Abstract
Inherited bone marrow failure syndromes, such as Fanconi anemia (FA) and Shwachman-Diamond syndrome (SDS), feature progressive cytopenia and a risk of acute myeloid leukemia (AML). Using deep phenotypic analysis of early progenitors in FA/SDS bone marrow samples, we revealed selective survival of progenitors that phenotypically resembled granulocyte-monocyte progenitors (GMP). Whole-exome and targeted sequencing of GMP-like cells in leukemia-free patients revealed a higher mutation load than in healthy controls and molecular changes that are characteristic of AML: increased G>A/C>T variants, decreased A>G/T>C variants, increased trinucleotide mutations at Xp(C>T)pT, and decreased mutation rates at Xp(C>T)pG sites compared with other Xp(C>T)pX sites and enrichment for Cancer Signature 1 (X indicates any nucleotide). Potential preleukemic targets in the GMP-like cells from patients with FA/SDS included SYNE1, DST, HUWE1, LRP2, NOTCH2, and TP53. Serial analysis of GMPs from an SDS patient who progressed to leukemia revealed a gradual increase in mutational burden, enrichment of G>A/C>T signature, and emergence of new clones. Interestingly, the molecular signature of marrow cells from 2 FA/SDS patients with leukemia was similar to that of FA/SDS patients without transformation. The predicted founding clones in SDS-derived AML harbored mutations in several genes, including TP53, while in FA-derived AML the mutated genes included ARID1B and SFPQ. We describe an architectural change in the hematopoietic hierarchy of FA/SDS with remarkable preservation of GMP-like populations harboring unique mutation signatures. GMP-like cells might represent a cellular reservoir for clonal evolution.
Keywords: Hematology, Oncology
Keywords: Bone marrow, Genetic diseases, Leukemias
The hematopoietic hierarchy in FA/SDS is characterized by markedly reduced hematopoietic stem cells/progenitor cells, with relative preservation of GMP-like cells harboring high mutation rates and a unique mutation signature.
Introduction
Myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) comprise a spectrum of hematopoietic disorders. Despite intensive chemotherapy and hematopoietic stem cell (HSC) transplantation, the overall survival of advanced MDS/AML remains low, approximately 60% in children and approximately 30% in adults (1). The outcome is further compromised by treatment-related, long-term adverse events (2).
Hematopoiesis is a complex developmental system that is organized as a hierarchy sustained by multipotent HSCs. Although typically depicted with increasingly restricted oligopotent and unipotent progenitors downstream of HSCs, recent studies demonstrate a reshaping of the architecture of human hematopoietic hierarchy between in utero fetal liver and adulthood time points (3–5). Transcriptional and functional analysis suggests that by adulthood, there is predominantly a 2-tier hierarchy of multipotent and unipotent human stem progenitor stem cells (HSPCs) (5).
AML is a heterogeneous disorder that derives from early HSPCs, which undergo malignant transformation to leukemic blasts and clonal expansion. Deep sequencing of leukemic samples extrapolated the existence of founding clones and derived subclones (6). AML is sometimes preceded by MDS. MDS is a clonal preleukemic disease state with cytopenia due to underproduction, abnormal differentiation, increased apoptosis, and varying degrees of leukemic blasts and carries a high risk of progression to leukemia. The incidence of both MDS and AML increases with age (7), but both can present in early childhood (8, 9).
Several cytogenetic abnormalities have been identified in bone marrow samples from patients with de novo MDS/AML, including –7, +8, and del(20q). Genes that are mutated and might be involved in MDS/AML evolution have been recently discovered, for example, RNA-splicing machinery (e.g., SRSF2, SF3B1, U2AF1), DNA methylation (e.g., IDH1, IDH2, TET2, DNMT3A), transcription factor (e.g., RUNX1), chromatin modification (e.g., EZH2, ASXL1), signal transduction (e.g., FLT3), RAS pathway (e.g., KRAS), cohesin complex (e.g., STAG2), and DNA repair (e.g., FANCL) genes (reviewed in ref. 10). These data advanced our knowledge about MDS/AML pathology; however, the mechanisms underlying clonal initiation and progression are largely unknown.
Although rare, inherited bone marrow failure syndromes (IBMFSs) provide an opportunity to study AML evolution and progression because of a high risk of MDS/AML (11, 12) and stepwise progression from nonmalignant hematopoietic phase, to MDS (13), and on to AML (14–16). We previously showed that by the age of 18 years, patients with the common IBMFSs Fanconi anemia (FA) and Shwachman-Diamond syndrome (SDS) have a 75% and 25% risk, respectively, of developing marrow cytogenetic abnormalities, MDS, or AML (11). AML secondary to MDS has a particularly poor outcome. Only a few studies that focused on clonal hematopoiesis in IBMFSs have been published. TP53 mutations were identified in some SDS patients with (17) or without MDS/AML (18). RUNX1 mutations have been detected in whole marrow cells from several patients with FA without transformation (19). CSFR3 (18, 20, 21) and RUNX1 (22) mutations have been detected in whole marrow cells from severe congenital neutropenia patients with and without MDS/AML. Further studies are necessary to decipher the cells that initiate transformation and why they abnormally accumulate mutations.
In this study, we aimed to discover cellular and molecular signatures underlying early clonal evolution when no clinical signs of MDS/AML are detected in 2 relatively prevalent IBMFSs that feature an initial marrow failure phase and frequently progress to MDS/AML: FA and SDS. FA is caused by germline mutations in 1 of 23 DNA repair genes collectively referred to as the FA pathway (23), and SDS is caused by germline mutations in genes that are involved in the late stage of 60S ribosome subunit maturation, SBDS (24), DNAJC21 (25), and EFL1 (26), but also in SRP54 (27), which is involved in the cotranslational protein-targeting pathway. We found that the granulocyte-monocyte progenitor–like (GMP-like) population is relatively preserved compared with marked exhaustion of other cell populations and carries a high mutation load and a unique trinucleotide mutation signature, suggesting that GMP-like cells are a reservoir for clonal evolution.
Results
HSCs and multipotent progenitors are markedly reduced in FA/SDS.
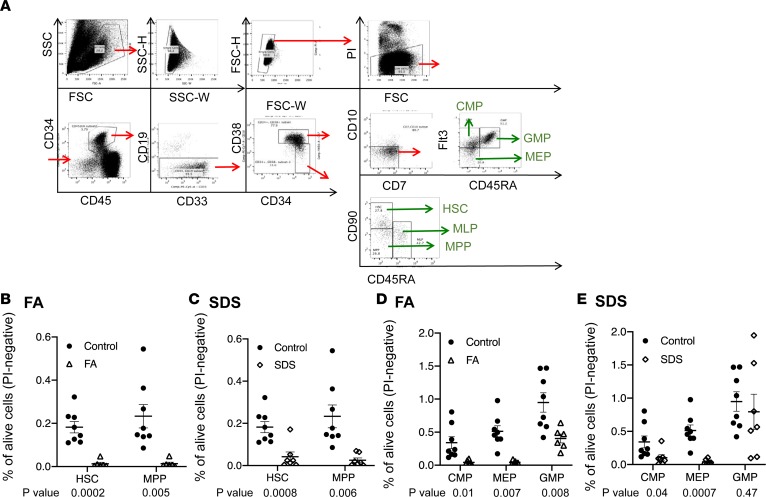
We and others showed global reduction in hematopoietic cells and in CD34+ cells in bone marrow from patients with FA (28) and SDS (29). We hypothesized that in both disorders defects begin within the most early hematopoietic cells and applied 12-paramater deep immunophenotyping profiling methodology based on recently developed approaches (refs. 5, 30, and Figure 1A). Cell numbers were normalized to the viable (propidium iodide–negative) cells in the sample. Within the CD34+CD38– primitive progenitor compartment and compared with healthy controls, the relative numbers of CD90+CD45RA– HSCs were reduced 14.1- and 4.6-fold in FA and SDS, respectively, and the CD90–/CD45RA– multipotent progenitors (MPPs) were reduced 17.7- and 7.8-fold in FA and SDS, respectively (Figure 1, B and C). Because most patients with FA/SDS included in this study had hypocellular bone marrow specimens (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131018DS1), we suggest that the average fold decrease in absolute numbers of patients’ HSPCs compared with healthy controls is likely higher than that of the above relative numbers.
Figure 1. Deep immunophenotyping revealed striking loss of most, but not all, HSCs and progenitors in bone marrow from patients with FA/SDS.
(A) Analytic strategy of bone marrow aspirate cells by immunophenotyping. (B and C) Comparison of multipotent cells between FA (n = 6), SDS (n = 7), and control (n = 8). The mean percentage of HSPCs among the viable bone marrow mononuclear cells is presented with SEM. (D and E) Comparison of oligopotent progenitors between FA, SDS, and control patients. The mean percentage of HSPCs among the viable bone marrow mononuclear cells is presented with standard error of the mean (SEM). PI, propidium iodide; Flt3, FMS-like tyrosine kinase 3; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte erythroid progenitor; HSC, hematopoietic stem cell; MLP, multilymphoid progenitor; MPP, multipotent progenitor. Student’s t test was used to compare between patients and controls. The same control data in C and E are also presented in B and D, respectively.
FA and SDS are characterized by variable levels of oligopotent hematopoietic progenitor loss.
CD34+CD38+ progenitors include the common myeloid progenitors (CMPs), megakaryocyte erythroid progenitors (MEPs), and GMPs. CMPs and MEPs were markedly and significantly reduced in the patients. CMPs were reduced 8.1- and 3.5-fold in FA and SDS, respectively. MEPs were reduced 12.3- and 15.5-fold in FA and SDS, respectively (Figure 1, D and E).
Unexpectedly, the reduction of HSCs did not result in universal reduction of all their downstream progenies. In SDS, MEPs represented the most affected population compared with CMPs or GMPs. In FA, MEPs and CMPs were markedly reduced compared with GMPs. Furthermore, in both SDS and FA, GMPs (CD34+/CD38+/FLT3+/CD45RA+) were least affected and relatively preserved, with only 1.5-fold reduction in SDS and 2.3-fold reduction in FA. In SDS, the percentages of GMPs were not significantly different from controls (Figure 1, D and E). Remarkably, when HSPC frequencies were normalized to the total number of CD34+ cells in the respective samples, the average percentage of SDS GMPs was a modest 1.56-fold higher than the average percentage of healthy controls’ GMPs (P = 0.03). In FA, the average percentage of GMPs was 1.15 times higher than that of controls, but the difference did not reach statistical significance (Supplemental Figure 1). These data about FA/SDS GMPs were surprising for both disorders, but particularly in SDS, because granulopoiesis is the most affected hematopoietic process in SDS (29, 31).
FA and SDS feature an abnormally high frequency of somatic variants in GMPs.
The IBMFSs are difficult to study genetically because there is a paucity of cells to work with. Therefore, we undertook genetic analysis to gain insight into the mutations present within the GMP population that seemed to be persisting more extensively than other progenitors. In addition, because of the relative abundance of GMP-like cells, we reasoned that they are more likely to carry mutations that confer a growth advantage than other progenitors that were markedly reduced.
We analyzed somatic tier 1 and 2 variants in GMP-like cells, as described in Methods. Bone marrow fibroblasts were used as a surrogate germline tissue. The cogency of variant detection was supported by a high congruence of mapped reads across the genome (Supplemental Figure 2) and per chromosome (Supplemental Figure 3). Analysis of a marrow fibroblast sample demonstrated that this congruence was seen between amplified and unamplified DNA before whole-exome sequencing (WES). Importantly, we consistently saw lower variant numbers when GMPs were compared to self fibroblasts versus fibroblasts from other subjects, which is expected given normal genomic variations between individuals (Supplemental Figure 4). There was a consistently higher number of variants in patients versus controls who were processed and analyzed in an identical fashion (see below). Detection of calls by MuTect2 and by other mutation caller software programs (Sterlka and VarScan) was also highly congruent (data not shown). In addition, there was no correlation between gene size and number of variants detected, which would be expected from random mutations along the genome. Also, we found no aberrantly high rates of C>T (G>A) errors in analysis of a GMP DNA sample compared with a blood DNA sample amplified by single-cell REPli-G whole-genome amplification kit and by VarScan mutation caller software (data not shown). Last, detecting variants by WES and the cancer gene panel showed high congruence (Supplemental Table 2).
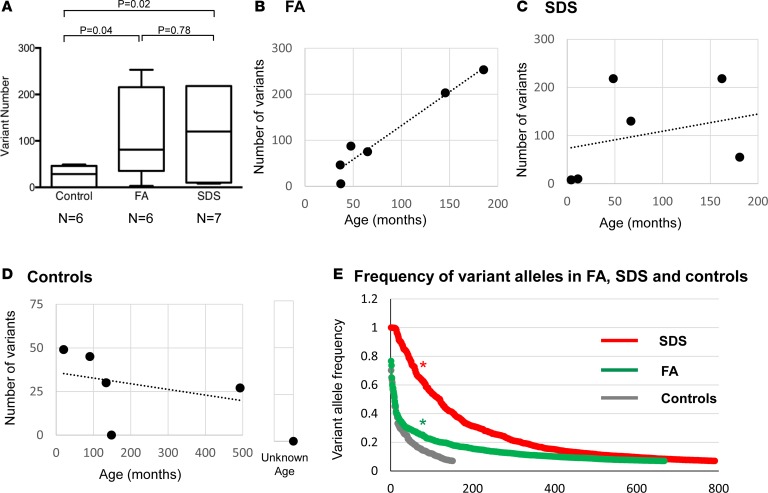
The numbers of somatic variants among FA patients (mean 111) and SDS patients (mean 108) were remarkably higher than that among control subjects (mean 25), whose samples were processed in the same way (P values of 0.04 and 0.02, respectively) (Figure 2A). All variants were rare (minor allele frequency ≤ 1%) or absent in the general population’s databases (data not shown). There was no significant age difference between FA/SDS patients and controls (P = 0.34, and P = 0.41, respectively). Interestingly, the frequency of variants in FA was not statistically different from SDS (Figure 2A).
Figure 2. Frequency of somatic variants in bone marrow samples from patients with SDS and FA and healthy control subjects.
(A) Comparison of average (±SEM) variant rate between FA (n = 6), SDS (n = 7), and healthy control subjects (n = 6). The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range. Results by Student’s t test are shown. P = 0.0695 by Kruskal-Wallis test with Dunn’s post hoc test when comparing the 3 subject groups. (B–D) Variant rate among controls, FA subjects, and SDS subjects organized according to ages. (E) Allele frequency of the various variants in controls, FA subjects, and SDS subjects. The groups were compared using the Wilcoxon’s signed-rank test. *P < 0.0001. The y axis represents the variant frequency and the x axis represents the variants arranged from those with the highest allele frequency to the lowest. In each group, each number may represent a different variant.
The total numbers of variants in each subject according to age at sampling are in Figure 2, B–D. A statistically significant correlation between mutation burden and age could not be accurately determined because a larger number of subjects in each group is required for this analysis. Importantly, the variants in SDS/FA appeared in significantly higher allele frequencies compared with those of controls (P < 0.0001) (Figure 2E).
Types of nucleotide change across patients.
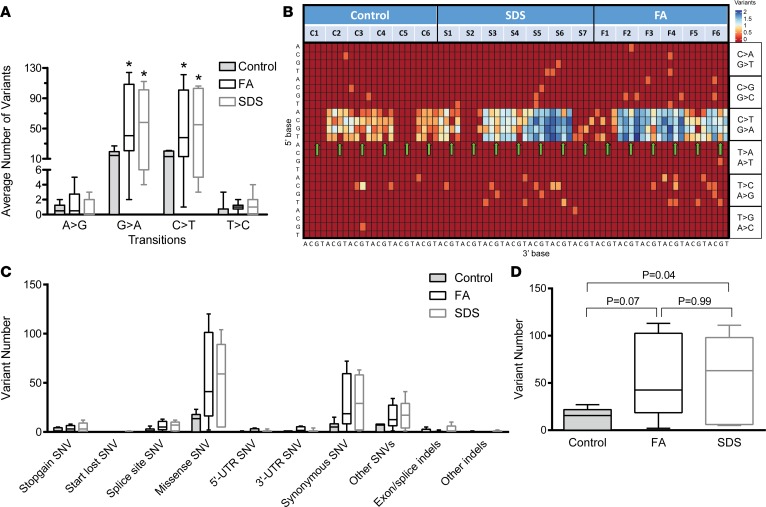
Because of their AML predisposition, we reasoned that mutations in FA/SDS GMP-like cells are characterized by previously published AML mutational patterns. Therefore, we used multiple analytical techniques to understand the mutational process and patterns underlying the high mutational load in FA/SDS. First, we determined the variants underlying transition changes (interchanges between purine bases or between pyrimidine bases; Figure 3A) and transversion changes (interchanges between purine and pyrimidine bases; Supplemental Figure 5). We found that the most abundant single nucleotide variants (SNVs) in all groups (FA, SDS, and controls) were as seen in AML (32) — namely G>A/C>T transitions, followed by A>G/T>C transitions and G>T/C>A, C>G/G>C transversions. Nevertheless, the proportions of G>A/C>T transitions in FA/SDS were significantly higher than those of control subjects (P < 0.05).
Figure 3. Patterns of single nucleotide and trinucleotide alterations among FA, SDS, and healthy control subjects.
(A) Average number (±SEM) of each transition (inside the CT purine group or inside the GA pyrimidine group) variant per subject among the FA, SDS, and healthy control groups. *P < 0.05 when comparing each patient group to the control group. P = 0.9313 for A>G; P = 0.0735 for G>A; P = 0.086 for C>T; P = 0.2586 for T>C by Kruskal-Wallis test with Dunn’s post hoc test when comparing the 3 subject groups. The average numbers of transversions (change from pyrine to pyrimidine or vice versa) are in Supplemental Figure 5. (B) Heatmap depicting trinucleotide SNV patterns. The heatmap depicts specific trinucleotide variants (SNV including the base immediately 3′ and 5′ to the SNV site). The 5′ base is shown on the y axis and the 3′ base on the x axis. Z score of the log-transformed values from 0 to 2 was used. To generate the heatmap, the number of each variant plus 1 was converted to log. (C) Percentage of SNVs and indels according to their damaging effects on the protein in each of the study subject groups. (D) Mean number of mutated genes in FA subjects, SDS subjects, and controls with SEM. Results of comparison between each patient group to controls by Student’s t test are shown. P = 0.069 by Kruskal-Wallis test when comparing the 3 subject groups. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
To gain further insight into the mutational processes in FA/SDS, we analyzed variants in the context of a trinucleotide change: the 6 options of nucleotide substitutions and the 16 combinations of bases immediately 3′ and 5′ to this variant. Overall, this resulted in a mutational signature that comprised 96 trinucleotide frames for each subject that are displayed in a heatmap in Figure 3B. All the subject groups showed a high C>T mutation rate regardless of the flanking 5′- and 3′-nucleotides, that is, Xp(C>T)pX sites. However, this propensity was much more prominent in patients with FA (P = 0.04) and SDS (P = 0.02) than in controls. The visualization of vertical rows on the heatmap suggests that the 3′ base has a greater influence on the mutational pattern. The vertical rows seen within the C>T region indicate that most patients have lower mutation rates at Xp(C>T)pG sites (arrows in Figure 3B) compared with other Xp(C>T)pX sites. This pattern was less prominent in healthy control subjects. The low number of mutations seen at Xp(C>T)pG sites may be attributed to the relatively low number of CpG sites in the genome and could be the result of the deamination of methylated cytosines (33). Last, there was a modestly increased mutational load at T>C sites in FA/SDS.
Different cancers generate mutations through distinct processes and leave their mark on the genome through a unique mutational signature (34). To identify the specific cancer trinucleotide signature of GMP-like cells from each subject, we first normalized variants to the relative contribution of each trinucleotide in the exome region using the DeconstructSigs R package and then compared our results to those in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. Normalization entails determining the amount of a certain trinucleotide variant relative to the amount of native trinucleotides occurring within the respective genome. De novo AML has previously been characterized by the COSMIC database to have a trinucleotide pattern contributed by Signatures 1 (spontaneous deamination of 5-methylcytosine) and 5 (transcriptional strand bias for T>C substitutions at ApTpX context). Because of a minimum 50-variant criterion for analysis, cancer signatures could be constructed from 9 of the 14 FA/SDS GMP-like cell samples but from none of the control subjects (Supplemental Figures 6–14). Importantly, the AML Signature 1 was more frequent (8 of the 9 patients) and more often the dominant signature (4 of the 9 patients) than other signatures (Supplemental Table 3).
The analysis of tier 1 and 2 SNVs and indels predicted varying degrees of damage to the encoded protein from stop-gain, frameshift, start-loss, splicing, and missense alterations to potentially less severe effects of 3′ UTR, 5′ UTR, and synonymous changes (Figure 3C). The distribution of mutation types for patients was similar to controls although the rates of mutations were higher.
Mutated genes and mutational trees.
To identify genes that might be involved in malignant myeloid transformation and to construct mutational trees, we selected genes with mutations that fulfilled the criteria described in Methods and had moderate to high software-predicted impact on the protein, namely, nonsense, splicing, frameshift, indel/in-frame, start-loss, and missense. The average number of mutated genes per subject was significantly higher in FA (61) and SDS (58) compared with controls (ref. 14 and Figure 3D) but was not statistically different when FA and SDS were compared.
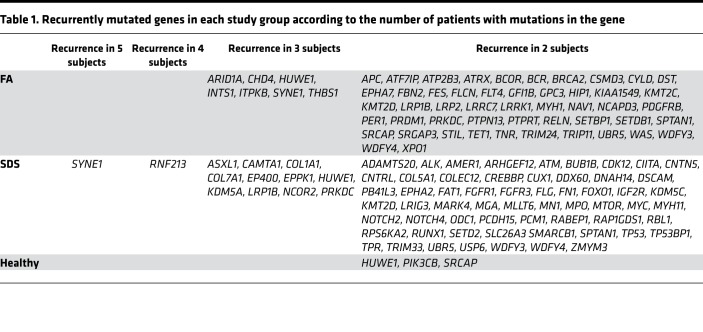
Importantly, there were a substantial number of genes with moderate- to high-impact mutations in more than 1 FA/SDS patient (Table 1). Commonly mutated cancer-related genes in both FA and SDS included the nuclear membrane gene SYNE1 and the ubiquitin E3 ligase gene HUWE1. Genetic mutations or dysregulation of these genes have previously been implicated in several solid tumors, such colon and gastric cancer, though not in leukemia. Several known MDS/AML driver genes were recurrently mutated in SDS (e.g., ASXL1, TP53, and CUX1) and FA (e.g., BCOR) (Supplemental Table 4). It is noteworthy that mutations in the TP53 binding protein 1 gene, TP53BP1 (p.Asp11Asn and p.Val687Ile), were seen in 2 SDS patients. The number of variants in mutated genes was not related to gene size (Supplemental Figure 15), indicating a nonrandom distribution of mutations.
Table 1. Recurrently mutated genes in each study group according to the number of patients with mutations in the gene.
Compilation of a dominant mutational tree in samples without clinical evidence of transformation was performed as described previously (35) in all FA (Supplemental Figure 16, A–F) and SDS samples (Supplemental Figure 17, A–H). The specific genes and variants in each clone are listed in Supplemental Table 5. In all samples there were mutations in known MDS/AML genes and in other cancer-related genes that have not previously been reported in MDS/AML to our knowledge. Interestingly, in 2 FA samples the founding clones harbored somatic mutations in MDS/AML-related genes (KDM6A in FA3 and FANCE in FA5), while in the rest of the FA samples, the founding clones harbored cancer-related genes that have not been previously associated with MDS/AML to our knowledge. TP53 mutations were part of the founding clones in 2 SDS patients (SDS1 and SDS5) (Supplemental Table 5) but in none of the samples of FA patients without leukemia. Other MDS/AML-related genes were identified in the founding clones in 3 other patients with SDS (Supplemental Table 5).
Analysis of MDS/AML-related gene pathways showed high rates of mutations in the transcription factors/regulation pathway, DNA repair/checkpoint gene pathway, and activated signaling molecules pathway in FA/SDS (Supplemental Figure 18).
Clonal landscape of AML samples in FA/SDS.
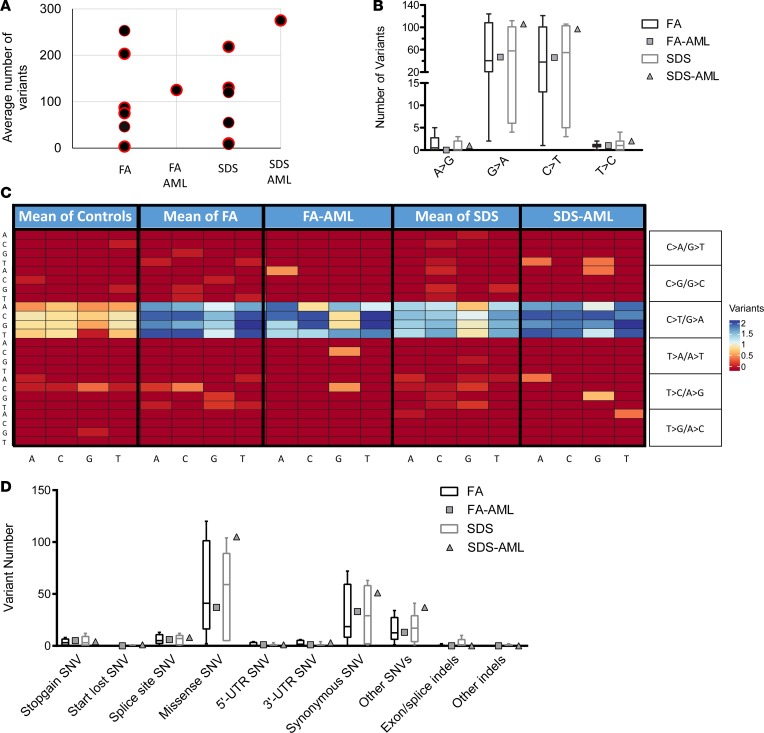
To gain insight into the relevance of variants and mutated genes detected in samples without transformation, we analyzed leukemic cells from 1 FA patient (FA7) with AML and 1 SDS patient (SDS7) with AML. Although only 2 AML cases from these rare disorders were available for the study, these anecdotes provide a unique opportunity to observe processes that appeared at 2 stages: before any clinical and standard laboratory evidence of transformation and at an ultimate catastrophic phase of leukemia. Blast cell samples were paired with marrow fibroblasts or T cells from the same subject, and somatic variants in blasts were analyzed as described in Methods. The mutation rate in SDS-derived AML (SDS/AML) blasts was slightly higher than the rates in all other SDS samples without transformation, but the number of variants in FA-derived AML (FA/AML) blasts was within the range of those in untransformed FA samples (Figure 4A).
Figure 4. Patterns of single nucleotide and trinucleotide alterations in FA/SDS-associated AML.
(A) Mutation rate in FA/SDS patients with AML or without AML. (B) Percentage of each type of transition mutation across FA/SDS patients with or without AML samples. Percentages of transversions are in Supplemental Figure 19. (C) Trinucleotide heatmap of patients with FA, FA/AML, SDS, and SDS/AML. The trinucleotide mutations are shown with the 5′ base on the y axis and the 3′ base on the x axis. (D) Types of mutations in AML versus non-AML samples. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
Similar to our findings in non-AML samples, both FA/SDS samples showed higher G>A/C>T transition rates than controls, the predominant mutation type in de novo AML (Figure 4B and ref. 34). The number of transversions was low (Supplemental Figure 19), and meaningful comparison between transformed and untransformed samples was impossible.
The trinucleotide heatmap depicting the variant change and adjacent 5′ and 3′ bases in non-AML and AML patients is in Figure 4C. All samples, including AML blasts and GMPs from subjects at no transformation, featured high mutation rates at Xp(T>C)pX sites. Importantly, AML Signature 1 was the predominant trinucleotide signature in FA/AML blasts (64%) and comprised a substantial faction in SDS/AML blasts also (22%) (Supplemental Figures 20 and 21).
Analysis of the potential impact of mutations on the protein showed a generally similar pattern in FA/SDS with AML samples compared to those without AML (Figure 4D).
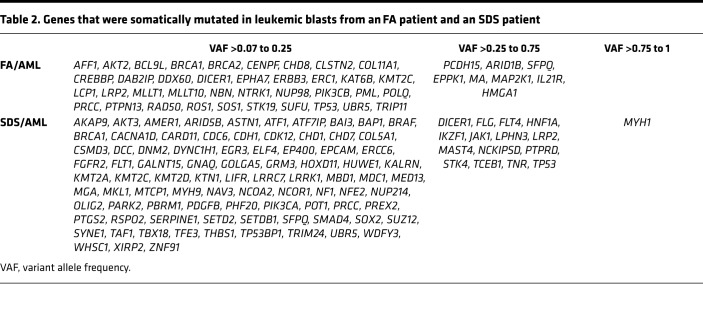
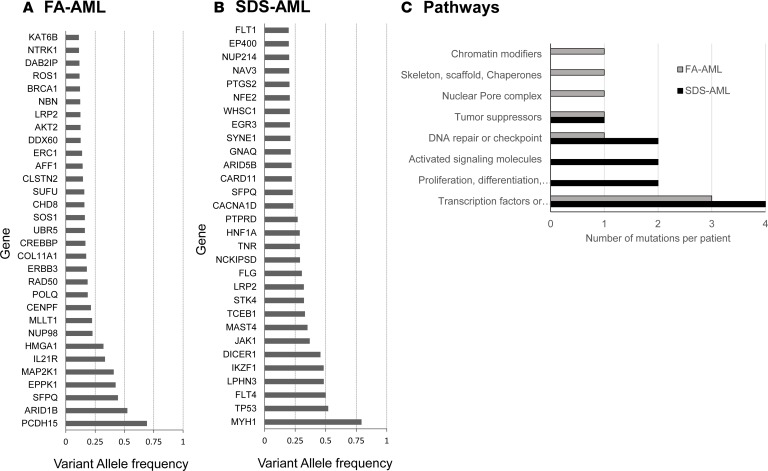
Cancer-associated genes with moderate- to high-impact mutations in AML samples are listed in Table 2. The genes with the highest VAF are in Figure 5, A and B. Several genes harbored variants with high frequency in FA/AML and were predicted to be part of the founding clone by mutational tree analysis (Supplemental Figure 22A and Supplemental Table 5). These genes were ARID1B, SFPQ, PCDH15, EPPK1, and MAP2K1. The founding clone gave rise to 3 subclones that included mutations in the MDS/AML genes NUP98, PML/BRCA1, and TP53/BRCA2, respectively. The first clone gave rise to an additional clone with mutations in the CREBBP MDS/AML-associated gene.
Table 2. Genes that were somatically mutated in leukemic blasts from an FA patient and an SDS patient.
Figure 5. Genes mutated in FA/SDS-associated AML.
(A) Top 30 genes mutated in AML cells from a patient with FA. (B) Top 30 genes mutated in AML cells from a patient with SDS. (C) Pathways, such as “proliferation, differentiation apoptosis” and “transcription factors or regulation,” that are disrupted in AML blasts from a patient with FA and in a patient with SDS.
The genes that appeared in highest allele frequency in SDS/AML included MYH1, TP53, FLT4, LPHN3, and DICER1 (Table 2). These genes were predicted to be part of the founding clone, which gave rise to 2 subclones (Supplemental Figure 22B and Supplemental Table 5). The mutated genes in 1 of the subclones included the MDS/AML gene PTPRD and other cancer genes (e.g., JAK1 and an additional mutation in DICER1). This subclone gave rise to additional clones harboring mutations in MDS/AML genes, such as BRAF and SETD2. The second subclone featured a mutation in SFPQ, and subsequent clones included mutations in NCOR1, SMAD4, NF1, and BRCA1. Similar to samples without AML (Supplemental Figure 18), in both FA/AML and SDS/AML, commonly mutated pathways included transcription factor or transcription factor regulation and DNA repair (Figure 5C).
Last, we evaluated whether genes with high- or moderate-impact mutations that appeared in patients without transformations were also mutated in the AML phase. In FA, 18 of the 255 genes that were part of clonal hematopoiesis in patients without MDS/AML appeared in the AML blasts (Supplemental Figure 23 and Supplemental Table 6). In SDS, 52 of the 282 genes that were part of clonal hematopoiesis in patients without transformation appeared in the AML blasts also (Supplemental Figure 23 and Supplemental Table 6).
Clonal evolution and progression observed in sequential samples.
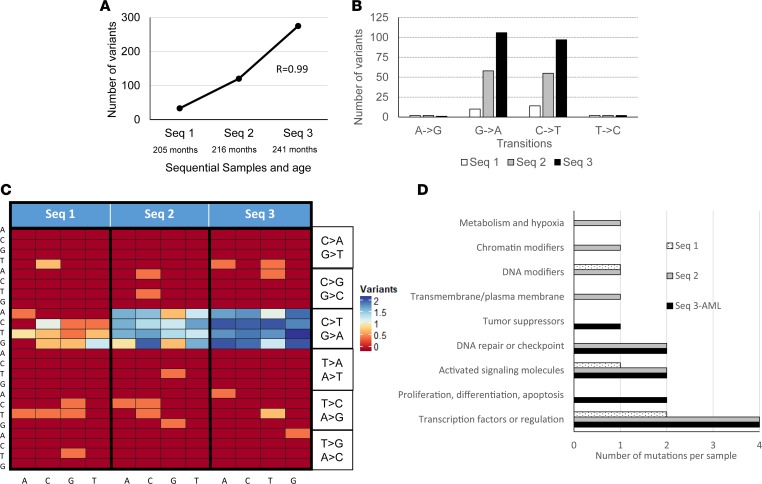
From the patient with SDS who developed leukemia, 2 additional samples 36 months and 25 months before the development of AML were available. The number of mutations grew prominently from stage to stage (Pearson’s r value of 0.99) (Figure 6A). The growth was more prominent than the age-related mutation increment we found in our SDS patient cohort (Figure 2C). Interestingly, there was a gradual increase in G>A (r = 0.99) and C>T transitions (R = 0.99938) but not in A>G or T>C transitions (Figure 6B). There was also a gradual accentuation of the trinucleotide signature (heatmap in Figure 6C). The number of transversions was low (Supplemental Figure 24) and did not show a conclusive pattern.
Figure 6. Transformational alterations in sequential samples from a nonmalignant to malignant state.
The figure displays results from 3 sequential samples from a patient with SDS. (A) Total number of mutations in each sequential sample. (B) Percentage of total transition mutations in each sequential sample. Percentages of transversions are in Supplemental Figure 24. (C) Changes in trinucleotide signature heatmap in each sequential sample. (D) Pathways affected in each sequential sample.
Construction of trinucleotide cancer signatures using the COSMIC database was feasible for the last 2 sequential samples. Interestingly, Signature 1 accounted for 9.2% of the mutational signature in the second sequential sample (Supplemental Figure 14) and increased to 22.2% at the stage of AML (Supplemental Figure 21).
Similar to the variant numbers, there was also a gradual increase in the number of genes with moderate- or high-impact mutations in each sequential sample: 15, 65, and 103, respectively (Table 3) (r = 0.945). Of the 15 genes with mutations in first sequential sample, 2 were mutated in the second and third samples. Of the 65 mutated genes in the second sequential sample, 10 were mutated in the third sample.
Table 3. Genes mutated in sequential samples from an SDS patient who eventually developed leukemia (sequential sample 3).
In each of the sequential samples, a dominant mutational tree could be constructed. However, as seen with bone marrow cytogenetic abnormalities in FA (36) and SDS (37), the dominant tree may arise and regress, and in each sequential sample a different dominant tree was apparent. The founding clone in sequential sample 1 harbored 13 genes with high- or moderate-impact mutations, including ARHGEF12 and NOTCH2; in sequential sample 2 there were 28 such genes, including IDH2 and MYH2; and in the third sample (AML) there were 6 such genes, including TP53. The known pathogenic mutation in TP53 (c.742C>T; p.Arg248Trp) was dominant in the AML stage (52%). It is noteworthy that with progression from sequential samples 1 to 3, the proportion of mutations in transcription factors, transcription factor regulation, activated signaling molecules and DNA repair, and checkpoint molecule pathways increased (Figure 6D).
Discussion
The present study focused on evaluating the cellular and molecular events before overt leukemia develops and their potential impact on malignant transformation. We report for the first time to our knowledge detailed analysis of the very early hematopoietic cells (HSCs, MPPs) and subsequent progenitors (CMPs, MEPs, GMPs) in FA and SDS. Most HSPCs were markedly reduced except for GMPs, which were much more frequently preserved. Molecular analysis of phenotypically GMP cells revealed a high number of somatic mutations compared with control subjects and genetic signatures that resembled those seen in AML. Using sequential SDS samples before and at AML stage, we were able to show that somatic nucleotide-level mutations develop and disappear very rapidly in this disorder, resembling observations related to some large clonal marrow cytogenetic abnormalities (36, 37). The reconstructed founding clone at the AML stage harbored mutations in several genes, including TP53.
The overrepresentation of immunophenotypic GMPs versus other myeloid progenitors in patients with FA/SDS suggests that these cells feature higher survival or growth properties and possibly harbor some of the initial transformational events that lead to MDS/AML. We cannot rule out the possibility that relative preservation of GMP-like cells reflects a general compensatory mechanism for bone marrow failure unrelated to leukemia risk. Although possible, it would be surprising that a compensatory mechanism targets GMPs regardless of whether the mostly affected lineage is granulocytic (SDS) or megakaryocytic/erythrocytic (FA). It is noteworthy that the initiating events may occur in earlier HSPCs, which then acquire the immunophenotype of GMPs. The markedly elevated somatic variants in FA/SDS GMP-like cells is in keeping with this hypothesis. It is possible that some of these mutations enhance proliferation or inhibit cell death, thereby conferring a growth advantage to these progenitors. For example, the TP53 mutation p.Arg248Trp seen in patients with SDS inactivates the protein and its proliferation-regulating properties. Future studies are necessary to decipher the mechanism underlying the relative preservation of GMP-like cells in FA/SDS bone marrow and whether it is related to increased proliferation, decreased apoptosis, or self-renewal.
Interestingly, despite different functions of FA genes from SDS genes, in both conditions GMPs were relatively more preserved, and there were no significant differences in the average number of somatic mutations. This raises the possibility that, at least in part, clonal evolution in bone marrow failure disorders does not depend on the direct biochemical sequela of the germline mutation and might be related to the consequent growth disadvantage of bone marrow cells, mitotic stress, and a drive for survival through growth-promoting somatic mutations.
The cause of an increased propensity for MDS/AML in IBMFSs and the mechanisms of leukemogenesis are unclear, and several hypotheses have been proposed (38, 39). Our findings of an increased mutation rate in GMP-like cells and their relative preservation provide a groundwork for research focusing on these questions. Several pathological processes have been identified in FA/SDS and may be considered while trying to explain an increased risk of somatic mutations. FA proteins are involved in correction of interstrand DNA cross-links (40) and telomere length maintenance (41), leading to chromosomal instability. There is also evidence for short telomeres and genomic instability in SDS (42, 43). These pathologies may lead to somatic structural chromosomal abnormalities that are commonly seen in SDS (37, 43, 44) and in FA (45, 46); however, they may not directly explain the increased numbers of SNVs seen in our study. In FA, DNA interstrand cross-links may lead to DNA double-strand breaks due to prolonged stalling of the replication fork or collapse. This may eventually lead to errors during repair or replication. Oxidative stress has been implicated in DNA damage and cancer development (47, 48) and is increased in both FA (49–51) and SDS (52, 53). In addition, the accelerated cell death and slow-growing cells in FA (49, 50, 54) and SDS (55–57) may lead to replicative stress, which can consequently increase the rate of randomly occurring mutations. Interestingly, it has been suggested that the slow-growing HSPCs in bone marrow failure disorders are under selective pressure for mutations that reverse their growth defect and ameliorate the restraints on proliferation (58, 59). Last, similar to AML (60) and MDS (61), SDS bone marrow stroma features increased angiogenesis (62). SDS bone marrow stroma has also been shown to be functionally impaired in humans (29) and in mice (63). In the latter study, deletion of Sbds in mouse mesenchymal stem cells resulted in DNA damage in HSPCs and in a proinflammatory response that was shown to contribute to leukemic transformation (63).
To our knowledge, there are no published data about the rate, type, and signature of somatic variants in GMPs from inherited leukemia predisposition syndromes, and only little information is available about somatic mutations in bone marrow samples from patients with FA (19) and SDS (17, 18). An explicit comparison between results from the present work to those from previously published studies on IBMFSs is challenging because of different methodologies and analytic approaches. Nonetheless, the number of variants in our study might be different from that reported in few published papers on FA/SDS, and there are several possible explanations for that. First, mutation rates in GMP-like cells have not previously been published. GMP-like cells were relatively preserved in FA/SDS, which might be attributed to a higher rate of somatic mutations that confer growth advantage. Second, published studies focused on mutations with high allele frequency. For example, in the study on somatic mutations in FA patients (19), mainly Sanger sequencing was used; the technique typically detects variant with allele frequency of over 10% to 20%. In the published WES data on 2 patients with SDS (18), few variants were reported; however, the authors focused on variants at the expected binomial distribution around 50%. Because of the analysis of highly purified progenitors and limited number of progenitors in FA/SDS, we used amplified DNA. Quality assessment of the data, paired analysis of amplified and unamplified DNA from control marrow fibroblasts across the genome (described in the Results section), and our internal robust methodology suggest that the trends seen herein are real and that significant bias by DNA amplification is unlikely.
The molecular changes found herein in FA/SDS GMP-like cells are reminiscent of those seen in AML, for example, abundance of G>A/C>T and G>T (32). G>A/C>T hypermutations have been attributed to the endogenous process of deamination at methylated cytosine sites (32). Importantly, this pattern was also dominant in FA/SDS with AML samples and steadily increased in sequential samples from a patient with SDS who eventually developed AML. Studies of sequential samples from additional cases are needed to determine whether gradual acquirement of this pattern is indeed part of the transformational process in FA/SDS.
The characterization of mutational signatures unveils a new hypothesized mutational etiology that could provide insight into the mutational processes underlying leukemic predisposition in FA/SDS. Per the COSMIC database, AML features a trinucleotide pattern contributed by Signatures 1 (spontaneous deamination of 5-methylcytosine and increased mutations at CpG sites) and 5 (transcriptional strand bias for T>C substitutions at ApTpX sites). To our knowledge, the COSMIC trinucleotide signature database has not been previously applied to FA/SDS bone marrow samples. Our results suggest that GMP-like cells are prone to developing an AML-type trinucleotide signature in FA/SDS. This hypothesis is solidified by finding Signature 1 in AML cells derived from patients with FA/SDS and by observing an increment in the proportion of Signature 1 in sequential samples from a patient with SDS who eventually developed AML. The predominance of Signature 1 indicates that deamination of methylated cytosines plays a role in the mutations seen in FA/SDS; however, mutational processes related to the other concomitant signatures may also be in play.
In most samples we were able to reconstruct a dominant mutational tree. However, most mutations were not part of the dominant mutational tree, suggesting that FA/SDS marrows contain multiple unrelated clones. Further, we cannot rule out a possibility that at the stage of AML, additional smaller, unrelated AML clones coexisted. Importantly, using sequential samples, we found that similar to large cytogenetic abnormalities that may appear and disappear with time in FA (36) and SDS (37), including del(20q10-11) and i(7q), SNVs may also appear and disappear, as described in 1 patient with severe congenital neutropenia (21). Our study further shows that most clones do not culminate in leukemia evolution, and despite a burst of evolving clones, most of them disappear and become outnumbered by new clones. This process probably continues until a combination of critical mutations appears in the same clone and drives progression toward MDS/AML.
It is noteworthy that the frequency of mutations in genes that are commonly mutated in de novo MDS/AML (e.g., DNMT3A, TET2 and SF3B1) was low in patients with FA/SDS, particularly in the ones who developed AML, suggesting that transformation in FA/SDS may use novel mechanisms. PCDH15 was mutated in FA/AML with high VAF (69%) and was predicted to be part of the founding clone of the dominant mutational tree. PCDH15 is a member of the cadherin superfamily, which encode integral membrane proteins that mediate calcium-dependent cell-cell adhesion. It is mutated in several solid cancers, including breast cancer, glioma, and lymphoma (64–66). The findings of mutations in this gene also in 2 SDS patients without AML (1 of them in the founding clone) suggest a potential pathogenic role.
It is noteworthy that SFPQ was mutated in both our patients with AML, in the founding clone in FA, and in a subclone in SDS. To our knowledge, SFPQ was previously reported to be mutated only in 1 subject with AML (67). A recent study suggested downregulation of SFPQ by miRNA-1296 in colorectal cancer as a mechanism for cell proliferation (68). The published mutation in a patient with AML was described as nonsynonymous without further details. The mutation in our patient with FA/AML was a missense variant in the N-terminal domain (p.Gly14Ser). The mutation in the patient with SDS/AML was a missense variant in the C-terminal domain (p.Glu699Lys). It is possible that loss of or aberrant SFPQ alters splicosome function and drives MDS/AML. Further studies are necessary to determine whether SPFQ mutations are more common in IBMFS-associated MDS/AML than in de novo MDS/AML and whether there is synergism between HSC loss and SFPQ in developing leukemia.
It is important to note that TP53 was mutated herein mainly in patients with SDS. It was mutated in the SDS/AML founding clone and in 2 SDS patients without transformation, indicating that it is indeed an early transformational event. It is interesting that in sequential samples, the TP53 mutation p.Arg248Trp (previously reported as pathogenic) was detected in the founding clone of SDS/AML but not in the founding clone in previous samples. This information supports the notion that early hematopoietic cells in IBMFSs have heightened tendency for clonal evolution, but most clones eventually subside and do not progress.
In summary, FA and SDS are characterized by a burst of clonal evolution. Although the molecular changes largely follow AML features, most hematopoietic clones do not progress, and at a leukemic stage only a few clones become predominant. The differences between clones that progress to leukemia and those that do not need to be further elucidated. Future studies should also evaluate the prognostic value of the identified molecular changes in this study and their potential use for early detection of irreversible transformation or therapeutic targets in FA and SDS. Last, because AML blasts from only 2 patients with FA/SDS were available for this study, the molecular data at the AML stage are anecdotal, and multicenter, collaborative efforts are required to collect a larger number of AML samples from these rare disorders to validate our observations.
Methods
Flow cytometry.
Bone marrow HSPC population sizes were evaluated by multiparametric immunophenotyping (Figure 1A), as previously described (5). Cell frequencies were normalized as previously described to the total bone marrow mononuclear cells (5, 69) and to total bone marrow CD34+ cells (70, 71).
DNA preparation for genomic studies.
To identify the spectrum of somatic mutations and affected genes, we analyzed DNA from phenotypical sorted GMP cells. DNA samples from 200 to 965 sorted GMPs were amplified by whole-genome amplification (REPli-G Mini Kit, QIAGEN) for 16 hours with adjustment of reagents to cell number as per the manufacturer’s instructions and as previously described (72–75).
To eliminate germline variants, we paired each subject’s data with his or her marrow fibroblast genome as a source of nonhematopoietic DNA. We enriched marrow fibroblasts by culturing marrow cells, removing floating hematopoietic cells, and passaging 3 to 5 times. Because of poor growth of passaged patient cells, DNA of marrow fibroblasts from close to half of the patients (and 1 healthy subject for quality control) was amplified, with no apparent effect on the number of filtered somatic variants (Supplemental Table 7) and no apparent bias toward specific nucleotide change (Supplemental Table 8). Furthermore, matched amplified and unamplified DNA from fibroblasts showed a high congruence of mapped reads across the genome and per chromosome (Supplemental Figures 2 and 3).
To study molecular events in AML samples, we sorted blast cells. In a case of an SDS patient with AML, amplified DNA from marrow myeloblasts underwent paired analysis with DNA from marrow fibroblasts. For an FA patient with AML, a peripheral blood sample was available, and amplified DNA from myeloblasts underwent paired analysis with amplified DNA from T cells.
WES.
DNA underwent exome enrichment by the Sure Select 50 Mb Human All Exon Capture Kit (Agilent Technologies) according to the manufacturer’s instructions and sequencing on the Illumina HigSeq2500 at The Centre for Applied Genomics (The Hospital for Sick Children) as previously described (25). The average reads per nucleotide among the analyzed subjects was 146 (range 116–189).
Next-generation sequencing cancer gene panel assay.
To augment mutation discovery by deep variant analysis and validate variants in cancer-related genes found by WES, we used a deep sequencing panel of 877 genes, which either were known cancer-related genes from the COSMIC database or are hypothesized to play a role in cancer (based on published expression in tumors, known function, or constitutive mutation in cancer predisposition syndromes). The total number of bases for nonoverlapping exons covered by the panel ± 10 bp is 3,012,823 bp. The panel was developed by our group as previously described (76). The average reads per nucleotide among the analyzed subjects was 1216 (range 775–2098).
Variant calling.
Somatic variant calling followed the bcbio pipeline (http://github.com/bcbio/bcbio-nextgen). The pipeline is used to identify somatic variants by comparing them to normal human genome alignments and annotating the mutations for subsequent analysis. The pipeline includes alignment of FASTQ files to the reference genome (GRCh37) using Burrows-Wheeler Aligner mem v0.7.17 (http://bio-bwa.sourceforge.net), duplication of marking using biobambam v2.0.87 (https://github.com/gt1/biobambam), and removal of low-complexity regions by bedtools v2.27.1 (https://github.com/arq5x/bedtools2).
GMP and marrow fibroblast FASTQ files were aligned and mapped separately to the reference genome to create binary alignment map (BAM) files, and both BAM files were then processed using MuTect v1.1.5 (http://www.broadinstitute.org/cancer/cga/mutect) for somatic point mutations and indels.
Variants from GMP WES and cancer panel sequencing were selected as true somatic variants if (a) they appeared in GMPs from both WES and the cancer panel, (b) the variant frequency in marrow fibroblasts was 0, (c) the variant comprised over 7% of the total reads for the respective nucleotides in GMPs (using this threshold, over 90% of the variants fulfilled all criteria in both WES and cancer panel) (Supplemental Table 2), and (d) the read depths by the cancer panel in GMPs and in marrow fibroblasts were over 50.
Analysis of somatic variants.
Somatic variants were classified into tiers as described (77). As conventionally done in cancer genomics analysis, we used only tier 1 and 2 variants, which are more likely to have a pathogenic effect than tier 3 and 4 variants.
The R package deconstructSigs (http://github.com/raerose01/deconstructSigs) was used to construct tumor signatures from somatic variants, to normalize signatures according to variant frequencies, and to compare them to known tumor signatures in COSMIC. A mutation signature was determined by comparing the total variant profile of a patient to the known variant profile of different cancers. For this analysis a minimum of 50 somatic variants per sample was required to construct a signature. ComplexHeatmap (http://bioconductor.org) was used to create a sample heatmap of somatic variants. Variant Effect Predictor (http://grch37.ensembl.org/info/docs/tools/vep/index.html) was used to annotate the mutations for functional consequence.
Mutational trees were reconstructed by the PhyloWGS software program as developed by Quaid Morris’s group (35) (http://github.com/morrislab/phylowgs). The program can reconstruct related clonal subpopulations in tumor samples from whole-genome sequencing/WES data. It is based on VAFs of the mutations and uses the Markov chain Monte Carlo procedure. It can construct mutational trees with or without data about copy number variants (78). Using this software, we designated marrow fibroblast cells as molecular group 0. Subsequent clones were ordered and numbered by the software program.
Statistics.
Descriptive analysis was used to characterize groups. Two-tailed Student’s t test was used to determine the statistical significance of differences between 2 means. To determine significant differences between multiple means, the nonparametric Kruskal-Wallis test was performed followed by Dunn’s post hoc test. Wilcoxon’s signed-rank test was used for testing whether 3 samples have different VAF distributions. P < 0.05 was considered significant. The statistical analyses were performed using Microsoft Excel, XLSTAT Version 2019.1.2 (Addinsoft), and GraphPad Prism v8. The bioinformatics software programs used in this study are described with the respective analyses in the Methods and Results sections.
Study approval.
Patients with SDS were eligible for the study if they fulfilled the international consensus diagnostic criteria (79) and had biallelic SBDS mutations. Patients with FA were eligible if they had a clinical diagnosis of FA and positive chromosome fragility testing. At the time of testing, most patients without leukemia had cytopenia and hypocellular bone marrow (Supplemental Table 1); no patient had clonal marrow cytogenetic abnormalities. Healthy control subjects were donors for bone marrow transplantation. The study was approved by the Research Ethics Board at The Hospital for Sick Children, and informed written consent was obtained from all enrolled subjects. Usage of a sample that had been cryopreserved in the Tissue Bank at The Hospital for Sick Children was done according to the Research Ethics Board’s regulations and approval. A total of 7 FA, 8 SDS, and 8 healthy control subjects were studied. The list of subjects and samples is in Supplemental Table 7.
Author contributions
SH contributed to study design, acquisition of data, and analysis and interpretation of data and assisted in writing the manuscript. BB contributed to study design, acquisition of data, and analysis and interpretation of data and drafted the article and revised it for important intellectual content. SZ contributed to study design, acquisition of data, and analysis and interpretation of data and assisted in writing the manuscript. HL contributed to study design, acquisition of data, and analysis and interpretation of data and assisted in writing the manuscript. S Abelson contributed to analysis and interpretation of data and reviewed/revised the manuscript for important intellectual content. RJK heads 1 of the Canadian Inherited Marrow Failure Registry site research teams that contributed acquisition of vital data and interpretation of data and reviewed/revised the manuscript for important intellectual content. S Abish heads 1 of the Canadian Inherited Marrow Failure Registry site research teams that contributed acquisition of vital data and interpretation of data and reviewed/revised the manuscript for important intellectual content. MR heads 1 of the Canadian Inherited Marrow Failure Registry site research teams that contributed acquisition of vital data and interpretation of data and reviewed/revised the manuscript for important intellectual content. VRB heads 1 of the Canadian Inherited Marrow Failure Registry site research teams that contributed acquisition of vital data and interpretation of data and reviewed/revised the manuscript for important intellectual content. RDB contributed to study design and analysis and interpretation of data. HM contributed to analysis, generation of figures, and interpretation of data. SD contributed to study design, interpretation of data, and review of the manuscript. AS developed the cancer panel used in this study, contributed to study design and analysis and interpretation of data, and assisted in writing the manuscript. JED contributed to study design and analysis and interpretation of data and assisted in writing the manuscript. YD contributed to study conception and design, acquisition of data, and analysis and interpretation of data and drafted and revised the article.
Supplementary Material
Acknowledgments
The work in the manuscript was supported by grants from the Canadian Institute of Health Research application number 377600, from the Canadian Institute of Health Research application number 378100, and from Fanconi Anemia Canada.
Version 1. 01/28/2020
In-Press Preview
Version 2. 02/27/2020
Electronic publication
Footnotes
SZ’s present address is: Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Ontario, Canada.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e131018.https://doi.org/10.1172/jci.insight.131018.
Contributor Information
Brian Bursic, Email: bbursic@uwo.ca.
Sasan Zandi, Email: sazandi@gmail.com.
Hongbing Li, Email: hongbing.li@sickkids.ca.
Sagi Abelson, Email: sagi.abelson@oicr.on.ca.
Robert J. Klaassen, Email: rklaassen@cheo.on.ca.
Sharon Abish, Email: sharon.abish@muhc.mcgill.ca.
Vicky R. Breakey, Email: breakev@mcmaster.ca.
Houtan Moshiri, Email: houtan.moshiri@sickkids.ca.
Santhosh Dhanraj, Email: santhoshloyola@gmail.com.
Richard de Borja, Email: richard.deborja@gmail.com.
Adam Shlien, Email: adam.shlien@utoronto.ca.
John E. Dick, Email: jdick@uhnresearch.ca.
Yigal Dror, Email: yigal.dror@sickkids.ca.
References
- 1.DeSantis CE, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.Schultz KA, et al. Health conditions and quality of life in survivors of childhood acute myeloid leukemia comparing post remission chemotherapy to BMT: a report from the children’s oncology group. Pediatr Blood Cancer. 2014;61(4):729–736. doi: 10.1002/pbc.24881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velten L, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19(4):271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurenti E, Göttgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553(7689):418–426. doi: 10.1038/nature25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notta F, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351(6269):aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saultz JN, Garzon R. Acute myeloid leukemia: a concise review. J Clin Med. 2016;5(3):E33. doi: 10.3390/jcm5030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogle CR, Craig BM, Rollison DE, List AF. Incidence of the myelodysplastic syndromes using a novel claims-based algorithm: high number of uncaptured cases by cancer registries. Blood. 2011;117(26):7121–7125. doi: 10.1182/blood-2011-02-337964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasle H, Wadsworth LD, Massing BG, McBride M, Schultz KR. A population-based study of childhood myelodysplastic syndrome in British Columbia, Canada. Br J Haematol. 1999;106(4):1027–1032. doi: 10.1046/j.1365-2141.1999.01645.x. [DOI] [PubMed] [Google Scholar]
- 9.Das U, et al. A single center experience in 266 patients of infantile malignancies. Pediatr Hematol Oncol. 2014;31(6):489–497. doi: 10.3109/08880018.2013.852644. [DOI] [PubMed] [Google Scholar]
- 10.Ganguly BB, Kadam NN. Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res Rev Mutat Res. 2016;769:47–62. doi: 10.1016/j.mrrev.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Cada M, et al. The impact of category, cytopathology and cytogenetics on development and progression of clonal and malignant myeloid transformation in inherited bone marrow failure syndromes. Haematologica. 2015;100(5):633–642. doi: 10.3324/haematol.2014.117457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandel K, Dror Y, Poon A, Freedman MH. A practical, comprehensive classification for pediatric myelodysplastic syndromes: the CCC system. J Pediatr Hematol Oncol. 2002;24(7):596–605. doi: 10.1097/00043426-200210000-00028. [DOI] [PubMed] [Google Scholar]
- 13.Hasle H, et al. A pediatric approach to the WHO classification of myelodysplastic and myeloproliferative diseases. Leukemia. 2003;17(2):277–282. doi: 10.1038/sj.leu.2402765. [DOI] [PubMed] [Google Scholar]
- 14. Dror Y. Genetic basis of inherited bone marrow failure syndromes. In: Ikehara K, ed. Advances in the Study of Genetic Disorders. Rijeka, Croatia: InTech Open Access Publisher; 2011:357–392. [Google Scholar]
- 15.Smith OP, Hann IM, Chessells JM, Reeves BR, Milla P. Haematological abnormalities in Shwachman-Diamond syndrome. Br J Haematol. 1996;94(2):279–284. doi: 10.1046/j.1365-2141.1996.d01-1788.x. [DOI] [PubMed] [Google Scholar]
- 16.Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood. 1994;84(5):1650–1655. [PubMed] [Google Scholar]
- 17.Lindsley RC, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia J, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia. Blood. 2018;131(4):408–416. doi: 10.1182/blood-2017-08-801985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quentin S, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117(15):e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- 20.Germeshausen M, Skokowa J, Ballmaier M, Zeidler C, Welte K. G-CSF receptor mutations in patients with congenital neutropenia. Curr Opin Hematol. 2008;15(4):332–337. doi: 10.1097/MOH.0b013e328303b9f6. [DOI] [PubMed] [Google Scholar]
- 21.Beekman R, et al. Sequential gain of mutations in severe congenital neutropenia progressing to acute myeloid leukemia. Blood. 2012;119(22):5071–5077. doi: 10.1182/blood-2012-01-406116. [DOI] [PubMed] [Google Scholar]
- 22.Skokowa J, et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123(14):2229–2237. doi: 10.1182/blood-2013-11-538025. [DOI] [PubMed] [Google Scholar]
- 23.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17(6):337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 24.Boocock GR, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 25.Dhanraj S, et al. Biallelic mutations in DNAJC21 cause Shwachman-Diamond syndrome. Blood. 2017;129(11):1557–1562. doi: 10.1182/blood-2016-08-735431. [DOI] [PubMed] [Google Scholar]
- 26.Stepensky P, et al. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in aShwachman-Diamond like syndrome. J Med Genet. 2017;54(8):558–566. doi: 10.1136/jmedgenet-2016-104366. [DOI] [PubMed] [Google Scholar]
- 27.Carapito R, et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J Clin Invest. 2017;127(11):4090–4103. doi: 10.1172/JCI92876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rackoff WR, et al. Prolonged administration of granulocyte colony-stimulating factor (filgrastim) to patients with Fanconi anemia: a pilot study. Blood. 1996;88(5):1588–1593. [PubMed] [Google Scholar]
- 29.Dror Y, Freedman MH. Shwachman-Diamond syndrome: An inherited preleukemic bone marrow failure disorder with aberrant hematopoietic progenitors and faulty marrow microenvironment. Blood. 1999;94(9):3048–3054. [PubMed] [Google Scholar]
- 30.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 31.Hashmi SK, et al. Comparative analysis of Shwachman-Diamond syndrome to other inherited bone marrow failure syndromes and genotype-phenotype correlation. Clin Genet. 2011;79(5):448–458. doi: 10.1111/j.1399-0004.2010.01468.x. [DOI] [PubMed] [Google Scholar]
- 32.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16:35. doi: 10.1186/s13059-015-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alter BP, Caruso JP, Drachtman RA, Uchida T, Velagaleti GV, Elghetany MT. Fanconi anemia: myelodysplasia as a predictor of outcome. Cancer Genet Cytogenet. 2000;117(2):125–131. doi: 10.1016/S0165-4608(99)00159-4. [DOI] [PubMed] [Google Scholar]
- 37.Dror Y, et al. Clonal evolution in marrows of patients with Shwachman-Diamond syndrome: a prospective 5-year follow-up study. Exp Hematol. 2002;30(7):659–669. doi: 10.1016/S0301-472X(02)00815-9. [DOI] [PubMed] [Google Scholar]
- 38.Dror Y. Shwachman-Diamond syndrome: implications for understanding the molecular basis of leukaemia. Expert Rev Mol Med. 2008;10:e38. doi: 10.1017/S1462399408000938. [DOI] [PubMed] [Google Scholar]
- 39.Cooper JN, Young NS. Clonality in context: hematopoietic clones in their marrow environment. Blood. 2017;130(22):2363–2372. doi: 10.1182/blood-2017-07-794362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–278. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar J, Liu Y. Fanconi anemia proteins in telomere maintenance. DNA Repair (Amst) 2016;43:107–112. doi: 10.1016/j.dnarep.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thornley I, Dror Y, Sung L, Wynn RF, Freedman MH. Abnormal telomere shortening in leucocytes of children with Shwachman-Diamond syndrome. Br J Haematol. 2002;117(1):189–192. doi: 10.1046/j.1365-2141.2002.03371.x. [DOI] [PubMed] [Google Scholar]
- 43.Valli R, et al. Shwachman-Diamond syndrome with clonal interstitial deletion of the long arm of chromosome 20 in bone marrow: haematological features, prognosis and genomic instability. Br J Haematol. 2019;184(6):974–981. doi: 10.1111/bjh.15729. [DOI] [PubMed] [Google Scholar]
- 44.Valli R, De Paoli E, Nacci L, Frattini A, Pasquali F, Maserati E. Novel recurrent chromosome anomalies in Shwachman-Diamond syndrome. Pediatr Blood Cancer. 2017;64(8):8. doi: 10.1002/pbc.26454. [DOI] [PubMed] [Google Scholar]
- 45.Cioc AM, Wagner JE, MacMillan ML, DeFor T, Hirsch B. Diagnosis of myelodysplastic syndrome among a cohort of 119 patients with fanconi anemia: morphologic and cytogenetic characteristics. Am J Clin Pathol. 2010;133(1):92–100. doi: 10.1309/AJCP7W9VMJENZOVG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tönnies H, Huber S, Kuhl JS, Gerlach A, Ebell W, Neitzel H. Clonal chromosomal aberrations in bone marrow cells of Fanconi anemia patients: gains of the chromosomal segment 3q26q29 as an adverse risk factor. Blood. 2003;101(10):3872–3874. doi: 10.1182/blood-2002-10-3243. [DOI] [PubMed] [Google Scholar]
- 47.Bavarva JH, Tae H, McIver L, Garner HR. Nicotine and oxidative stress induced exomic variations are concordant and overrepresented in cancer-associated genes. Oncotarget. 2014;5(13):4788–4798. doi: 10.18632/oncotarget.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dizdaroglu M. Oxidatively induced DNA damage: mechanisms, repair and disease. Cancer Lett. 2012;327(1-2):26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O’Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat Med. 2001;7(7):814–820. doi: 10.1038/89937. [DOI] [PubMed] [Google Scholar]
- 50.Li J, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117(11):3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumari U, Ya Jun W, Huat Bay B, Lyakhovich A. Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi anemia cells. Oncogene. 2014;33(2):165–172. doi: 10.1038/onc.2012.583. [DOI] [PubMed] [Google Scholar]
- 52.Ambekar C, Das B, Yeger H, Dror Y. SBDS-deficiency results in deregulation of reactive oxygen species leading to increased cell death and decreased cell growth. Pediatr Blood Cancer. 2010;55(6):1138–1144. doi: 10.1002/pbc.22700. [DOI] [PubMed] [Google Scholar]
- 53.Sen S, et al. The ribosome-related protein, SBDS, is critical for normal erythropoiesis. Blood. 2011;118(24):6407–6417. doi: 10.1182/blood-2011-02-335190. [DOI] [PubMed] [Google Scholar]
- 54.Huang F, et al. The Fanconi anemia group C protein interacts with uncoordinated 5A and delays apoptosis. PloS one. 2014;9(3):e92811. doi: 10.1371/journal.pone.0092811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dror Y, Freedman MH. Shwachman-Diamond syndrome marrow cells show abnormally increased apoptosis mediated through the Fas pathway. Blood. 2001;97(10):3011–3016. doi: 10.1182/blood.V97.10.3011. [DOI] [PubMed] [Google Scholar]
- 56.Rujkijyanont P, et al. SBDS-deficient cells undergo accelerated apoptosis through the Fas-pathway. Haematologica. 2008;93(3):363–371. doi: 10.3324/haematol.11579. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe K, Ambekar C, Wang H, Ciccolini A, Schimmer AD, Dror Y. SBDS-deficiency results in specific hypersensitivity to Fas stimulation and accumulation of Fas at the plasma membrane. Apoptosis. 2009;14(1):77–89. doi: 10.1007/s10495-008-0275-9. [DOI] [PubMed] [Google Scholar]
- 58.Warren AJ. Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul. 2018;67:109–127. doi: 10.1016/j.jbior.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanley N, Olson TS, Babushok DV. Recent advances in understanding clonal haematopoiesis in aplastic anaemia. Br J Haematol. 2017;177(4):509–525. doi: 10.1111/bjh.14510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Padró T, et al. Increased angiogenesis in the bone marrow of patients with acute myeloid leukemia. Blood. 2000;95(8):2637–2644. [PubMed] [Google Scholar]
- 61.Pruneri G, et al. Angiogenesis in myelodysplastic syndromes. Br J Cancer. 1999;81(8):1398–1401. doi: 10.1038/sj.bjc.6693515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung EW, et al. Shwachman-Diamond syndrome: an inherited model of aplastic anaemia with accelerated angiogenesis. Br J Haematol. 2006;133(5):558–561. doi: 10.1111/j.1365-2141.2006.06069.x. [DOI] [PubMed] [Google Scholar]
- 63.Zambetti NA, et al. Mesenchymal Inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Cell Stem Cell. 2016;19(5):613–627. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 64.Yap YS, et al. Elucidating therapeutic molecular targets in premenopausal Asian women with recurrent breast cancers. NPJ Breast Cancer. 2018;4:19. doi: 10.1038/s41523-018-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nikas JB. Independent validation of a mathematical genomic model for survival of glioma patients. Am J Cancer Res. 2016;6(6):1408–1419. [PMC free article] [PubMed] [Google Scholar]
- 66.Rouget-Quermalet V, et al. Protocadherin 15 (PCDH15): a new secreted isoform and a potential marker for NK/T cell lymphomas. Oncogene. 2006;25(19):2807–2811. doi: 10.1038/sj.onc.1209301. [DOI] [PubMed] [Google Scholar]
- 67.Dolnik A, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120(18):e83–e92. doi: 10.1182/blood-2011-12-401471. [DOI] [PubMed] [Google Scholar]
- 68.Tao Y, Ma C, Fan Q, Wang Y, Han T, Sun C. MicroRNA-1296 facilitates proliferation, migration and invasion of colorectal cancer cells by targeting SFPQ. J Cancer. 2018;9(13):2317–2326. doi: 10.7150/jca.25427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manso BA, et al. Bone marrow hematopoietic dysfunction in untreated chronic lymphocytic leukemia patients. Leukemia. 2019;33(3):638–652. doi: 10.1038/s41375-018-0280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Townsley DM, et al. Eltrombopag added to standard immunosuppression for aplastic anemia. N Engl J Med. 2017;376(16):1540–1550. doi: 10.1056/NEJMoa1613878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong WM, Dolinska M, Sigvardsson M, Ekblom M, Qian H. A novel Lin-CD34+CD38- integrin α2- bipotential megakaryocyte-erythrocyte progenitor population in the human bone marrow. Leukemia. 2016;30(6):1399–1402. doi: 10.1038/leu.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 2019;25(1):103–110. doi: 10.1038/s41591-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt M, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28(12):2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 75.Aubry MC, et al. Chromosomal rearrangements and copy number abnormalities of TP63 correlate with p63 protein expression in lung adenocarcinoma. Mod Pathol. 2015;28(3):359–366. doi: 10.1038/modpathol.2014.118. [DOI] [PubMed] [Google Scholar]
- 76.Shlien A, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 77.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16:35. doi: 10.1186/s13059-015-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dror Y, et al. Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome. Ann N Y Acad Sci. 2011;1242:40–55. doi: 10.1111/j.1749-6632.2011.06349.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.