 [DOI] [PubMed] [Google Scholar]
- Shen H, Slifka MK, Matloubian M, Jensen ER, Ahmed R, & Miller JF (1995). Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc Natl Acad Sci U S A, 92(9), 3987–3991. doi: 10.1073/pnas.92.9.3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, & Pamer EG (2011). Monocyte recruitment during infection and inflammation. Nat Rev Immunol, 11(11), 762–774. doi: 10.1038/nri3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivick KE, Desbien AL, Glickman LH, Reiner GL, Corrales L, Surh NH, … McWhirter SM (2018). Magnitude of Therapeutic STING Activation Determines CD8(+) T Cell-Mediated Anti-tumor Immunity. Cell Rep, 25(11), 3074–3085.e3075. doi: 10.1016/j.celrep.2018.11.047 [DOI] [PubMed] [Google Scholar]
- Slobedman B, Barry PA, Spencer JV, Avdic S, & Abendroth A (2009). Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol, 83(19), 9618–9629. doi: 10.1128/jvi.01098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, & Medzhitov R (2006). Type I interferons in host defense. Immunity, 25(3), 373–381. doi: 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Sun JC, Williams MA, & Bevan MJ (2004). CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol, 5(9), 927–933. doi: 10.1038/ni1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, … Beutler B (2006). The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol, 7(2), 156–164. doi: 10.1038/ni1297 [DOI] [PubMed] [Google Scholar]
- Theisen E, McDougal CE, Nakanishi M, Stevenson DM, Amador-Noguez D, Rosenberg DW, … Sauer JD (2018). Cyclooxygenase-1 and −2 Play Contrasting Roles in Listeria-Stimulated Immunity. J Immunol, 200(11), 3729–3738. doi: 10.4049/jimmunol.1700701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen E, & Sauer JD (2016). Listeria monocytogenes and the Inflammasome: From Cytosolic Bacteriolysis to Tumor Immunotherapy. Curr Top Microbiol Immunol, 397, 133–160. doi: 10.1007/978-3-319-41171-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva MS, Sijts AJ, & Pamer EG (1995). Listeriolysin is processed efficiently into an MHC class I-associated epitope in Listeria monocytogenes-infected cells. J Immunol, 155(11), 5227–5233. [PubMed] [Google Scholar]
- von Koenig CHW, Finger H, & Hof H (1982). Failure of killed Listeria monocytogenes vaccine to produce protective immunity. Nature, 297(5863), 233–234. doi: 10.1038/297233a0 [DOI] [PubMed] [Google Scholar]
- von Moltke J, Ayres JS, Kofoed EM, Chavarria-Smith J, & Vance RE (2013). Recognition of bacteria by inflammasomes. Annu Rev Immunol, 31, 73–106. doi: 10.1146/annurev-immunol-032712-095944 [DOI] [PubMed] [Google Scholar]
- Waite JC, Leiner I, Lauer P, Rae CS, Barbet G, Zheng H, … Dustin ML (2011). Dynamic Imaging of the Effector Immune Response to Listeria Infection In Vivo. 7(3), e1001326. doi: 10.1371/journal.ppat.1001326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SE, Duong H, Mao DP, Armstrong A, Rajan J, Miao EA, & Aderem A (2011). Generation of a Listeria vaccine strain by enhanced caspase-1 activation. Eur J Immunol, 41(7), 1934–1940. doi: 10.1002/eji.201041214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, & Kurachi M (2015). Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol, 15(8), 486–499. doi: 10.1038/nri3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CE, Archer KA, Rae CS, Sauer JD, Woodward JJ, & Portnoy DA (2012). Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol, 113, 135–156. doi: 10.1016/b978-0-12-394590-7.00002-6 [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, & Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science, 328(5986), 1703–1705. doi: 10.1126/science.1189801 [DOI] [PMC free article] [PubMed] [Google Scholar]


