Abstract
Accumulation of amyloid β protein (Aβ) due to increased generation and/or impaired degradation plays an important role in Alzheimer’s disease (AD) pathogenesis. In this report, we describe the identification of rare coding mutations in the endothelin-converting enzyme 2 (ECE2) gene in 1 late-onset AD family, and additional case-control cohort analysis indicates ECE2 variants associated with the risk of developing AD. The 2 mutations (R186C and F751S) located in the peptidase domain in the ECE2 protein were found to severely impair the enzymatic activity of ECE2 in Aβ degradation. We further evaluated the effect of the R186C mutation in mutant APP–knockin mice. Overexpression of wild-type ECE2 in the hippocampus reduced amyloid load and plaque formation, and improved learning and memory deficits in the AD model mice. However, the effect was abolished by the R186C mutation in ECE2. Taken together, the results demonstrated that ECE2 peptidase mutations contribute to AD pathogenesis by impairing Aβ degradation, and overexpression of ECE2 alleviates AD phenotypes. This study indicates that ECE2 is a risk gene for AD development and pharmacological activation of ECE2 could be a promising strategy for AD treatment.
Keywords: Neuroscience
Keywords: Alzheimer’s disease, Genetic variation
Variants in Abeta-degrading enzyme ECE2 gene are associated with AD development, and activation of ECE2 could be a promising therapeutical strategy for AD.
Introduction
Alzheimer’s disease (AD) accounts for two-thirds of dementia in the elderly. The majority of cases are sporadic late-onset AD (LOAD) and less than 1% are genetic forms of early-onset AD (EOAD). Mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) genes cause inherited familial AD. Genetic factors such as ApoE-ε4 have been demonstrated to contribute to the development of sporadic LOAD (1). In recent years, genome-wide association studies (GWAS), massive parallel sequencing (MPS), and whole-exome sequencing (WES) also identified additional candidate genes and loci for AD, including clusterin (CLU), sortilin-related receptor 1 (SORL1), bridging integrator 1 (BIN1), triggering receptor expressed on myeloid cells 2 (TREM2), ATP-binding cassette subfamily A member 7 (ABCA7), etc. (2–9). However, the genetic contribution to AD development is far from clear and new candidate genes remain to be identified.
Deposition of amyloid β protein (Aβ) leading to the formation of neuritic plaques in the brain is a hallmark of AD neuropathology (10–12). Aβ is generated from APP by β- and γ-secretase cleavages (13, 14). Several proteases including endothelin-converting enzymes (ECE), insulin-degrading enzyme (IDE), angiotensin-converting enzyme (ACE), and neprilysin (MME) have been shown to degrade Aβ in vivo and in vitro (15–18). Recent studies revealed that several susceptibility variants in these Aβ-degrading genes are associated with LOAD, which strongly suggested the critical role of Aβ metabolism in AD pathogenesis (19–22).
Here, we performed WES in a large LOAD pedigree without any known AD causative gene mutations and found that the R186C mutation encoded in the ECE2 gene was segregated with disease status in 1 branch of the pedigree. Furthermore, an excess burden of ECE2 deleterious coding mutations that contributed to AD pathogenesis was found through screening whole ECE2 gene exomes in a small cohort of Han Chinese ancestry. Subsequently, functional studies demonstrated that ECE2 mutations impaired its enzymatic activity in degrading Aβ in HEK293 cells. We further demonstrated that the total Aβ load in AD model mice was reduced, and impaired learning and memory functions were rescued by overexpression of wild-type but not mutant ECE2 protein. In summary, our study demonstrates that ECE2 rare variants may increase AD risk through interfering with the degradation of Aβ degradation, which suggests that ECE2 is a potential target for AD treatment.
Results
The clinical phenotypes of the family with LOAD.
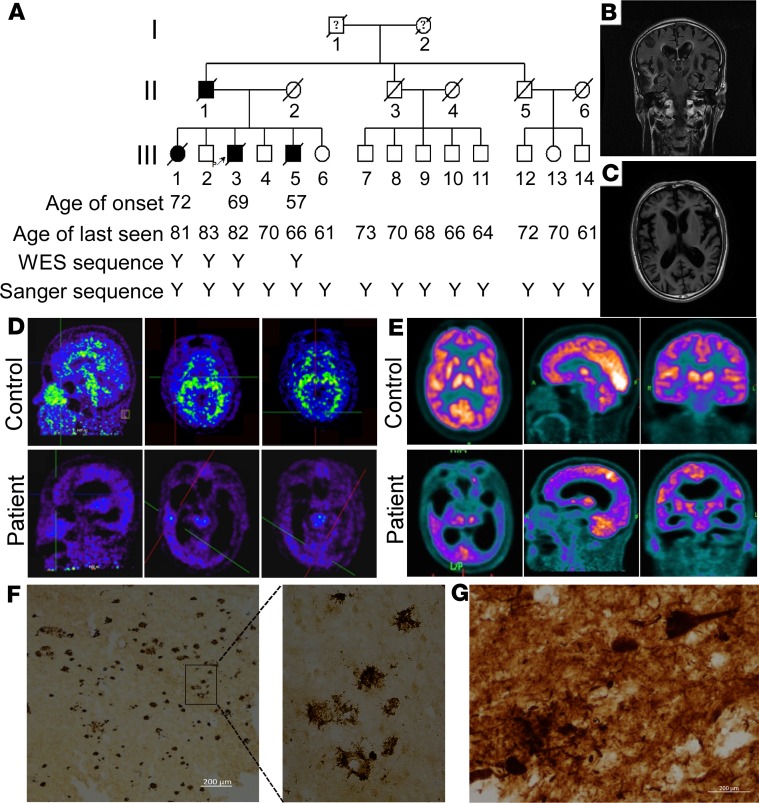
We identified a large family with 4 AD patients over 2 generations (family A) (Figure 1A). All affected individuals and healthy members of the third generation were assessed in at least 1 medical history review and thorough neurological examination by 2 experienced neurologists, and the clinical features of the 3 patients are summarized in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135119DS1). The average age at onset of the patients was 66 ± 7.9 years, and the average disease duration was 11 ± 1.7 years. The proband (III:3) began to develop episodic memory loss at the age of 69. He often repeated words, misplaced things, and had difficulty in remembering the names of friends. Over the next 7 years, his symptoms worsened with a gradual loss of calculation abilities, orientation in time and space, and perceptivity. He also presented with personality changes characterized by irritability, depression, and barely talked to others. He relied entirely on others for care after 8 years of onset, including bathing, dressing, eating and other daily tasks. He died 10 years after the onset of the disease. The clinical features of patients III:1 and III:5 were similar to those of the proband, with progressive cognitive decline and personality changes. Neurological examination of the 3 patients revealed no myoclonic jerks, seizures, extrapyramidal or upper motor neuron signs. Structural magnetic resonance imaging (MRI) showed widespread cortical and hippocampal atrophy in patient III:3 (Figure 1, B and C), and lacunar stroke and medial temporal lobe atrophy in patient III:5 (Supplemental Figure 1). The proband had increased 11C-Pittsburgh compound B (PiB) accumulation in the frontal lobe, temporal lobe, and occipital lobe by PET imaging (Figure 1D). 18F-fluorodeoxyglucose (18F-FDG) PET examination showed hypometabolic activity in temporoparietal, frontal, and occipital cortices of patient III:3 (Figure 1E). Senile plaques and neurofibrillary tangles were clearly detected in the proband by autopsy (Figure 1, F and G). There was no accumulation of α-synuclein in these brain tissues (Supplemental Figure 2). The patients had a clear set of symptoms and pathological phenotypes of LOAD. Patients III:7–III:14 had no subjective complaints about cognitive decline, and neurological examination and neuropsychological tests (Mini-Mental State Exam [MMSE] and Montreal Cognitive Assessment [MoCA]) were also normal (Supplemental Table 2).
Figure 1. Pedigree and clinical features.
(A) Pedigree of family A with Alzheimer’s disease. Subjects in the family were identified by number in 3 generations I, II, and III. Open symbols = unaffected; filled symbols = affected; symbols with a question mark = phenotype unknown; symbols with a diagonal line = deceased subjects; squares = male; circles = female; arrow indicates the proband; Y indicates the analyzed subject. (B and C) Structural magnetic resonance imaging (MRI) of patient III: 3. Coronal T2 flair MRI (B) and axial T1-weighted MRI (C) showed widespread atrophy in cortex and hippocampus. (D) 11C-Pittsburgh compound B (PiB) PET. Compared with the control (III:4, top row), patient III:3 (bottom row) showed increased PiB signal accumulation in frontal lobe (left), temporal lobe (middle), and occipital lobe (right). (E) 18F-fluorodeoxyglucose (18F-FDG) PET. Axial (top, left), sagittal (top, middle), and coronal (top, right) images from the control III:4; axial (bottom, left), sagittal (bottom, middle), and coronal (bottom, right) images from patient III:3. Hypometabolic activity in temporal-parietal, frontal, and occipital cortices of patient III:3. (F) Amyloid plaques in brain tissue sections from patient III:3 were immunostained with anti-Aβ antibody 6E10. (G) Phospho-tau staining in brain tissue sections from patient III:3. Scale bars: 200 μm.
Exome sequencing identified ECE2 as a candidate risk gene for AD.
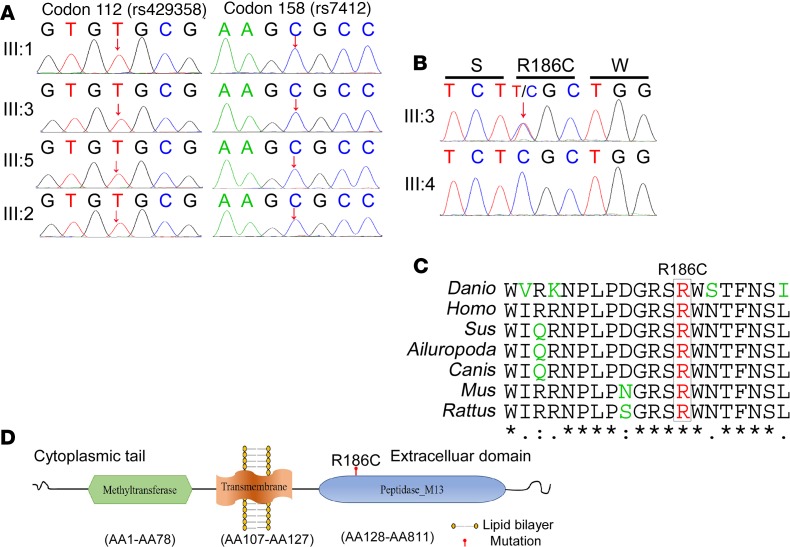
Initial genotyping results showed that the proband AD patient carried no pathogenic mutations in the APP, PSEN1, PSEN2, GRN, MAPT, NOTCH3, PPNR, or C9ORF72 gene (Supplemental Figure 3). The APOE genotypes of the 3 patients (III:1, III:3, and III:5) and 1 control (III:2) were all ε3/ε3, which means that the APOE genotype cannot account for the phenotype of this pedigree (Figure 2A). To further determine the genetic contributions, exome sequencing was performed on the 3 affected individuals (III:1, III:3, and III:5) and 1 control (III:2) in family A (Supplemental Table 3). After SNP and indel calling, we identified over 66 million reads/person, of which 98% mapped to the exome. Variants identified with a quality score lower than 20 and a depth of coverage lower than 5 were removed. No previously reported AD-associated pathogenic mutation was identified in this family by screening the Human Gene Mutation Database (HGMD) and ClinVar database. Because family A showed possible autosomal dominant inheritance, the dementia-associated mutation in this family should not be common in the general population. We then removed the nonsynonymous/splice acceptor and donor site/insertions or deletions (NS/SS/indel) variants with minor allele frequency (MAF) greater than 0.5% in the following databases: Genome Aggregation Database (gnomAD), ExAc database, the 1000 Genomes Project, the 8 previously exome-sequenced HapMap samples (HapMap 8), the Beijing Genomics Institute (BGI) in-house exome variant data set, and the PVFD database (variant frequency in normal population database). More than 2200 rare variants were identified per individual. Twenty variants were shared by the 3 affected individuals and absent in the unaffected individual within the family. Four variants were further removed due to their known associated clinical symptoms not matching the phenotype of family A (Supplemental Table 4). Genotyping the remaining 16 variants in 10 controls (III:4, III:6, and III:7–III:14) of family A using Sanger sequencing excluded an additional 13 variants because they did not cosegregate with disease status. Finally, 3 variants in 3 genes segregated with disease were selected as candidate mutations (Supplemental Table 5). The potential pathogenicity of the identified missense variants was evaluated by ReVe Score (23), PolyPhen-2 (Polymorphism Phenotyping v2; http://genetics.bwh.harvard.edu/pph/) (24), Sorting Intolerant from Tolerant (SIFT) (http://sift.jcvi.org/) (25), and Mutation Taster (26). The variants, D378N in YEATS2 and H462Y in APEH, were evaluated to be “tolerable” by ReVe and “Polymorphism” by Mutation Taster. The variant rs368866385 (Arg186Cys, ECE2, NM_001100120.1) with the highest ReVe score and evaluated to be damaging by 3 in silico prediction software packages was picked up as a promising candidate AD risk variant (Figure 2B). Arginine 186 is located in the M13 peptidase domain of ECE2 and a highly conserved residue across species (Figure 2, C and D).
Figure 2. ApoE genotyping and bioinformatics analysis of R186C in the ECE2 gene.
(A) Sanger sequencing of ApoE genotyping of codon 112 (rs429358) and codon 158 (rs7412). The arrow indicates the position of the single-nucleotide polymorphism. (B) Sanger sequencing of the ECE2 gene of the proband III:3 (up) and control III:4 (bottom) in family A with the c.556C>T, p.186R>C mutation. (C) R186 is a highly conserved residue across species. (D) Schematic diagram of the ECE2 protein. R186 is located in the M13 peptidase domain of ECE2.
Gene-based association test confirmed ECE2 as a promising candidate risk gene for AD.
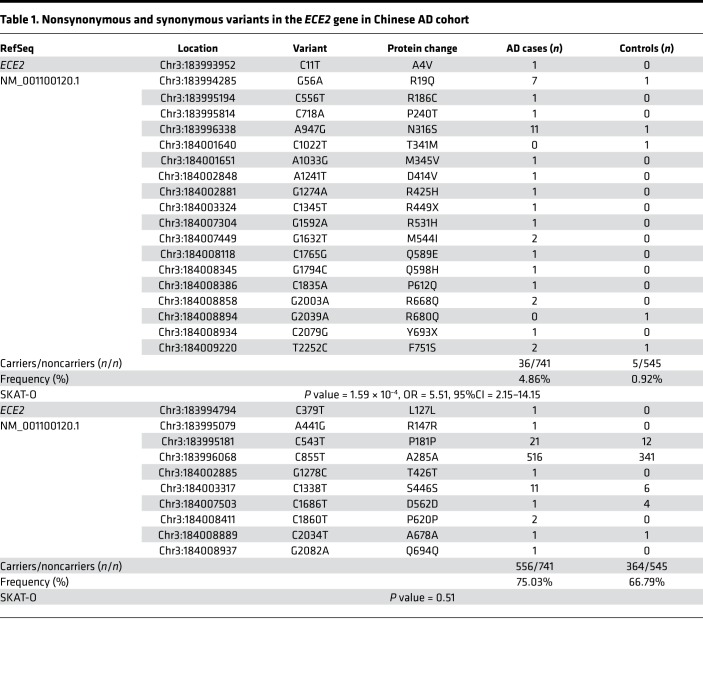
To determine if there are additional AD-associated variants in the ECE2 gene, we sequenced the coding region of ECE2 in 741 AD cases and 545 controls. Among the 741 cases, 72 were diagnosed with AD through cerebrospinal fluid testing (n = 33), PET-CT (n = 38), or autopsy (n = 1). We identified 29 high-quality SNPs that met predefined quality criteria. None of the polymorphic ECE2 coding variants was in linkage disequilibrium in our data sets. No subject carried more than 1 ECE2 coding variant. A total of 19 nonsynonymous variants and 10 synonymous variants in the ECE2 coding region have been identified. Gene-based burden analyses were performed using the sequence kernel association test – optimal (SKAT-O) (SNP set; ref. 27). In our AD data set, 4.86% of the cases (36 of 741) and only 0.92% of the controls (5 of 545) carried ECE2 nonsynonymous variants (P = 1.59 × 10–4; OR = 5.51, 95%CI = 2.15–14.15) (Table 1). When using more strict criteria to extract likely damaging variants (ReVe value > 0.7), the SKAT-O result remained significant (P = 0.01; OR = 9.71, 95%CI = 1.27–74.48) (Supplemental Table 6). In contrast, we did not observe a difference in synonymous variants for AD cases in our data set compared with controls (1.12-fold, P = 0.51). Our results indicate that the association between nonsynonymous variant burden in ECE2 and AD risk is positive, which further confirmed that ECE2 is a promising candidate risk gene for AD.
Table 1. Nonsynonymous and synonymous variants in the ECE2 gene in Chinese AD cohort.
ECE2R186C and ECE2F751S mutants reduced Aβ degradation in vitro.
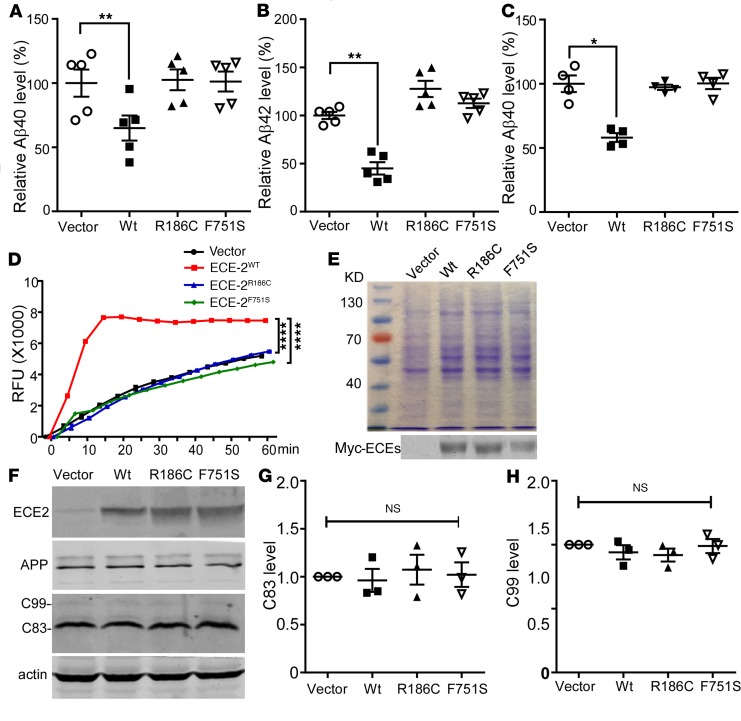
ECE2 was reported to degrade Aβ (28), and ECE2 homozygous knockout mice display significantly elevated endogenous Aβ40 and Aβ42 (29). As shown in Supplemental Table 6, F751S is the only variant predicted to be damaging by bioinformatics analysis, and is carried by both AD patients and normal control; therefore, further biological experiments were warranted. To investigate the effect of ECE2 R186C and F751S mutations on Aβ degradation, the Swedish-mutant-APP stable cell line 20E2 was transfected with ECE2 variants. Compared with empty vector control, overexpression of wild-type ECE2 (ECE2WT) significantly decreased the levels of secreted Aβ40 and Aβ42 in conditioned media to 64.93% ± 9.77% and 45.10% ± 6.38%, respectively (P < 0.01) (Figure 3, A and B), whereas ECE2R186C and ECE2F751S had no effect on Aβ40 and Aβ42 levels (P > 0.05). In cell extracts, ECE2WT also showed markedly reduced intracellular Aβ40 levels, to 58.07% ± 3.48% (P < 0.05), while the intracellular Aβ40 levels in the ECE2R186C- and ECE2F751S-transfected cells were similar to that in the control group (P > 0.05) (Figure 3C). The data suggested that unlike ECE2WT, overexpression of ECE2R186C and ECE2F751S mutants had no effect on Aβ degradation in 20E2 cells.
Figure 3. Effect of ECE2 mutants on Aβ levels and in vitro enzymatic activity.
20E2 cells were transfected with vector, ECE2WT, ECE2R186C, or ECE2F751S constructs for 72 hours. Levels of Aβ40 and Aβ42 from conditioned medium and cell lysates were measured using sandwich ELISA. (A) Aβ40 level in the conditioned media of 20E2 cells (n = 5). (B) Aβ42 level in the conditioned media of 20E2 cells (n = 5). (C) Aβ40 level in cell lysates of 20E2 cells (n = 4). For in vitro enzymatic activity test, HEK293 cells were transiently transfected with vector, ECE2WT, ECE2R186C, or ECE2F751S constructs for 48 hours, and the proteins were purified under native conditions. (D) McaBk2 peptide (10 μM) in 0.2 M sodium acetate buffer (pH 5.5) was incubated with purified vector, ECE2WT, ECE2R186C, or ECE2F751S proteins at 37°C for 1 hour. Relative fluorescent units (RFU) were recorded at Ex/Em = 320 nm/405 nm (n = 3). (E) Identification of purified proteins by Coomassie staining (upper) and Western blotting (bottom) (n = 3). (F) 20E2 cells were transfected with vector, ECE2WT, ECE2R186C, or ECE2F751S plasmids, and cell lysates were blotted for APP, C-terminal fragments (C83 and C99), and ECE2. Full-length APP and APP C-terminal fragments were detected with C20 antibody. ECE2 variants were detected by 9E10 antibody. C83 (G) and C99 (H) quantified and expressed as the ratio of C83 or C99 level in vector-expressing cells (n = 3). All results are expressed as mean ± SEM. Statistical significance was determined by ANOVA followed by Bonferroni’s multiple-comparisons test.
R186C and F751S mutations are located in the M13 peptidase domain of the ECE2 protein. Immunohistochemical analyses showed that these 2 mutations did not alter distribution and subcellular location of the mutant ECE2 proteins (Supplemental Figures 4 and 5). To examine whether the mutations impair ECE2’s metalloprotease activity, leading to the deficiency in Aβ degradation, the activity of ECE2 variants was measured using an McaBk2 assay (30). The results showed that the relative fluorescence units (RFU) value of ECE2WT was significantly higher than that of ECE2R186C and ECE2F751S (Figure 3D, P < 0.0001), indicating ECE2WT degraded the substrate much faster. There was no significant difference in RFU value between ECE2R186C, ECE2F751S, and empty vector control, indicating that R186C and F751S diminished ECE2’s enzymatic activity (Figure 3, D and E). To investigate whether the ECE2R186C and ECE2F751S mutants had effects on Aβ production, the levels of APP and its C-terminal fragments were assayed by Western blot with C20 antibody. No significant difference was detected in C83 and C99 level between groups, suggesting that overexpressed ECE2WT, ECE2R186C, and ECE2F751S did not alter APP processing to generate Aβ (Figure 3, F–H). These data indicated that both mutations R186C and F751S caused loss of ECE2’s ability to degrade Aβ, resulting in Aβ accumulation.
ECE2WT, not ECE2R186C, reduced plaque formation in AD model mice.
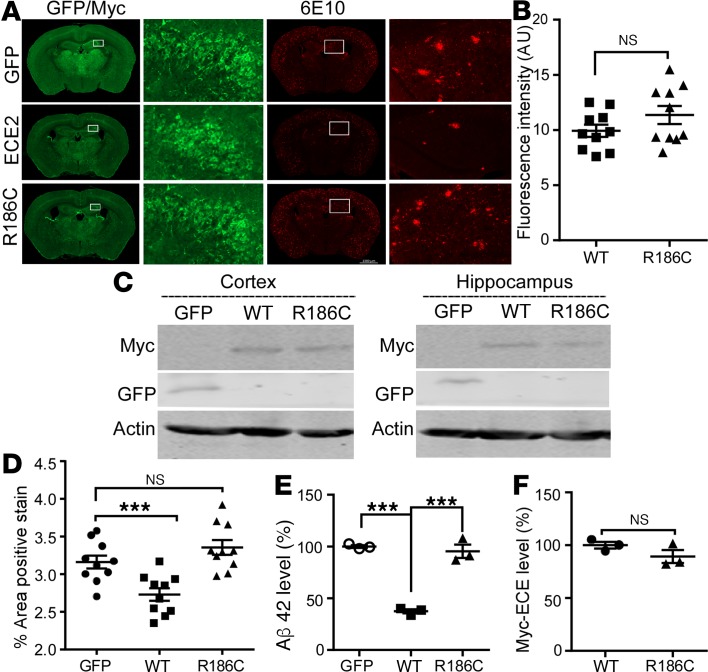
To investigate the effects of ECE2WT and the ECE2R186C mutant on amyloid degradation and plaque formation, adeno-associated viruses (AAVs) carrying human ECE2WT or ECE2R186C were injected bilaterally into the ventricles of neonatal APPNL-G-F/NL-G-F-knockin mice on postnatal day 0 (P0). At 5 months after injection, the exogenous expression of Myc-tagged ECE2 proteins was detected throughout the brain, with dense labeling apparent in cerebral cortex, hippocampus, and subcortical nuclei (Figure 4, A and C). The average immunofluorescence intensity of ECE2WT was similar to ECE2R186C in whole brain sections (P > 0.05, Figure 4B). Overexpression of ECE2WT significantly reduced plaque formation in rAAV9-ECE2WT–injected mice compared with rAAV9-GFP–injected mice (2.73% ± 0.08% vs. 3.16% ± 0.09%, P < 0.01) (Figure 4, A and D). However, APPNL-G-F/NL-G-F-knockin mice injected with rAAV9-ECE2R186C exhibited plaque loads similar to those in mice that received rAAV9-GFP (P > 0.05), and an increased plaque formation when compared with rAAV9-ECE2WT–injected mice (3.36% ± 0.10% vs. 2.73% ± 0.08%, P < 0.001) (Figure 4, A and D). ELISAs confirmed that ECE2WT overexpression resulted in a significant decrease in Aβ42 levels of 63% ± 1.9% in the hippocampal tissues from rAAV9-ECE2WT–injected mice compared with GFP control (P < 0.001) (Figure 4E). No differences in Aβ42 level were observed between mice that received ECE2R186C and GFP control virus injection (P > 0.05). The expression level of ECE2WT and ECE2R186C in bilateral hippocampus was verified to be similar by Western blot (P > 0.05) (Figure 4F). These data demonstrated that the expression of ECE2WT reduced the amyloid plaque formation but ECE2R186C abolished ECE2’s Aβ-hydrolyzing function and its inhibitory effect on plaque formation in the brains of APPNL-G-F/NL-G-F-knockin mice in vivo.
Figure 4. Overexpressed ECE2WT, but not ECE2R186C mutant, reduced the total amyloid load in APPNL-G-F/NL-G-F-knockin mice.
Intracranial injection of rAAV9-GFP-Vector, rAAV9-ECE2WT, or rAAV9-ECE2R186C into the cerebral lateral ventricle of neonatal APPNL-G-F/NL-G-F-knockin mice at P0. (A) GFP, ECE2WT, and ECE2R186C expression in bilateral cortex, hippocampus, and subcortical area; Aβ plaques were detected by 6E10 in APPNL-G-F/NL-G-F-knockin mouse brain sections injected with GFP, ECE2WT, or ECE2R186C viruses (n = 10). Scale bar: 1000 μm. (B) Quantification of average immunofluorescence intensities of ECE2WT and ECE2R186C proteins in APPNL-G-F/NL-G-F-knockin mouse brain sections after subtracting the image background (n = 10). (C) Cortex and hippocampus tissue lysates were blotted for GFP, ECE2WT, and ECE2R186C (n = 3). (D) Percentages of plaques were quantified as shown in the scatter diagram (n = 10). (E) Level of Aβ42 in bilateral hippocampus of APPNL-G-F/NL-G-F-knockin mice by ELISA. ELISA readout for Aβ40 was not higher than background noise (n = 3). (F) Quantification of ECE2WT and ECE2R186C expression in bilateral hippocampus (n = 3). All results are expressed as mean ± SEM. Two-tailed Student’s t test was used to analyze the difference between 2 groups, and multiple comparisons were analyzed by ANOVA followed by Bonferroni’s multiple-comparisons test.
ECE2WT, not ECE2R186C, rescued the cognitive deficits in AD model mice.
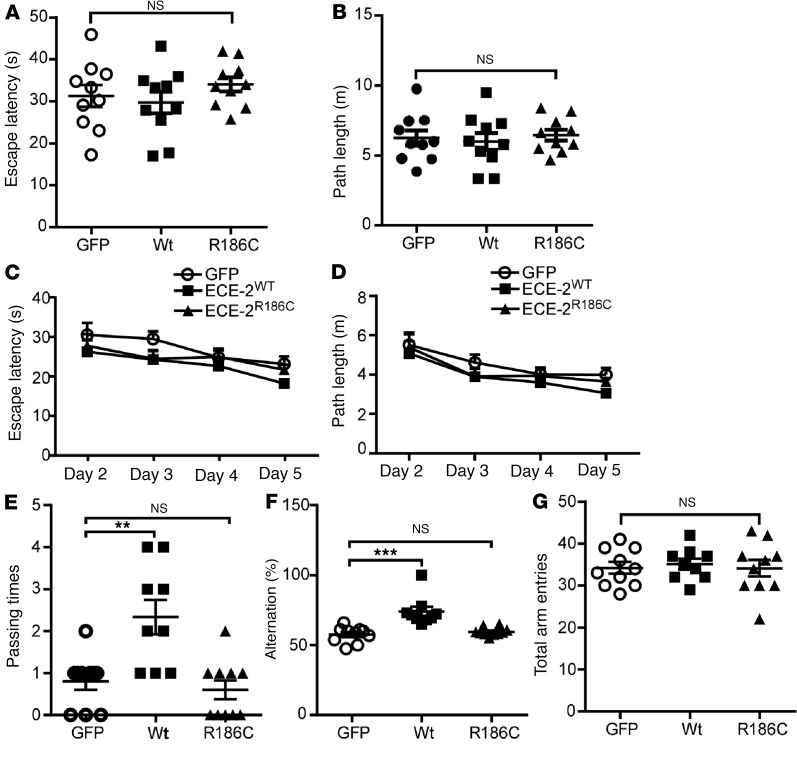
To examine the effects of ECE2WT and the ECE2R186C mutant on learning and memory in vivo, a Morris water maze test for spatial learning and memory and a Y-maze test for working memory were performed on the virus-injected APPNL-G-F/NL-G-F-knockin mice at 5 months of age. In the visible platform test of the Morris water maze test, no significant differences were observed in escape latency (P > 0.05) (Figure 5A) and path length (P > 0.05) (Figure 5B) between the GFP, ECE2WT, and ECE2R186C groups, indicating that all mice had similar motility and vision function. In the hidden platform tests, the rAAV9-ECE2WT–injected mice had relatively shorter escape latency and path length than the rAAV9-ECE2R186C–injected mice and the GFP control, but no significant differences were observed (Figure 5, C and D). However, in the probe trial, the rAAV9-ECE2WT–injected mice traveled significantly more times into the third quadrant where the hidden platform was previously placed than control mice (2.33 ± 0.41 vs. 0.80 ± 0.20, P < 0.05) (Figure 5E). In contrast, the rAAV9-ECE2R186C–injected mice had similar passing times compared with rAAV9-GFP–injected control mice (P > 0.05). We also performed a Y-maze test on the mice, and the results showed that the rAAV9-ECE2WT–injected mice exhibited a significantly higher percentage of spontaneous alternation than the rAAV9-GFP–injected mice (74.00% ± 3.47% vs. 57.53% ± 1.78%, P < 0.001), whereas rAAV9-ECE2R186C–injected mice and rAAV9-GFP–injected mice performed similarly in the Y-maze test (59.57% ± 0.95% vs. 57.53% ± 1.78%, P > 0.05) (Figure 5F). No difference between total arm entries was observed between these groups (P > 0.05) (Figure 5G). Taken together, these data demonstrated that overexpression of ECE2WT protein, but not ECE2R186C, alleviated learning and memory deficits in APPNL-G-F/NL-G-F-knockin mice.
Figure 5. Overexpressed ECE2WT, but not ECE2R186C mutant, alleviated cognitive deficits in APPNL-G-F/NL-G-F-knockin mice.
(A–E) A Morris water maze test consisted of 1 day of visible platform trials and 4 days of hidden platform trials, plus a probe trial 24 hours after the last hidden platform trial. Animal movement was traced and recorded by ANY-maze tracking software. In the visible platform tests, mice in each group exhibited a similar (A) escape latency and (B) swimming path length to escape onto the visible platform. In hidden platform tests, mice were trained with 5 trials per day for 4 days. ECE2WT group mice showed shorter latency (C) and path length (D) to escape onto the visible platform on the third and fourth days, but without statistically significant differences. (E) In the probe trial on the sixth day, ECE2WT group mice traveled into the third quadrant, where the hidden platform was previously placed, significantly more times than GFP and ECE2R186C groups. (F and G) The Y-maze test was performed. ECE2WT-injected mice exhibited significantly higher percentage alternation than GFP-injected mice, whereas ECE2R186C-injected mice and GFP-injected mice performed similarly (F). No difference between arm entries was observed between groups (G). Data represented as mean ± SEM (for behavior tests, n = 10 GFP, 9 ECE2WT, and 10 ECE2R186C). Significance was assessed by 1-way ANOVA with Newman-Keuls post hoc test.
Discussion
Recent studies showed that low-frequency coding variants not detected by GWAS may be a source of functional variants, with significant effects on LOAD risk. Next-generation sequencing technology provides powerful tools for identifying such rare variations. However, the frequency of rare coding variants is relatively low, and the impact of a single rare variation is often moderate in disease progression, which poses challenges to sample size and statistical method. One solution is to use highly selected pedigrees with increased genetic risk for disease to identify candidate risk genes, followed by a targeted resequencing of the gene in a case-control cohort. Here, we used WES to identify the ECE2 R186C mutation segregating with AD phenotypes in one branch of a LOAD pedigree. Although we cannot exclude the possibility that other variants may also contribute to the AD feature of this family, ECE2 R186C was the only variant we identified to be an AD-associated gene variant. Mutation analysis by using 7 in silico missense prediction programs (SIFT, Polyphen-2_HDIV, Polyphen-2_HVAR LRT, MutationTaster, Provean, and ReVe) suggests that this mutation would potentially result in altered ECE2 protein function. Additionally, this mutation was absent in 545 Chinese controls with normal cognition, and showed an ultrarare allele frequency in a public database (0.0003 and 0 in gnomAD East Asian and African American groups, respectively). Subsequently, our study showed that the R186C mutation impaired ECE2’s ability to degrade Aβ. By analyzing all ECE2 rare coding mutations identified in an AD case-control cohort of Han Chinese ancestry, the SKAT-O test revealed that nonsynonymous rare coding mutations in the ECE2 gene were enriched in AD patients by 5-fold compared with control, whereas synonymous mutations in the ECE2 gene were comparable between AD cases and the control cohort (P = 0.51). This observation provided robust evidence that ECE2 is an AD risk gene, and nonsynonymous mutations may play an important role in AD pathogenesis. Although a targeted exome and genome sequencing analysis for APP-Aβ metabolism genes in a cohort from North America and the United Kingdom did not identify an obvious correlation between ECE2 and AD pathogenesis (22), this difference might be due to the different ethnicities and small sample size of the 2 projects. Indeed, investigations on a larger sample size from multiple populations will be required in a future study to clarify the role of ECE2 in AD pathogenesis.
ECE family member proteins include ECE1 and -2, and encode membrane-bound metalloproteases mediating big endothelin conversion into vasoactive endothelins (31, 32). ECE2 is a type II transmembrane protein with a short N-terminal cytoplasmic domain, a transmembrane domain, and a long M13 peptidase domain within the C-terminal extracellular region (33). The mutations R186C and F751S identified in the AD patients were located in the M13 domain that contains an active peptidase domain. Consistent with previous reports (18, 34), we showed that overexpression of ECE2WT possessed sufficient Aβ-degrading ability to reduce Aβ40 and Aβ42 levels. However, overexpression of the R186C or F751S mutant could not reduce Aβ40 and Aβ42 levels. In vitro enzymatic activity analysis revealed that the R186C or F751S mutation diminished the enzymatic activity of ECE2, contributing to the impaired Aβ degradation. Furthermore, our in vivo studies demonstrated that the R186C mutation caused loss of ECE2 function in the mouse model, which indicated that ECE2R186C protein might lead to Aβ accumulation to form neuritic plaques and cognitive deficits.
It has been demonstrated that both Aβ40 and Aβ42 could enhance vascular tone by inducing endothelin 1 (ET-1) expression in rat aorta rings (35), which indicated that Aβ might reduce blood flow in cerebral vessels, leading to Aβ accumulation, cerebral vessel destruction, and finally neurodegeneration (36–40). Given that ECE2 inactivation could lead to Aβ accumulation in the central nervous system, it is possible that mutant ECE2 reduces cerebral blood flow via an Aβ-mediated ET-1 pathway and finally impairs cognitive function. Additionally, it has been reported that ECE2 is mainly expressed in somatostatin-expressing (SOM+) interneurons (41), which are essential for cognitive function. In addition, Aβ aggregates may be toxic to SOM+ interneurons (42–44) and SOM innervation is significantly impaired during AD (45). These results suggested that Aβ accumulation induced by ECE2 mutations may lead to the loss of SOM+ interneurons and cognitive deficits in AD. Therefore, it is important to explore the effect of ECE2 mutations on cerebral blood flow and the functions of SOM+ interneurons in the mutation carriers, which might provide valuable insights into the ECE2-associated AD pathogenesis.
We have identified an AD risk gene, ECE2, and provided evidence that low-frequency variants exert an intermediate to high effect on facilitating AD pathogenesis. Our study suggested that ECE2 is a potential therapeutic target for AD treatment.
Methods
A LOAD family for candidate gene discovery.
Family A was a 3-generation Chinese pedigree with 4 members presenting as possible autosomal dominant inheritance (Figure 1A). All family members of generation III completed a clinical assessment, neurological examination, and neuropsychological tests (including MoCA and MMSE). Patients III-3 and III-5 had blood tests to measure thyroid hormones, vitamin B12, and treponema pallidum hemagglutination test (TPHA). Patients III-3 and III-5 also underwent MRI to evaluate brain atrophy and other conditions, while only patient III-3 received 18F-FDG/PiB-PET scans. Brain autopsy was performed on patient III-3 after death within 3 hours. Immunohistochemistry was performed using the following antibodies: tau (AT8, Invitrogen, catalog MN1020); β-amyloid (6E10, Sigma-Aldrich, catalog A1474); and α-synuclein (LB509, Abcam, catalog ab27766).
Exome sequencing.
Exome-sequencing was performed as previously reported (46, 47). Genomic DNA from patients was sheared by the Covaris system, then hybridized to NimbleGen’s SeqCap EZ exome library v2.0 for enrichment according to the manufacturer’s instructions. The enriched library targeting the exome was sequenced on the Illumina HiSeq 2000 platform for paired-end reads with read length of 90 bp. The reads were aligned to the reference coding sequence using the BWA (Burrows Wheeler Aligner) software. After alignment, the output files were used to perform sequencing coverage and depth. Genome Analysis Toolkit (GATK) software was used to detect single-nucleotide variants (SNVs) and indels. Variants identified with a quality score lower than 20 and a depth of coverage lower than 5 were removed. The HGMD (http:// www.hgmd.cf.ac.uk/ac) and ClinVar database (48) were used to screen reported mutations in previously published studies.
Participants for targeted resequencing study.
The study recruited 741 AD patients (445 female and 296 male) with a mean age at onset of 66.20 ± 10.65 years from the Memory Clinic of Xiangya Hospital. Three hundred twelve (42.10%) AD patients carried the ApoE-ε4 allele. All patients were assessed in a thorough neurologic examination and neuropsychological assessment by 2 experienced neurologists. A diagnosis of probable or definite AD was determined following the NINCDS-ADRDA criteria (49). Ninety-six of 741 patients (12.96%) had a positive family history for AD, but only the probands were included in the SKAT-O analysis. AD patients carrying known mutations in APP, PSEN1, PSEN2, GRN, MAPT, NOTCH3, C9ORF72, or PPNR were excluded. Five hundred forty-five (261 female and 284 male) controls were recruited from the partners of patients visiting our Memory Clinic or geographically matched community-recruited volunteers. The mean age at inclusion was 68 ± 7.36 years, and 73 (13.39%) of the control subjects were ApoE-ε4 positive. Subjective memory complaints, neurological or psychiatric antecedents, and familial history of neurodegeneration were ruled out in the controls by a systematic medical interview. Cognitive screening was done in the controls with the MMSE (50, 51).
Targeted resequencing of the ECE2 gene.
Resequencing of ECE2 exons was performed on 741 AD cases and 545 controls using targeted capture and MPS. The baits, a pool of 18,167 individually synthesized 5′-biotinylated 120-bp RNA oligonucleotides, covered the untranslated regions (UTRs), coding regions, and exon-intron boundaries of 93 dementia-related genes, including APP, PSEN1, PSEN2, GRN, MAPT, NOTCH3, PPNR, ECE2, etc. Target enrichment was done with HaloPlex technology (Agilent Technologies). After quality control, captured library sequencing was carried out on Illumina HiSeq × Ten Analyzers following the manufacturer’s standard sequencing protocols for 150 cycles per read to generate paired-end reads. Image analysis, error estimation, and base calling were performed using Illumina Pipeline software to generate raw data. We used AfterQC (52) to generate clean reads for further analysis. The clean reads (with a length of 150 bp) derived from targeted sequencing and filtering were then aligned to the human genome reference (hg19) using BWA software (53). GATK software was used to detect SNVs and indels. Both variant and reference genotypes were called using the same parameters. When reaching a quality score greater than 20 and coverage greater than 5×, a variant or reference gene call was made. Numbering of variations at the gDNA level was based on the GenBank accession number NC_000003.11, transcript level on NM_001100120.1.
Gene-based analysis with SKAT-O.
The SKAT-O test (27, 54) was implemented in R using SKAT v1.0.9 to determine the difference in the aggregate burden of ECE2 gene variants between AD cases and controls. An empirical P value was derived from the distribution of null results based on 10,000 permutation trials in which case/control assignment was randomized. SKAT-O statistics were performed on 2 subsets of variants that were created based on impact of the variants. One is synonymous variants, the other one is nonsynonymous variants, including frameshift, missense, in-frame deletion, and nonsense variants. Also, any bias introduced in the test due to an unbalanced case-control set was compared to observations from the synonymous variants class that was used to set the background expectation.
Plasmids and virus.
Human ECE2 variant 5, preferentially expressed in brain (33), was subcloned into the pcDNA4/myc-His mammalian expression vector (Invitrogen) to generate pcDNA4/Myc-HisA-ECE2-WT. Plasmids carrying the ECE2 R186C or F751S mutation were generated by PCR-based site-directed mutagenesis using pcDNA4/Myc-HisA-ECE2-WT as a backbone according to the manufacturer’s instructions. All constructs were verified by Sanger sequencing. The WT and R186C mutant ECE2 with Myc tag were introduced into the pFB-AAV9-CAG-WPRE-SV40pA vector under the control of the chicken β-actin promoter for AAV virus production. The recombinant viruses were generated and purified by Virovek. Infectious rAAV particles were expressed as vector genomes (vg/mL).
Cell culture and transfection.
HEK293 cells (RRID: CVCL_0045) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM L-glutamine, and 100 U/mL penicillin-streptomycin sulfate (Invitrogen). HEK293 cells were stably transfected with human Swedish mutant APP695 to generate 20E2 cells (55). 20E2 cells were cultured in HEK293 medium containing 75 μg/mL G418 sulfate. All cells were maintained at 37°C in an incubator containing 5% CO2. For transfection, cells were grown to approximately 70% confluence and transfected with 2 μg of plasmid DNA per 35-mm plate and 4 μL of Lipofectamine-plus Reagent (Invitrogen), according to the manufacturer’s instructions.
Intraventricular injection.
APPNL-G-F/NL-G-F-knockin mice were obtained from Takaomi C. Saido of the RIKEN Brain Science Institute (Wako, Japan)(56). Experiments were performed on APPNL-G-F/NL-G-F-knockin neonates. Within 12 hours of birth, neonates were intracranially injected with 1 μL (2 × 1013 vg/mL) of the viral solution into each ventricle.
Western blot and immunohistochemical analysis.
Cells were lysed in cold RIPA-DOC buffer (0.1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 0.15 M NaCl, 0.05 M Tris-HCl, pH 7.2) plus protease inhibitor cocktail (Roche). Cell lysates were analyzed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) with 16% Tris-tricine or 7.5% Tris-glycine gels, and then transferred to Immobilon-FL PVDF membranes (Millipore). Antibody 9E10 (Abcam, catalog ab32) was used to detect Myc-tagged proteins. APP and its C-terminal fragment products were detected with C20 antibody (57). β-Actin was detected with monoclonal antibody AC-15 (Sigma-Aldrich). The membranes were incubated with IRDye 800CW–labeled goat anti-mouse (Li-COR Biosciences, 926-32212) or anti-rabbit secondary antibodies (Li-COR Biosciences, 925-32213) and scanned on the Odyssey system (LI-COR Biosciences). For amyloid plaque detection, the mouse brains were fixed in 4% paraformaldehyde and sectioned at 30 μm in a Leica cryostat. Monoclonal antibody 6E10 (Sigma-Aldrich, catalog A1474) against amino acids 1–16 of human Aβ was used for plaque detection. Monoclonal antibody 9E10 was used to detect ECE2 expression. 4′,6-Diamidino-2-phenylindole (DAPI) was used for visualization of nuclei. Images were acquired using an inverted fluorescence microscope (Zeiss Axio Observer). A total of 24 coronal brain slices (180 μm apart; every sixth section) per mouse and a total of 10 animals per group were used for staining. Plaques were quantified by percentage of staining area per slice for each mouse. For quantification of WT and R186C mutant ECE2 in brain sections, the average signal intensity of Myc-tagged protein was measured by ImageJ (NIH) with subtraction of mean background intensity.
Aβ ELISA.
Conditioned medium from cell culture was collected, and protease inhibitors and 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF) (Roche) were added to prevent Aβ degradation. For intracellular Aβ detection, cells were lysed in cold cell extraction buffer (1× PBS with 0.1% Triton X-100 and protease inhibitors), briefly sonicated, and cleared by centrifugation at 12000 g at 4°C for 15 minutes. For mouse brain tissue Aβ42 levels, bilateral hippocampus was homogenized in 5 M guanidine-HCl buffer with 50 mM Tris, pH 8.0. Samples were diluted 10-fold with cold PBS with 1× protease inhibitor cocktail and centrifuged at 16,000 g for 20 minutes at 4°C before application to the ELISA plate. Quantification of Aβ40 and Aβ42 in these fractions was performed using a Human Aβ ELISA kit (Invitrogen) according to the manufacturer’s instructions.
ECE2 variants preparation and McaBk2 assay for ECE2 activity.
HEK293 cells were transiently transfected with pcDNA4/Myc-HisA-ECE2WT, pcDNA4/Myc-HisA-ECE2R186C, or pcDNA4/Myc-HisA-ECE2F751S constructs for 48 hours, and the recombinant proteins were further purified using DEAD ion exchange and Talon metal affinity beads (Clontech, catalog 635504) under native conditions. ECE2 enzymatic activity routinely was assayed with the synthetic quenched fluorescent substrate McaBk2 (10 μM) in 0.2 M sodium acetate buffer, pH 5.5, containing 0.01% detergent C12E8 (octaethylene glycol monododecyl ether, Sigma-Aldrich). The rate of substrate hydrolysis was monitored on a FluoroMax plate reader (Horiba) with excitation at 320 nm and emission at 405 nm.
Morris water maze and Y-maze tests.
The mice were assessed in behavioral tests at 5 months of age. The Morris water maze test was performed as previously described (12). In the visible platform test performed on the first day, the mice were tested for 5 continuous trials, with an intertrial interval of 60 minutes. In the hidden platform tests on the following 4 days, a 10-cm-diameter platform was placed in the southwestern (SW) quadrant of the pool. Mice were trained for 5 trials, with an intertrial interval of 60 minutes. Each mouse was allowed 60 seconds to search for the platform. On the last day of the test, the platform was removed and the mice were given 60 seconds to locate where the platform was originally placed. Mouse movement was recorded by automated video tracking (ANY-maze, Stoelting). Y-maze testing occurred in a Y-shaped maze (Stoelting Co.) with 3 gray plastic arms at a 120° angle from each other, each 5 cm wide and 35 cm in length, and shielded with 10 cm high walls. The apparatus was positioned 60 cm above the floor. Each arm was differentiated using visual cues attached to the inner top portion of the wall: white (white squares), red (red triangles), or blue (blue dots). In this test, each mouse was placed in the center of the maze facing toward one of the arms and then allowed to explore freely for 8 minutes. Tracking of animals was achieved automatically using ANY-maze Video Tracking Software (version 4.63). The maze was cleaned before commencing with the next mouse. The spontaneous alternation was analyzed as a measure of spatial working memory. Percentage spontaneous alternation was calculated as the percentage of the number of alternations performed out of the total number of alternations possible, based on the number of arm entries per mouse.
Statistics.
The number of times each experiment was repeated was at least 3 times as indicated in the figure legends. All results were expressed as mean ± SEM. Two-tailed Student’s t test was used to analyze the difference between 2 groups, and multiple comparisons were analyzed by ANOVA followed by Bonferroni’s multiple-comparisons test unless otherwise specified in figure legends. A significance of P < 0.05 was set for acceptance: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant.
Study approval.
Human subject study was approved by the institutional ethics board of Xiangya Hospital. All subjects provided written informed consent for both study enrollment and sample collection. All animal experiments were conducted in accordance with institutional guidelines, with ethical approval from the UBC ACC (The University of British Columbia, Animal Care Center).
Author contributions
LS and WS conceived and designed the experiments. X. Liao, FC, ZS, and Y. Zhang performed the experiments. X. Liao, FC, ZS, Y. Zhang, Juelu Wang, BJ, JG, Jinchen Li, X. Liu, LG, Y. Zhou, Junling Wang, XY, HJ, KX, JL, BT, LS, and WS analyzed and contributed reagents/materials/analysis tools. X. Liao and WS wrote the manuscript. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) Operating Grant MOP-142487 to WS, the National Natural Science Foundation of China (no. 81671075), the National Key Research and Development Programs of China (no. 2017YFC0840104), and Provincial Key Plan for Research and Development of Hunan (no. 2017SK2031). WS was the holder of the Tier 1 Canada Research Chair in Alzheimer’s Disease. X. Liao is the recipient of the China Scholarship Council graduate student award. Y. Zhang is the recipient of Michael Smith Foundation for Health Research Post-Doctoral Fellowship Award.
Version 1. 02/27/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e135119.https://doi.org/10.1172/jci.insight.135119.
Contributor Information
Xinxin Liao, Email: xinliao_86@126.com.
Fang Cai, Email: caifang@mail.ubc.ca.
Zhanfang Sun, Email: sunzhanfang@163.com.
Yun Zhang, Email: yun31@alumni.ubc.ca.
Juelu Wang, Email: jlgracewang@gmail.com.
Bin Jiao, Email: jbin0911@163.com.
Jifeng Guo, Email: guojifeng2003@163.com.
Jinchen Li, Email: lijinchen@csu.edu.cn.
Xixi Liu, Email: liuxixi_677@qq.com.
Lina Guo, Email: 756399240@qq.com.
Yafang Zhou, Email: nuoyafangzhouzyf@126.com.
Junling Wang, Email: wjling8002@126.com.
Xinxiang Yan, Email: xxyan1268@126.com.
Hong Jiang, Email: jianghong73868@126.com.
Kun Xia, Email: xiakun@sklmg.edu.cn.
Jiada Li, Email: lijiada@sklmg.edu.cn.
Beisha Tang, Email: bstang7398@163.com.
Lu Shen, Email: shenlu2505@126.com.
Weihong Song, Email: weihong@mail.ubc.ca.
References
- 1.Zhang Y, Song W. Islet amyloid polypeptide: Another key molecule in Alzheimer’s pathogenesis? Prog Neurobiol. 2017;153:100–120. doi: 10.1016/j.pneurobio.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Lee JH, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68(3):320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, et al. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29(2):293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elias-Sonnenschein LS, Bertram L, Visser PJ. Relationship between genetic risk factors and markers for Alzheimer’s disease pathology. Biomark Med. 2012;6(4):477–495. doi: 10.2217/bmm.12.56. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel-Smith MK, et al. A rare mutation in UNC5C predisposes to late-onset Alzheimer’s disease and increases neuronal cell death. Nat Med. 2014;20(12):1452–1457. doi: 10.1038/nm.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368(2):107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellenguez C, et al. Contribution to Alzheimer’s disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol Aging. 2017;59:220 e1–220 e9. doi: 10.1016/j.neurobiolaging.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 8. doi: 10.1038/s41380-018-0112-7. Bis JC, et al. Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. [published online ahead of print] Mol Psychiatry. 2018; doi: 10.1038/s41380.018.0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120(3):885–890. doi: 10.1016/S0006-291X(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 11.Ly PT, et al. Inhibition of GSK3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest. 2013;123(1):224–235. doi: 10.1172/JCI64516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun X, et al. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A. 2006;103(49):18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Y, et al. Amyloid-β protein (Aβ) Glu11 is the major β-secretase site of β-site amyloid-β precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Aβ Asp1 contributes to Alzheimer pathogenesis. Eur J Neurosci. 2013;37(12):1962–1969. doi: 10.1111/ejn.12235. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S, et al. BACE1 cleavage site selection critical for amyloidogenesis and Alzheimer’s pathogenesis. J Neurosci. 2017;37(29):6915–6925. doi: 10.1523/JNEUROSCI.0340-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata N, et al. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292(5521):1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 16.Hu J, Igarashi A, Kamata M, Nakagawa H. Angiotensin-converting enzyme degrades Alzheimer amyloid beta-peptide (A beta); retards A beta aggregation, deposition, fibril formation; and inhibits cytotoxicity. J Biol Chem. 2001;276(51):47863–47868. doi: 10.1074/jbc.M104068200. [DOI] [PubMed] [Google Scholar]
- 17.Carty NC, et al. Adeno-associated viral (AAV) serotype 5 vector mediated gene delivery of endothelin-converting enzyme reduces abeta deposits in APP + PS1 transgenic mice. Mol Ther. 2008;16(9):1580–1586. doi: 10.1038/mt.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller LK, et al. Mice deficient in endothelin-converting enzyme-2 exhibit abnormal responses to morphine and altered peptide levels in the spinal cord. J Neurochem. 2011;119(5):1074–1085. doi: 10.1111/j.1471-4159.2011.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton G, et al. The role of ECE1 variants in cognitive ability in old age and Alzheimer’s disease risk. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(6):696–709. doi: 10.1002/ajmg.b.32073. [DOI] [PubMed] [Google Scholar]
- 20.Ertekin-Taner N, et al. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum Mutat. 2004;23(4):334–342. doi: 10.1002/humu.20016. [DOI] [PubMed] [Google Scholar]
- 21.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassi C, et al. Influence of coding variability in APP-abeta metabolism genes in sporadic Alzheimer’s disease. PLoS One. 2016;11(6):e0150079. doi: 10.1371/journal.pone.0150079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, et al. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46(15):7793–7804. doi: 10.1093/nar/gky678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012;91(2):224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckman EA, Reed DK, Eckman CB. Degradation of the Alzheimer’s amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276(27):24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- 29.Eckman EA, Watson M, Marlow L, Sambamurti K, Eckman CB. Alzheimer’s disease beta-amyloid peptide is increased in mice deficient in endothelin-converting enzyme. J Biol Chem. 2003;278(4):2081–2084. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- 30.Mzhavia N, Pan H, Che FY, Fricker LD, Devi LA. Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of regulatory peptides. J Biol Chem. 2003;278(17):14704–14711. doi: 10.1074/jbc.M211242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, et al. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994;78(3):473–485. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]
- 32.Shimada K, Takahashi M, Turner AJ, Tanzawa K. Rat endothelin-converting enzyme-1 forms a dimer through Cys412 with a similar catalytic mechanism and a distinct substrate binding mechanism compared with neutral endopeptidase-24.11. Biochem J. 1996;315(Pt 3):863–867. doi: 10.1042/bj3150863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda S, Emoto N, Alimsardjono H, Yokoyama M, Matsuo M. Molecular isolation and characterization of novel four subisoforms of ECE-2. Biochem Biophys Res Commun. 2002;293(1):421–426. doi: 10.1016/S0006-291X(02)00252-8. [DOI] [PubMed] [Google Scholar]
- 34.Pacheco-Quinto J, Eckman EA. Endothelin-converting enzymes degrade intracellular β-amyloid produced within the endosomal/lysosomal pathway and autophagosomes. J Biol Chem. 2013;288(8):5606–5615. doi: 10.1074/jbc.M112.422964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crawford F, Suo Z, Fang C, Mullan M. Characteristics of the in vitro vasoactivity of beta-amyloid peptides. Exp Neurol. 1998;150(1):159–168. doi: 10.1006/exnr.1997.6743. [DOI] [PubMed] [Google Scholar]
- 36.Jagust WJ. Neuroimaging in dementia. Neurol Clin. 2000;18(4):885–902. doi: 10.1016/S0733-8619(05)70231-0. [DOI] [PubMed] [Google Scholar]
- 37.Niwa K, Kazama K, Younkin L, Younkin SG, Carlson GA, Iadecola C. Cerebrovascular autoregulation is profoundly impaired in mice overexpressing amyloid precursor protein. Am J Physiol Heart Circ Physiol. 2002;283(1):H315–H323. doi: 10.1152/ajpheart.00022.2002. [DOI] [PubMed] [Google Scholar]
- 38.Kalaria RN. Cerebral vessels in ageing and Alzheimer’s disease. Pharmacol Ther. 1996;72(3):193–214. doi: 10.1016/S0163-7258(96)00116-7. [DOI] [PubMed] [Google Scholar]
- 39.Niwa K, Kazama K, Younkin SG, Carlson GA, Iadecola C. Alterations in cerebral blood flow and glucose utilization in mice overexpressing the amyloid precursor protein. Neurobiol Dis. 2002;9(1):61–68. doi: 10.1006/nbdi.2001.0460. [DOI] [PubMed] [Google Scholar]
- 40.Palmer JC, Baig S, Kehoe PG, Love S. Endothelin-converting enzyme-2 is increased in Alzheimer’s disease and up-regulated by Abeta. Am J Pathol. 2009;175(1):262–270. doi: 10.2353/ajpath.2009.081054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacheco-Quinto J, Eckman CB, Eckman EA. Major amyloid-β-degrading enzymes, endothelin-converting enzyme-2 and neprilysin, are expressed by distinct populations of GABAergic interneurons in hippocampus and neocortex. Neurobiol Aging. 2016;48:83–92. doi: 10.1016/j.neurobiolaging.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos B, et al. Early neuropathology of somatostatin/NPY GABAergic cells in the hippocampus of a PS1xAPP transgenic model of Alzheimer’s disease. Neurobiol Aging. 2006;27(11):1658–1672. doi: 10.1016/j.neurobiolaging.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 43.Ma K, McLaurin J. alpha-Melanocyte stimulating hormone prevents GABAergic neuronal loss and improves cognitive function in Alzheimer’s disease. J Neurosci. 2014;34(20):6736–6745. doi: 10.1523/JNEUROSCI.5075-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albuquerque MS, et al. Regional and sub-regional differences in hippocampal GABAergic neuronal vulnerability in the TgCRND8 mouse model of Alzheimer’s disease. Front Aging Neurosci. 2015;7:30. doi: 10.3389/fnagi.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martel G, Dutar P, Epelbaum J, Viollet C. Somatostatinergic systems: an update on brain functions in normal and pathological aging. Front Endocrinol (Lausanne) 2012;3:154. doi: 10.3389/fendo.2012.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang JL, et al. Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain. 2011;134(pt 12):3493–3501. doi: 10.1093/brain/awr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang JL, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133(pt 12):3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 48.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- 50.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Jia J, Yang Z. Mini-mental state examination in elderly Chinese: A population-based normative study. J Alzheimers Dis. 2016;53(2):487–496. doi: 10.3233/JAD-160119. [DOI] [PubMed] [Google Scholar]
- 52.Chen S, Huang T, Zhou Y, Han Y, Xu M, Gu J. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics. 2017;18(suppl 3):80. doi: 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S, Fuchsberger C, Kim S, Scott L. An efficient resampling method for calibrating single and gene-based rare variant association analysis in case-control studies. Biostatistics. 2016;17(1):1–15. doi: 10.1093/biostatistics/kxv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18(13):1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- 56.Saito T, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17(5):661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 57. doi: 10.1038/s41380-018-0101-x. Zhang S, et al. A presenilin-1 mutation causes Alzheimer disease without affecting Notch signaling [published online June 18, 2018]. Mol Psychiatry. doi: 10.1038/s41380.018.0101-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.