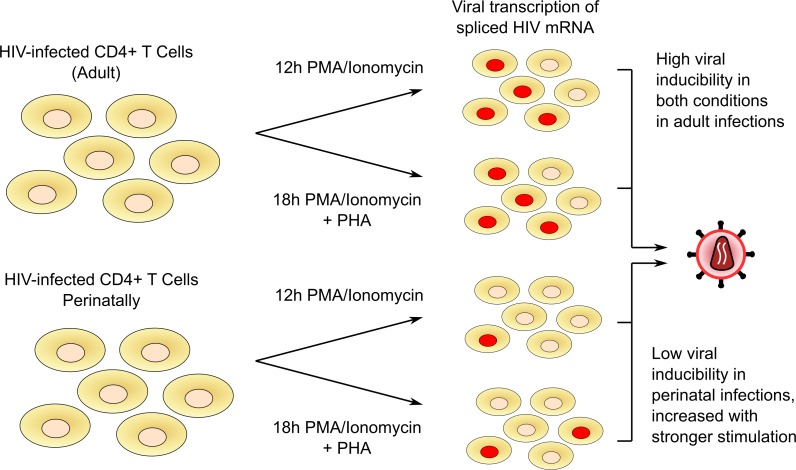
Abstract
The HIV latent reservoir in resting memory CD4+ T cells precludes cure. Therapeutics to reactivate and eliminate this reservoir are in clinical trials in adults, but not yet in pediatric populations. We determined, ex vivo, the inducibility of the latent reservoir in perinatal infection as compared with adult infections using the Tat/rev induced limiting dilution assay (TILDA), in which a single round (12 hours) of CD4+ T cell stimulation with PMA/ionomycin maximally activates T cells and leads to proviral expression with multiply spliced HIV RNA production. Markers of immune activation and exhaustion were measured to assess interactions with inducibility. Although rates of T cell activation with PMA/ionomycin were similar, the latent reservoir in perinatal infection was slower to reactivate and of lower magnitude compared with adult infection, independent of proviral load. An enhanced TILDA with the addition of phytohemagglutin and a duration of 18 hours augmented proviral expression in perinatal but not adult infection. The baseline HLA-DR+CD4+ T cell level was significantly lower in perinatal compared with adult infections, but not correlated with induced reservoir size. These data support the hypothesis that there are differences in kinetics of latency reversal and baseline immune activation in perinatal compared with adult infections, with implications for latency reversal strategies toward reservoir clearance and remission.
Keywords: AIDS/HIV, Immunology
Keywords: Molecular biology, T cells
The latent reservoir in perinatal infection reactivates at a lower magnitude than adult infections by detection of multiply-spliced HIV RNA.
Introduction
Nearly 1.7 million children under the age of 15 are living with HIV worldwide, with an estimated 160,000 new infections in children in 2018 (1). Treatment of HIV with combination antiretroviral therapy (ART) in both children and adults can lead to durable virologic suppression for decades but does not eradicate the infection due to early establishment of a latent HIV reservoir in resting memory CD4+ T cells (2–5). Within the latent reservoir, HIV persists as a transcriptionally silent, replication-competent, integrated provirus that is modulated at the transcriptional and posttranscriptional levels (6, 7), and by its site of integration (8, 9). The latent reservoir is highly heterogeneous and present in the various memory CD4+ T cell subsets including central, transitional, and effector memory CD4+ T cells, which differ in cell survival and reactivation propensity (10–13).
Identifying strategies to eliminate the latent reservoir is a rapidly evolving field of HIV therapeutics with hopes for long-term ART-free remission (14), and these include reversing latency to reactivate latent proviruses in order to promote clearance of HIV-infected cells (referred to as the “shock and kill” approach) (15, 16). Ex vivo studies of the latency reversal effects of various biologics on HIV-infected resting CD4+ T cells have informed the use of these agents in clinical trials in adults (17–19); similar studies in perinatal infections have not begun.
Typically, the effectiveness of latency reversal agents (LRAs) are determined by their capacity to induce unspliced cellular HIV transcripts (20), and their downstream effects on proviral latency by changes in reservoir size as measured by a quantitative viral outgrowth assay (QVOA) or proviral DNA loads (21). PCR-based DNA assays allows quantification of total proviral load but may overestimate the size of the reservoir due to the overabundance of defective genomes generated in the course of HIV infection (22). A recently developed Droplet Digital PCR–based (ddPCR-based) assay, the intact proviral DNA assay (IPDA), allows quantitation of intact and defective genomes and overcomes many limitations of total proviral DNA assays, but does not provide information on the inducibility of the proviruses within the latent reservoir (23).
The QVOA has defined the long-term stability of the latent reservoir in adult (t½ of 44 months) (24, 25) and perinatal HIV infection (2), and offers the advantage of quantifying the clinically relevant proviruses that can be induced to produce infectious virions (26–28). However, this assay is limited by the large blood volumes required to obtain sufficient CD4+ T cells to detect the inducible replication-competent proviral reservoir; its high cost and laborious nature, with 2–3 weeks of coculture; and that a fraction of intact proviruses require multiple rounds of T cell activation to be induced. An alternate approach to measuring the size of the inducible latent reservoir is with the Tat/rev induced limiting dilution assay (TILDA) as developed by Procopio et al. (29), which quantifies the inducible, transcriptionally competent reservoir ex vivo within 3 days. With TILDA, a 12-hour CD4+ T cell activation protocol with PMA and ionomycin in the presence of antiretroviral drugs leads to proviral expression as measured by production of the multiply spliced HIV mRNA transcripts tat and rev. With TILDA, the size of the inducible latent reservoir was found to be 48-fold greater than that detected by QVOA in adult infections, with the caveat that the replication competence of reactivated provirus cannot be fully assessed (29).
In adults with HIV infections, it is estimated that only 1.5% of proviruses in resting CD4+ T cells can be reactivated ex vivo following T cell activation with anti-CD3/CD28 costimulation (30). Understanding of the kinetics and fraction of the latent reservoir established in perinatal infection that can be reactivated is lacking but critical for informing strategies aimed at eliminating the reservoir through proviral reactivation. In this study, we sought to determine the permissivity and fraction of the latent reservoir susceptible to reactivation in adolescents with perinatal infections compared with participants infected during adulthood and their correlates using TILDA, in order to advance latency reversal strategies for this population.
Results
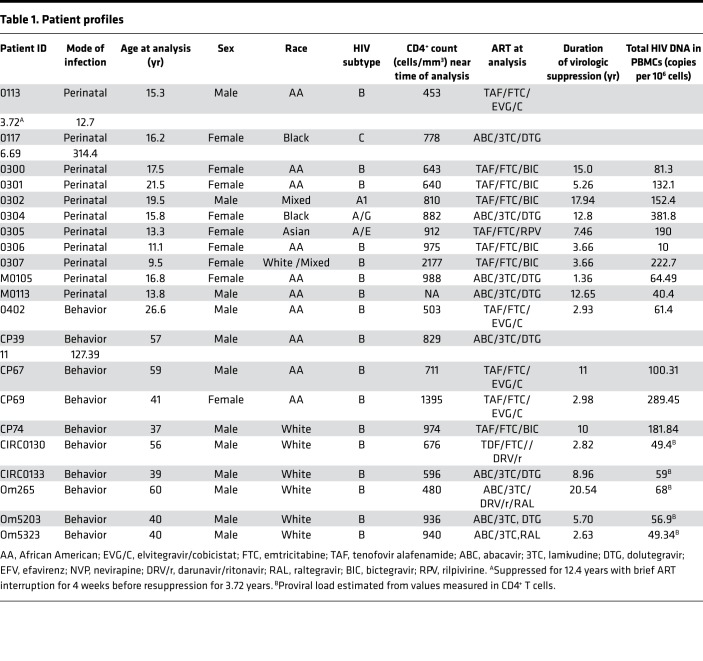
A total of 11 adolescents with perinatal infections and 10 adult participants were included in the analysis. Table 1 summarizes the demographic characteristics, duration of virologic suppression, antiretroviral regimens, and viral biomarker profiles of the study participants. 64% (7 of 11) of the perinatally infected participants were infected with subtype B HIV, whereas 100% (10 of 10) of the adult participants were subtype B infected. The median age of the perinatally infected cohort was 15.8 years (IQR 13.3–17.5); 64% (n = 8) were Black or African American, 18% (n = 1) Asian, and 18% (n = 2) White or mixed race; 73% (n = 8) were female, and the median duration of virologic suppression was 6.7 years (IQR 3.7–12.8). The median age of the adult participants was 40.5 years (IQR 38.5–57.5); 60% (n = 6) were White, 40% (n = 4) were Black, and 90% (n = 9) were male. Their median duration of virologic suppression was 7.3 years (IQR 2.9–11.0).
Table 1. Patient profiles.
Overall, there was a large variation in the size of the proviral reservoir as measured by total HIV DNA concentrations in PBMCs (Figure 1A and Table 1), with no significant difference between the 2 groups. The median HIV DNA concentrations were 132.1 (IQR 40.4–222.7) and 66.7 (IQR 57.7–141.0) copies per million PBMCs in the perinatal and adult infections, respectively (P = 0.51). The median HIV DNA concentration in the participants with subtype B perinatal infections was 64.5 (IQR 12.7–132.1) copies per million PBMCs and not significantly different from the adult infections (P = 0.54).
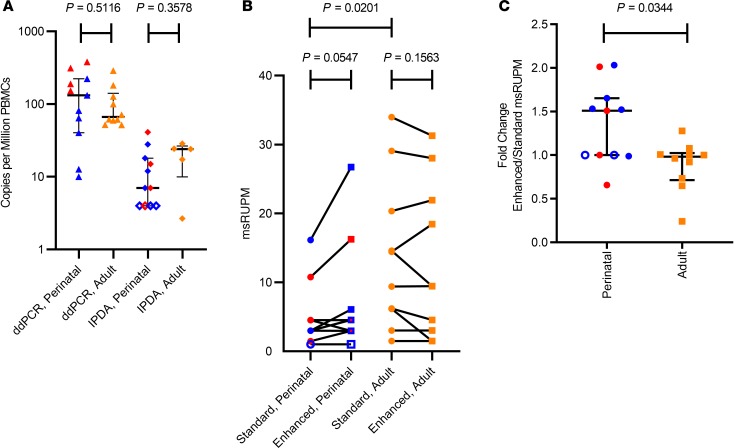
Figure 1. The viral reservoir in perinatal infection is more resistant to reactivation than in adult infection.
(A) Proviral load as quantified by both ddPCR measuring GAGLTR only and double-positive intact droplets as measured by IPDA (23). Perinatally infected samples are shown in blue (subtype B) and red (non–subtype B). Adult samples are labeled in orange; open symbols indicate samples that were undetectable by the specified assay. (B) Size of the inducible reservoir quantified as multiply spliced HIV RNA–producing units per million CD4+ T cells (msRUPM) in participants with perinatal (n = 11) and adult (n = 10) infection as measured by standard (circles) and enhanced TILDA (squares). (C) Fold change in the size of the inducible reservoir with the enhanced stimulation condition in perinatal and adult infections. Bars represent median and IQR. The significance of differences between populations or stimulation conditions was calculated using nonparametric 2-tailed unpaired Mann-Whitney U tests and paired Wilcoxon’s tests.
With standard TILDA conditions, an induced HIV reservoir was detected in 9 of 11 (82%) perinatally infected samples and 10 of 10 (100%) adult-infected samples. In the samples from adolescents with subtype B perinatal infections, 71% (5 of 7) had detectable induced proviral reservoirs, and all non–subtype B participants (4 of 4, 100%) had detectable induced reservoirs. Using the standard TILDA assay, the median size of the induced reservoir in those with perinatal infections was 2.99 multiply spliced RNA–producing units per million (msRUPM) CD4+ T cells (IQR 1.47–4.52), significantly lower than in adults (median = 11.92; IQR 5.37–22.52, P = 0.020) (Figure 1B). When only subtype B–infected participants were compared, the difference in the induced reservoir size with standard TILDA remained significant (P = 0.016). We then used a modified TILDA (enhanced TILDA), with the addition of phytohemagglutinin (PHA) and a duration of 18 instead of 12 hours to augment detection of proviral reactivation. With enhanced TILDA conditions, there was a slight but nonsignificant increase in msRUPM in individuals with perinatal infections (median msRUPM = 4.52, IQR = 2.96–6.08, P = 0.055). In contrast, there was a slight decrease in msRUPM in the adult infections (median = 9.46, IQR = 2.63–23.44, P = 0.16) (Figure 1B). The size of the induced latent reservoir, under Enhanced TILDA, in the individuals with perinatal infections was no longer statistically significantly different from that of adult participcants (P = 0.16).
To determine the individual effect of the enhanced TILDA conditions on the induced reservoir size, a fold change calculation was performed for each sample stimulated with standard and enhanced conditions. A consistent increase in induced provirus with enhanced compared with standard TILDA was detected in samples from individuals with perinatal infections (Figure 1C). With the enhanced stimulation, the size of the induced latent reservoir increased in the participants with perinatal infections by a median of 1.51-fold (IQR 1.00–1.65), whereas no fold change in the induced reservoir in adult infection was observed (median, 0.98-fold; IQR 0.71–1.03); the difference in fold change in msRUPM between adolescents with perinatal infections and adult participants was significant (P = 0.034). This difference remained significant when the comparison was restricted to subtype B perinatal infections (median fold change in perinatal subtype B, 1.52, IQR 1.00–1.65; median in adults, 0.98; IQR 0.71–1.03; P = 0.029).
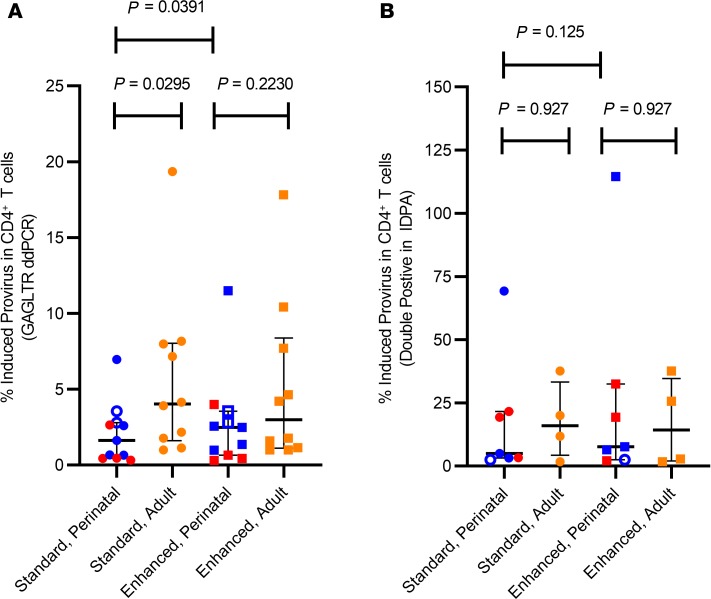
To determine the fraction of proviral genomes that were induced with standard and enhanced TILDA, the msRUPM was normalized to the concentration of HIV-infected cells, estimated as copies of gag long terminal repeats (GAGLTRs) per million CD4+ T cells. The median proportion of induced provirus in the population with perinatal infection using standard TILDA conditions was 1.62% (IQR 0.45%–2.80%), significantly lower than the proportion in adults (median, 4.03%; IQR 1.61%–8.03%; P = 0.030) (Figure 2A). With the enhanced TILDA, the proportion of induced proviral genomes increased significantly to a median of 2.49% in the perinatally infected participants (IQR 0.65%–3.56%, P = 0.039), but decreased in adult infections (median, 2.99%; IQR 1.11%–8.38%; P = 0.31) (Figure 2A). When the proportions of induced proviral genomes of only HIV subtype B perinatal samples was assessed, the median percent induced provirus in HIV subtype B samples remained low at 1.62% (IQR 0.66%–4.77%), and increased slightly but not significantly to 2.49% (IQR 1.17%–7.03%) with enhanced TILDA (P = 0.13).
Figure 2. A small fraction of proviruses retained in the latent reservoir in perinatal and adult HIV infection are induced to produce multiply-spliced HIV RNA transcripts.
(A) Percentage of proviruses in CD4+ T cells that were induced under standard and enhanced TILDA conditions in perinatal (n = 11, subtype B in blue and non–subtype B in red) and adult (n = 10, in orange) infections, normalized to GAGLTR copies per calculated CD4+ T cells. (B) Percentage of proviruses in CD4+ T cells that were induced under standard and enhanced TILDA conditions in perinatal (n = 7) and adult (n = 4) infections, normalized to double-positive intact copies per calculated CD4+ T cells as detected by IPDA. Five samples from perinatal infections were excluded from the IPDA analysis due to undetectable double-positive droplets, and thus could be used to normalize the TILDA. Open symbols represent individuals with nondetectable inducible reservoir. Bars represent median and IQR. The significance of the difference in percent induced provirus was determined using nonparametric 2-tailed unpaired Mann-Whitney U tests and paired Wilcoxon’s tests.
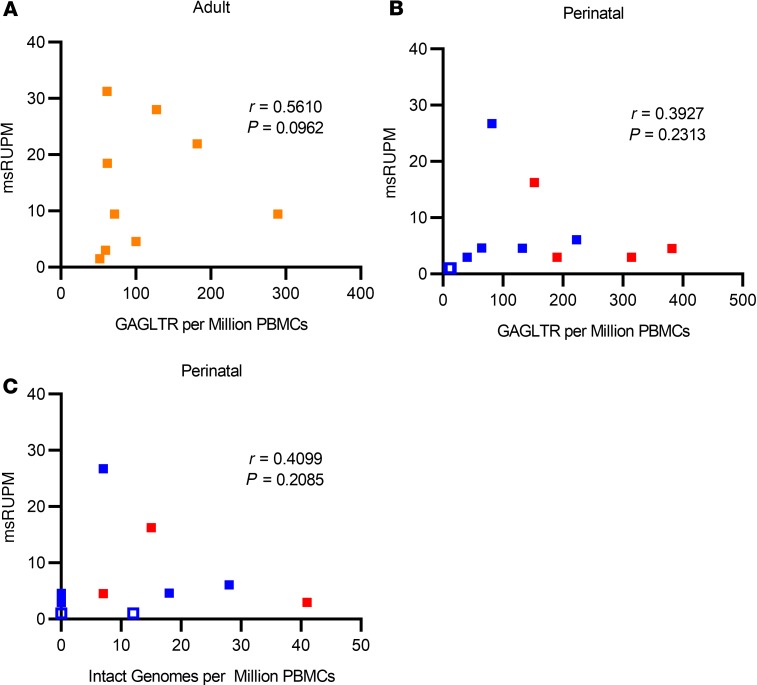
Given that the total HIV proviral DNA assay detects both intact and 3′ defective proviral genomes, correlations between msRUPM and the double-positive “intact” proviral reservoir size as measured by IPDA were also examined (Figure 1A). The median concentration of double-positive proviruses in perinatal infection (n = 11) was 7 per million PBMCs (IQR undetectable–18), 18.9-fold lower than total HIV DNA concentrations. In the 4 adult participants with IPDA measurements, the median double-positive intact proviral genomes was 4.52 per million PBMCs (IQR 2.96–6.082), 12-fold lower than the total HIV DNA concentrations. Since the multiply spliced HIV RNA transcripts detected in the TILDA assays were likely derived from intact proviral genomes, we also examined the fraction of induced provirus in the standard and enhanced TILDAs relative to double-positive proviral genomes when normalized to CD4+ T cell counts (Figure 2B). Among the 7 perinatal participants with detectable double-positive provirus, the median fraction of induced double-positive intact proviral genomes increased to 4.99% with the standard TILDA (IQR 3.21%–21.56%) and 7.64% with the enhanced TILDA (IQR 2.54%–32.53%), although the difference between the standard and enhanced was no longer statistically significant (P = 0.13), likely due to the small sample size. In adult participants, the median fraction of induced double-positive provirus was also increased to 15.95% with the standard TILDA (IQR 4.26%–33.26%) and 14.24% with the enhanced TILDA (IQR 2.01–34.65%). No significant correlation between msRUPM and the total HIV DNA concentrations was found in perinatal or adult infections (Figure 3, A and B). The induced reservoir size in perinatal infections also did not correlate with the size of the double-positive proviral genomes (r = 0.41, P = 0.21) (Figure 3C).
Figure 3. Plot of enhanced TILDA against ddPCR proviral DNA assays.
Multiply spliced RNA producing units per million PBMCs (msRUPM) plotted against GAGLTR copies per million PBMCs in (A) adult (n = 10, orange) individuals and (B) perinatally infected (n = 11, subtype B in blue and non–subtype B in red) individuals. Open data points represent individuals with undetectable msRUPM. (C) msRUPM plotted against double-positive intact proviruses per million PBMCs using IPDA. There was no correlation between TILDA and total DNA ddPCR, or between TILDA and IPDA. Spearman’s rank correlation coefficients and the corresponding P values are shown in the graphs.
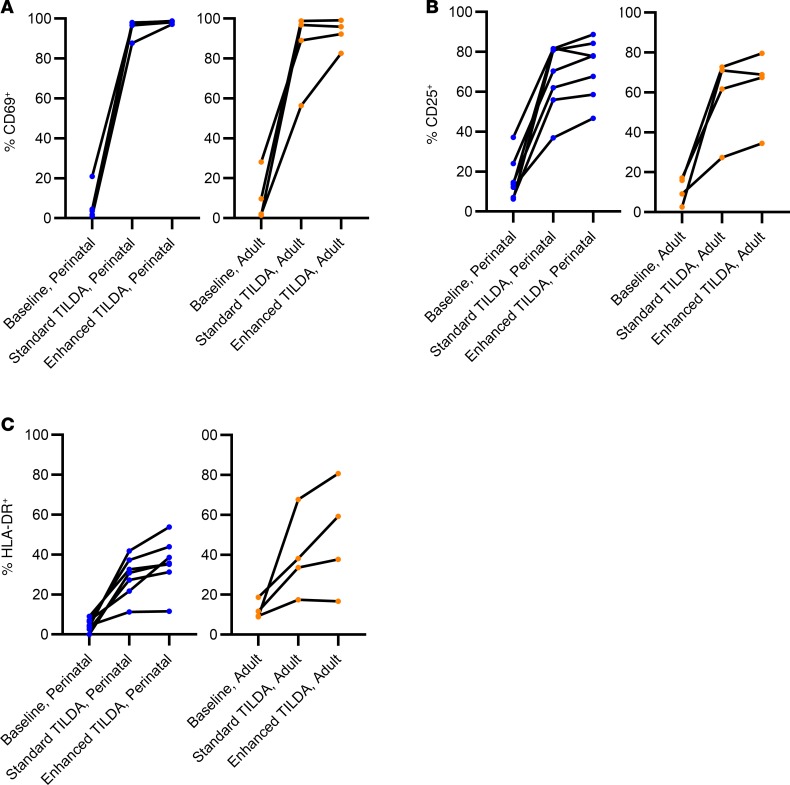
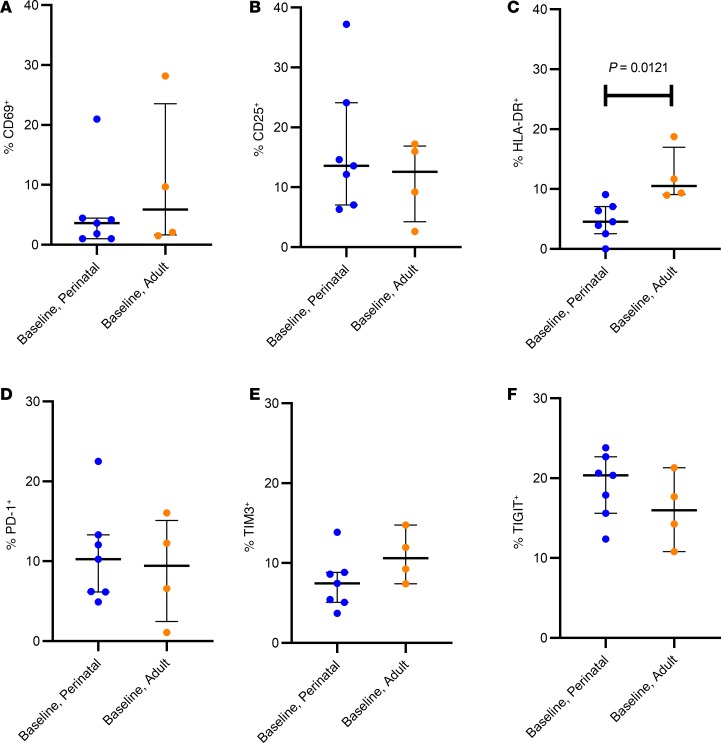
T cell immune activation, as measured by CD69 (an early marker of cell activation), CD25 (intermediate activation), and HLA-DR (late activation) at baseline, and following standard and enhanced TILDA was quantified in the subset of 11 study participants (7 adolescents with perinatal infections and 4 adult participants). We found that despite lower reactivation rates of HIV proviruses in perinatal infections, CD4+ T cells in pediatric and adult participants responded similarly to T cell activation stimuli, as determined by cell surface expression of CD69, CD25, and HLA-DR (Figure 4). CD69 expression uniformly increased from a median of 3.62% (IQR 1.03%–4.45%) in unstimulated pediatric samples to 96.72% (IQR 96.62%–98.75%) with standard TILDA and to 98.42% (IQR 98.22%–98.75%) with enhanced TILDA. A similar trend was observed in the adult samples, with a median baseline CD69 expression of 5.88% (IQR 1.645–23.55), and more variation in the range of CD69 expression with standard TILDA (median = 92.95, IQR 64.6–98.29) and enhanced conditions (median, 94.10; IQR 85.05–98.37; Figure 4A). Likewise, when comparing perinatal and adult infection within each stimulation condition, no significant difference in the intermediate (CD25) or late (HLA-DR) activation markers was noted with either standard or enhanced TILDA (Figure 4, B and C). At baseline, no significant difference in the proportion of CD69+ or CD25+ cells in unstimulated CD4+ T cells from pediatric and adult study participants on ART was found (Figure 5, A and B, and Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134105DS1). T cell surface exhaustion marker expression on CD4+ T cells has been previously reported to correlate with the size of the latent reservoir in adults (31, 32). To determine whether CD4+ T cells had lower expression of exhaustion markers in those with perinatal infections compared with adult infections, PD-1, TIM3, and TIGIT cell surface staining was performed and showed no difference in expression between perinatal and adult infections (Figure 5, D–F, and Supplemental Tables 1 and 2). The proportion of HLA-DR+CD4+ T cells was significantly lower in perinatal (median, 4.55%; IQR 2.56%–7.07%) compared with adult infection (median, 10.52%; IQR 9.07%–16.99%; P = 0.012; Figure 5C). However, the size of the induced reservoir was not correlated with any of the immune activation markers at baseline in either perinatal or adult populations.
Figure 4. Percentage of CD4+ T cells displaying activation markers.
(A) CD69, (B) CD25, and (C) HLA-DR expression during baseline. Standard and enhanced TILDA stimulation conditions in perinatal (left, n = 7) and adult (right, n = 4) infections.
Figure 5. Baseline levels of activation markers.
Comparison of the activation markers (A) CD69, (B) CD25, (C) HLA-DR and the exhaustion markers (D) PD-1, (E) TIM3, and (F) TIGIT between perinatal (n = 7) and adult (n = 4) infections as measured by flow cytometry. Bars represent median and IQRs. The tests for the differences in percent cells with the shown surface markers was conducted using a nonparametric 2-tailed unpaired Mann-Whitney U test.
Discussion
In this study, we identified that the induced latent reservoir for HIV in CD4+ T cells of adolescents with perinatal infections on long-term effective ART was significantly more resistant to reactivation despite similar proviral loads and rates of cellular activation in response to PMA/ionomycin in standard TILDA conditions. The addition of PHA, a T cell receptor signaling molecule, and a longer duration of stimulation of 18 hours (enhanced TILDA) led to a 1.5-fold increase in the detected inducible reservoir in the perinatally acquired compared with the adult infections, where no increase in msRUPM was observed. More than 96% of CD4+ T cells from those with perinatal infections expressed CD69 after 12 hours of T cell stimulation with PMA and ionomycin. The proportion of CD25+ and HLA-DR+ CD4+ cells also increased similarly in both study populations under the standard and enhanced conditions, confirming similar rates of T cell activation (33). These comparable patterns of activation markers exclude kinetics of T cell activation as the major mechanism for the quantitative differences in multiply spliced HIV RNA expression we observed in perinatal and adult infections.
Previous studies have shown that perinatally infected individuals treated early and with long-term virologic control can have exceedingly low levels of HIV-infected cells in the circulation, whereas those with delayed control of HIV replication in childhood have larger established pools of HIV-infected reservoir cells (34, 35). We found no correlation between proviral load, as measured by total HIV DNA that includes both defective and intact proviral genomes, and the size of the inducible reservoir in both perinatal and adult infections. The proviral reservoir size as measured by double-positive intact proviruses also did not correlate with the size of the induced reservoir, although this was limited by the sample size. An overall increase in the fraction of induced reservoir was observed in both perinatal and adult infections, confirming the relevance of measuring the double-positive intact proviruses in reservoir studies. This finding is consistent with the notion that total proviral load measurements significantly overestimate the inducible viral reservoir due to the preponderance of defective genomes (36, 37). In addition, studies have shown that not all intact proviruses can be induced with a single round of T cell stimulation (37), highlighting differences in permissivity of the latent reservoir to reactivation. The lack of correlation between the induced reservoir size and double-positive intact genomes is also compounded by the finding that only 70% of double-positive cells were free of lethal defects, as illustrated through near full-length sequencing, which can also lead to overestimation of intact proviral reservoir size (23). Altogether, these data confirm that in both perinatal and adult infections, only a small fraction of CD4+ T cells bear intact proviral genomes that are susceptible to virus reactivation and production of infectious virions, although detection of inducible virus in perinatally infected cells can be enhanced through combination T cell stimuli. The difference in the kinetics of HIV reactivation between the 2 populations, with the increase in multiply spliced HIV RNA transcription following enhanced stimulation conditions in perinatal infection, may reflect intrinsic differences in cellular activation pathways that influence proviral gene expression in perinatal infections, which requires further investigation.
Nevertheless, the TILDA has potential for measuring efficacy of HIV cure interventions, including latency reversal strategies, in pediatric and adult infections, as it requires a limited number of CD4+ T cells (2 million CD4+ T cells) and has a turn-around time on the order of 3 days compared with weeks for the QVOA, making it feasible for clinical trials. With standard TILDA, we found that the percentage of induced proviruses in perinatal infections at 1.6% were significantly lower than the 4% we detected in adult infections. Furthermore, with PMA/ionomycin combined with PHA in the enhanced TILDA, the fraction of induced proviruses increased in perinatal infections by 2.5%. When the double-positive intact proviruses were used, rather than total HIV DNA load, the size of the induced reservoir was increased even further to 7.64% with the enhanced TILDA. Taken together, the enhanced TILDA combined with measurements of double-positive intact proviruses improved the detection of the fraction of inducible proviral reservoir in perinatal infection, with implications for use of this assay in clinical trials of reservoir elimination in pediatric participants.
The median fraction of provirus in CD4+ T cells that are induced following T cell stimulation to produce intracellular unspliced RNA in adult infections has been estimated at approximately 7.5% (20). These findings highlight the overabundance of transcriptionally incompetent proviruses persisting in both perinatal and adult infections. Although it is possible for spliced tat/rev to be transcribed from replication-deficient virus, it has been reported that 97% of non-intact genomes have defects in the tat and rev genes (23). Thus, the transcription of spliced tat/rev from a defective genome is unlikely.
Our finding of enhanced HIV reactivation with the enhanced TILDA is also consistent with the finding by Hosmane et al. that a fraction of latent proviruses in adult infections (an estimated 40%) requires multiple rounds of stimulation to produce infectious HIV (38). The percentage of noninduced intact provirus appears to be amplified in perinatal infections, and these differences in kinetics between adult and perinatal infection may be due to epigenetic effects, transcriptional interference, or orientation of HIV integration on proviral gene expression in perinatal infections (39–42). Studies on integration sites of proviral genomes have shown that long-term virologic suppression is associated with changes in the composition of the viral reservoir, favoring sites associated with maintenance of deep viral latency (8, 43, 44). The immune-selective pressure driving the establishment of the latent reservoir may be differentially affected by the underdeveloped immune system of infancy (45). Other factors that can account for the differences in reactivation are the immune landscape in which HIV infection is established, including hyporesponsiveness due to higher proportions of T regulatory cells in early infancy and childhood (46–48); as well as differences in the distribution of the latent HIV reservoir in different memory T cell subsets (49, 50). We previously reported that the reservoir predominated in effector memory CD4+ T cells in perinatally infected adolescents (51) — as opposed to central and transitional memory CD4+ T cells as reported in adults (11) — highlighting differences in the composition of the reservoir in perinatal and adult infections.
It was previously shown that PMA/ionomycin and PHA have varying effects on cellular activation and HIV reactivation in cell line models (52). Although CD4+ T cells from both adult and perinatal infections were effectively activated by both the standard stimulation conditions as determined by high cell surface marker expression of CD69, this was insufficient to maximally induce proviral gene expression in perinatal infections. PMA combined with plant lectins (PHA) mediates T cell activation through different pathways: PMA activation is mediated mainly through PKC, whereas PHA requires a functional TCR to cause an increase in intracellular calcium for the downstream cellular activation effects. With TILDA, PMA/ionomycin was shown to cause rapid onset of multiply spliced HIV RNA transcription in adults (29). Previous studies have shown that the sequences in the long terminal repeat (LTR) targeted by PMA differ from those mediated by TCR agonists such as PHA, whereby both NF-κB sites and the upstream U3 sequences, which contain the binding site for NFAT, are required for activation of the LTR (53). Here, we identified that the combination of PMA/ionomycin with PHA for 18 hours enhanced detection of the inducible reservoir in perinatal but not in adult infections. This raises the possibility that NF-κB/NFAT binding mediated with PMA/ionomycin is insufficient to maximally stimulate HIV transcription in perinatal infections, for which alternate pathways triggered via TCR-mediated signaling maybe operative, with implications for latency reversal strategies for this population toward eradication of reservoir cells. In addition to the cellular conditions, the different stimulation conditions may influence the stochastic nature of TAT protein production, a key requirement for latency reversal (54). Nevertheless, we have established that the standard TILDA is insufficient to reliably quantify the inducible proviruses in perinatal infections, but can be enhanced with longer duration of stimulation and with the addition of PHA to the PMA/ionomycin without compromising cell viability, supporting the potential use of the enhanced TILDA to measure the inducible latent reservoir in perinatal infections. Furthermore, we identified the importance of attention to HIV subtype and consideration of subtype-specific primers in widespread application of molecular assays monitoring the inducible reservoir. The study participants with non–subtype B infection required subtype-specific primers to detect multiply spliced HIV RNA; the currently published primer sets are specific for HIV subtypes B and A/E (29, 55).
A faster rate of reactivation of HIV from latency in adult infections could also have been a result of higher baseline levels of immune activation compared with individuals with perinatal infections, which may serve to prime the HIV provirus for faster reactivation kinetics following a 12-hour stimulation with PMA/ionomycin. Indeed, at baseline, the median proportion of CD4+ T cells expressing HLA-DR was significantly higher in the adult-acquired infection compared with perinatal infections. This higher state of immune activation may also alter epigenetic repression, making the proviruses more permissive to virus-mediated transcription. A recent study reported that children with perinatally acquired infection have genes that were differentially hypermethylated when compared with age-matched uninfected children, especially within the HLA genes (56). In adults with HIV infection, a hypomethylated profile when compared with HIV-negative adults has been reported (57). There are other intrinsic differences between the 2 study populations that likely contribute to the differences in viral reactivation profiles. Specifically, the immune system in neonates when HIV reservoirs are established mainly consists of naive T cells, which gradually mature to effector and central memory T cells (58, 59). The establishment of latency in these cell populations, which are different from the memory CD4+ T cell population in adults, may result in a more deeply repressed provirus. In addition, the population of regulatory T cells is relatively large in early life (10%–30% of CD4+ T cells) and contracts in adulthood (60); these cells have been previously shown to be a viral reservoir (61).
Our study limitations include a relatively small sample size, differences in ART regimens, limited cell numbers to allow study of all conditions systematically and full assessment of double-positive proviral load in adult samples, and sex differences between the 2 groups (mostly females with perinatal and males with adult infections) — all of which can impact the quantitative measure of the inducible reservoir. Sex differences in HIV persistence markers have been reported, with higher levels of T cell activation, exhaustion states, and baseline HIV cell–associated RNA in men compared in men with women (62, 63). Despite these limitations, we identified that the latent reservoir in perinatal infections is slower to reactivate and is of a lower magnitude than with infection in adulthood, an effect that is independent of proviral load and kinetics of cellular activation. We also demonstrate that several modifications to the TILDA, with increased duration of T cell stimulation (18 hours) and combination with PHA, led to an increase in the inducibility of the proviral reservoir persisting in perinatal infections, without affecting cell viability. Additionally, we show that inclusion of measures that quantify the double-positive intact proviruses further enhanced the quantification of the proportion of induced proviruses persisting in patients on long-term ART. Overall, the study highlights differences in the kinetics and magnitude of latency reversal in perinatal compared with adult infections, for which future mechanistic studies will be important. The study findings have the potential to impact the assessment of latency reversal strategies and assays to measure efficacy of cure strategies in perinatal infections.
Methods
Study population.
Study participants with perinatal HIV infection were recruited from the Johns Hopkins Pediatric and Adolescent HIV/AIDS Program and the University of Maryland Division of Pediatric Immunology and Adolescent Medicine (Table 1). The inclusion criteria for participation of participants with perinatal HIV infections were: confirmed perinatal infection; and known adherence to ART with virologic suppression for at least 1 year, defined as undetectable plasma viral load (<20 copies/mL). Intermittent viremia with 2 or fewer consecutive low-level (<400 copies/mL) detectable viral load measures during the course of virologic suppression was allowed. The adult study participants were recruited through the Reservoir Characterization Core (RCC) of the Martin Delaney BELIEVE Collaboratory and the John G. Bartlett Specialty Practice Clinic at Johns Hopkins University School of Medicine (JHUSOM). The virologic inclusion criteria used to identify adult study participants were similar to those used with the perinatal HIV infections.
Isolation of PBMCs from whole blood.
For the study participants recruited at JHUSOM, whole blood was collected in EDTA-containing tubes and processed within 24 hours of collection. PBMCs were isolated by Ficoll-Hypaque gradient centrifugation (GE Healthcare) and cryopreserved in 90% FBS containing 10% DMSO (MilliporeSigma) in 10-million-cell aliquots and stored in liquid nitrogen following slow freezing at –80°C for a minimum of 4 hours. A separate aliquot of 5 million cells was also preserved as a cell pellet for HIV DNA quantitation by ddPCR. For the adult participants in the RCC Study of the BELIEVE Collaboratory, deidentified cryopreserved PBMCs were shipped on dry ice overnight and stored in liquid nitrogen until further use.
TILDA and enhanced TILDA.
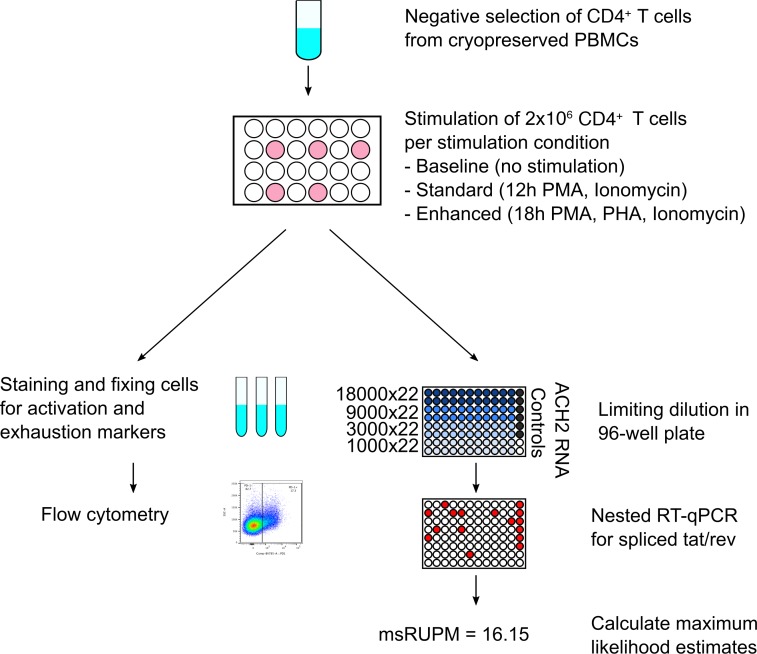
Following a thaw using 50% FBS and 50% RPMI 1640, CD4+ T cells were purified from PBMCs using a negative bead enrichment system (Miltenyi Biotec). The size of the inducible latent reservoir in perinatal infection was first assessed with standard TILDA (Figure 6). The size of the reservoir in CD4+ T cells that can be induced to produce the multiply spliced HIV transcripts tat and rev following 12-hour stimulation with PMA and ionomycin and in the presence of antiretroviral drugs was measured by reverse transcriptase PCR (RT-PCR) followed by quantitative PCR (qPCR) and estimated using maximum likelihood methods. The frequency of viral RNA-producing cells was then quantified as msRUPM (29, 64). Each sample and each condition were tested with the standard limiting dilution assay as published by Procopio et al. (29); purified CD4+ T cells were plated, following the T cell stimulation, in a serial dilution consisting of 22 replicates at each of the following dilutions: 18,000, 9000, 3000, and 1000 cells (total 682,000 CD4+ T cells; Figure 6). Positive controls had purified RNA from ACH2 cells, which contain a single copy of HIV that can be induced to produced spliced viral mRNA (65, 66).
Figure 6. Schematic of experimental conditions summarizing CD4+ T cell selection and stimulation and subsequent assay conditions.
Cryopreserved PBMCs are thawed and enriched for CD4+ T cells by negative bead enrichment. Cells are subsequently aliquoted at 2 million cells per 1 mL of medium and rested for 2–6 hours. Following the rest period, each sample is distributed into individual wells as (i) unstimulated, for subsequent flow studies for baseline measurements, (ii) stimulated, using PMA and ionomycin for 12 hours (standard TILDA), or (iii) stimulated, with PMA, PHA, and ionomycin for 18 hours (enhanced TILDA). After stimulation, cells are washed and either fixed and stained for flow cytometry, or plated in a limiting dilution scheme for direct lysis and RT-qPCR followed by qPCR as previously published.
A modified TILDA (enhanced TILDA) that includes the addition of PHA at 1 μg/mL with the PMA/ionomycin and for 18 hours instead of 12 hours was developed to augment detection of proviral reactivation (Figure 6 and ref. 67). Additionally, given that our study cohort was diverse with respect to country of origin, HIV subtyping was conducted on all study participants and HIV subtype–specific primer and probe sets were designed and optimized specifically for subtypes A1, C, and A/G (Supplemental Table 3) for RT-PCR, and qPCR. The already published primer and probe sets were used for subtypes B and A/E (29, 55).
HIV DNA quantitation.
Genomic DNA was isolated from 5 million PBMC pellets using the QIAamp DNA Blood Midi Kit (QIAGEN), but modified as reported by Strain et al. to include additional column washes and an overnight ethanol precipitation step to maximize HIV DNA purity and yield for ddPCR (68). Eight replicates of 1000 ng genomic DNA were assayed for total HIV DNA concentrations with the QX200 Auto Droplet Generator and Digital Droplet Reader (Bio-Rad) (69). Proviral load is expressed as GAGLTR copies per million PBMCs, with cell numbers determined using RPP30 as the housekeeping gene. Due to the insufficiency of cells for a separate determination of the proviral load in CD4+ T cells, the HIV-infected CD4+ T cell load was approximated by dividing the GAGLTR copies per million PBMCs by the percentage of CD4+ T cells present in the sample at the time of analyses. For samples that were insufficient to assay for the proportion of CD4+ T cells, a factor of 0.30 was used based on published values (22). The percent-induced provirus in CD4+ T cells was then calculated by dividing the msRUPM of each sample by this calculated GAGLTR per million CD4+ T cells. HIV DNA concentrations were also determined using the recently published intact proviral DNA assay, which differentiates double-positive intact proviral genomes from single positives containing hypermutations, or 3′ and 5′ deletions (23). Genomic DNA from an average of 300,000 cells was assayed for intact proviral DNA and reported as double-positive intact HIV DNA per million cells assayed. IPDA on the perinatal samples was performed at Accelevir Diagnostics and on the adult cohort at the RCC.
Flow cytometric analyses of immune activation and exhaustion markers.
Cells were thawed and processed for flow cytometry in tandem with the TILDA. After negative enrichment for CD4+ T cells, aliquots of 500,000 unstimulated cells were incubated with antiretrovirals to serve as baseline control for flow cytometric analyses of markers of immune activation and exhaustion; and an aliquot of 1 million cells from both standard and enhanced TILDA stimulation conditions was used for flow cytometric analyses (Figure 6).
The activation phenotype of cultured CD4+ T cells was evaluated using 2 antibody panels (Supplemental Table 4). The panel included “backbone” markers (live/dead FVS780, CD3 APC-R700, CD4 BV421, CD8 BV510, CD14 BV510, CD19 BV510) with either T cell activation markers (CD69 FITC, CD25 -PE and HLA-DR–BB515) or exhaustion molecules (PD-1–BV786, TIM-3–PE, TIGIT–PerCP-eF710) (BD Biosciences; eBioscience). After stimulation, cells were counted, washed with PBS twice, and stained with live/dead antibody for 15 minutes at room temperature. Cells were then washed with PBS twice, stained for 30 minutes with an antibody cocktail of the cell surface markers, washed twice with stain buffer (PBS/2% FBS), and fixed with 1% paraformaldehyde. Samples were immediately analyzed on an LSR II flow cytometer using FACSDiva software version 10 (BD Biosciences) through the Johns Hopkins Becton Dickinson Immune Function Laboratory. Cell phenotypic data were analyzed using FlowJo software (version 10.2, Tree Star). Each activation and exhaustion marker had an appropriate isotype control in the fluorescence-minus-one control, which was used to set gates for each population in FlowJo (Supplemental Figure 1).
Statistics.
All comparisons were performed using nonparametric tests due to small sample sizes. Tests for differences between paired measurements (i.e., 2 or more measurements within the same participant; for example, the standard versus enhanced TILDAs within adults) were conducted using 2-tailed Wilcoxon’s signed-rank test. For unpaired data, such as comparing perinatal with adult participants, 2-tailed Mann-Whitney U 2-sample rank-sum test was used. Correlations were quantified by the Spearman’s rank correlation coefficient and the corresponding P values reported. P values less than 0.05 were considered statistically significant. GraphPad Prism version 8.0.0 was used for data compilation and statistical analyses. All error bars in graphs show median and IQR.
Study approval.
This study was approved by the Johns Hopkins Medicine Office of Human Subjects Research Institutional Review Boards as study number NA_00087629. Written informed consent was received from all participants before inclusion in the study.
Author contributions
DP, JHR, and AD designed the study. AA, ACS, and TA organized sample collection from the Johns Hopkins Pediatric and Adolescent HIV/AIDS Program. VT and RBJ aided in data acquisition. AD, JHR, LP, and TN performed the experiments. DP and AD wrote the manuscript. LVS and AD conducted statistical analyses. BASN oversaw statistical analyses. All coauthors edited and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank the participants and their families for contributing to the research; Bonnie Addison and Gerard Quarles for recruitment of patients and acquisition of clinical data; and Joel Blankson for recruitment of adult patients for our study. IPDA on the perinatally infected participants’ samples was performed by Accelevir Diagnostics. We also thank Joe Szewczyk and Ya Hui Chen for help with GAGLTR ddPCR and subtyping. This research was supported by the NIH (R01 HD080474 [to DP], PO1 AI131365 [to DP], U01 1U01AI135941, [to DP]), the BELIEVE Collaboratory (1UM1AI26617), EPIICAL (16108367), International Maternal Pediatric Adolescent Clinical Trials Laboratory Center (UM1AI106716), and the Johns Hopkins Center for AIDS Research (P30 AI094189). The following reagent was obtained through the AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: ACH-2 cells from Thomas Folks (catalog 349).
Version 1. 01/30/2020
In-Press Preview
Version 2. 02/27/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(4):e134105.https://doi.org/10.1172/jci.insight.134105.
Contributor Information
Adit Dhummakupt, Email: adhumma1@jhmi.edu.
Jessica H. Rubens, Email: jhodge7@jhmi.edu.
Thuy Anderson, Email: tander34@jhmi.edu.
Laura Powell, Email: llarson3@jhmi.edu.
Bareng A.S. Nonyane, Email: bnonyan1@jhmi.edu.
Lilly V. Siems, Email: lilly.v.k.siems@gmail.com.
Aleisha Collinson-Streng, Email: acolli14@jhmi.edu.
R. Brad Jones, Email: rbjones@med.cornell.edu.
Vicki Tepper, Email: vtepper@som.umaryland.edu.
Allison Agwu, Email: ageorg10@jhmi.edu.
Deborah Persaud, Email: dpers@jhmi.edu.
References
- 1. [No authors listed]. 2018 Global HIV Statistics — Fact Sheet for World AIDS Day 2019. UNAIDS. https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf Accessed February 20, 2020.
- 2.Persaud D, et al. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J Clin Invest. 2000;105(7):995–1003. doi: 10.1172/JCI9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 4.Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71(3):2059–2071. doi: 10.1128/JVI.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khoury GD, et al. The molecular biology of HIV latency. In: Zhang L, Lewin SR, eds. HIV Vaccines and Cure. Singapore: Springer; 2018:187–212. [Google Scholar]
- 7.Sarracino A, et al. Posttranscriptional regulation of HIV-1 gene expression during replication and reactivation from latency by nuclear matrix protein MATR3. mBio. 2018;9(6):e02158-18. doi: 10.1128/mBio.02158-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes SH, Coffin JM. What integration sites tell us about HIV persistence. Cell Host Microbe. 2016;19(5):588–598. doi: 10.1016/j.chom.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray AJ, Kwon KJ, Farber DL, Siliciano RF. The latent reservoir for HIV-1: how immunologic memory and clonal expansion contribute to HIV-1 persistence. J Immunol. 2016;197(2):407–417. doi: 10.4049/jimmunol.1600343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardons M, et al. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. 2019;15(2):e1007619. doi: 10.1371/journal.ppat.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee GQ, Lichterfeld M. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS. 2016;11(4):383–387. doi: 10.1097/COH.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. Latency reversal and viral clearance to cure HIV-1. Science. 2016;353(6297):aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbato JM, Purves HV, Lewin SR, Rasmussen TA. Between a shock and a hard place: challenges and developments in HIV latency reversal. Curr Opin Virol. 2019;38:1–9. doi: 10.1016/j.coviro.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deeks S. Towards an HIV cure. J Int AIDS Soc. 2014;17(4 Suppl 3):19479. doi: 10.7448/IAS.17.4.19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contreras X, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284(11):6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009;23(14):1799–1806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cillo AR, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 2015;23(4):192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson S, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruner KM, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566(7742):120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 25.Crooks AM, et al. Precise Quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenbloom DIS, et al. Assessing intra-lab precision and inter-lab repeatability of outgrowth assays of HIV-1 latent reservoir size. PLoS Comput Biol. 2019;15(4):e1006849. doi: 10.1371/journal.pcbi.1006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 28.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Procopio FA, et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2(8):874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho YC, Laird GM, Siliciano RF. Measuring reversal of HIV-1 latency ex vivo using cells from infected individuals. Proc Natl Acad Sci U S A. 2014;111(19):6860–6861. doi: 10.1073/pnas.1405194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fromentin R, et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4+ T cells from ART-suppressed individuals. Nat Commun. 2019;10(1):814. doi: 10.1038/s41467-019-08798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fromentin R, et al. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog. 2016;12(7):e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caruso A, et al. Flow cytometric analysis of activation markers on stimulated T cells and their correlation with cell proliferation. Cytometry. 1997;27(1):71–76. doi: 10.1002/(SICI)1097-0320(19970101)27:1<71::AID-CYTO9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 34.Uprety P, et al. Human immunodeficiency virus type 1 DNA decay dynamics with early, long-term virologic control of perinatal infection. Clin Infect Dis. 2017;64(11):1471–1478. doi: 10.1093/cid/cix192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uprety P, et al. Cell-associated HIV-1 DNA and RNA decay dynamics during early combination antiretroviral therapy in HIV-1-infected infants. Clin Infect Dis. 2015;61(12):1862–1870. doi: 10.1093/cid/civ688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med. 2017;214(4):959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarracino A, Marcello A. The relevance of post-transcriptional mechanisms in HIV latency reversal. Curr Pharm Des. 2017;23(28):4103–4111. doi: 10.2174/1381612823666170803102355. [DOI] [PubMed] [Google Scholar]
- 40.Shan L, et al. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol. 2011;85(11):5384–5393. doi: 10.1128/JVI.02536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taura M, Song E, Ho YC, Iwasaki A. Apobec3A maintains HIV-1 latency through recruitment of epigenetic silencing machinery to the long terminal repeat. Proc Natl Acad Sci U S A. 2019;116(6):2282–2289. doi: 10.1073/pnas.1819386116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson EM, Maldarelli F. The role of integration and clonal expansion in HIV infection: live long and prosper. Retrovirology. 2018;15(1):71. doi: 10.1186/s12977-018-0448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Einkauf KB, et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest. 2019;129(3):988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roider J, et al. Increased regulatory T-cell activity and enhanced T-cell homeostatic signaling in slow progressing HIV-infected children. Front Immunol. 2019;10:213. doi: 10.3389/fimmu.2019.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goulder PJ, Lewin SR, Leitman EM. Paediatric HIV infection: the potential for cure. Nat Rev Immunol. 2016;16(4):259–271. doi: 10.1038/nri.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez DR, Permar SR, Fouda GG. contrasting adult and infant immune responses to HIV infection and vaccination. Clin Vaccine Immunol. 2016;23(2):84–94. doi: 10.1128/CVI.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabe H, Lundell AC, Andersson K, Adlerberth I, Wold AE, Rudin A. Higher proportions of circulating FOXP3+ and CTLA-4+ regulatory T cells are associated with lower fractions of memory CD4+ T cells in infants. J Leukoc Biol. 2011;90(6):1133–1140. doi: 10.1189/jlb.0511244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persaud D, et al. Influence of age at virologic control on peripheral blood human immunodeficiency virus reservoir size and serostatus in perinatally infected adolescents. JAMA Pediatr. 2014;168(12):1138–1146. doi: 10.1001/jamapediatrics.2014.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhn L, et al. Age at antiretroviral therapy initiation and cell-associated HIV-1 DNA levels in HIV-1-infected children. PLoS ONE. 2018;13(4):e0195514. doi: 10.1371/journal.pone.0195514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luzuriaga K, et al. HIV type 1 (HIV-1) proviral reservoirs decay continuously under sustained virologic control in HIV-1-infected children who received early treatment. J Infect Dis. 2014;210(10):1529–1538. doi: 10.1093/infdis/jiu297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spina CA, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9(12):e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong-Starkesen SE, Luciw PA, Peterlin BM. Signaling through T lymphocyte surface proteins, TCR/CD3 and CD28, activates the HIV-1 long terminal repeat. J Immunol. 1989;142(2):702–707. [PubMed] [Google Scholar]
- 54.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS. A hardwired HIV latency program. Cell. 2015;160(5):990–1001. doi: 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colby DJ, et al. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med. 2018;24(7):923–926. doi: 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiau S, et al. Distinct epigenetic profiles in children with perinatally-acquired HIV on antiretroviral therapy. Sci Rep. 2019;9(1):10495. doi: 10.1038/s41598-019-46930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gross AM, et al. Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell. 2016;62(2):157–168. doi: 10.1016/j.molcel.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tobin NH, Aldrovandi GM. Immunology of pediatric HIV infection. Immunol Rev. 2013;254(1):143–169. doi: 10.1111/imr.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Broncano P, et al. Early antiretroviral therapy in neonates with HIV-1 infection restricts viral reservoir size and induces a distinct innate immune profile. Sci Transl Med. 2019;11(520):eaax7350. doi: 10.1126/scitranslmed.aax7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar BV, Connors TJ, Farber DL. Human T cell development, localization, and function throughout life. Immunity. 2018;48(2):202–213. doi: 10.1016/j.immuni.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cesana D, et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat Commun. 2017;8(1):498. doi: 10.1038/s41467-017-00609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scully EP, et al. Sex-based differences in human immunodeficiency virus type 1 reservoir activity and residual immune activation. J Infect Dis. 2019;219(7):1084–1094. doi: 10.1093/infdis/jiy617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scully EP. Sex differences in HIV infection. Curr HIV/AIDS Rep. 2018;15(2):136–146. doi: 10.1007/s11904-018-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis. 2015;2(4):ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clouse KA, et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142(2):431–438. [PubMed] [Google Scholar]
- 66.Folks TM, et al. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc Natl Acad Sci USA. 1989;86(7):2365–2368. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim M, et al. A primary CD4(+) T cell model of HIV-1 latency established after activation through the T cell receptor and subsequent return to quiescence. Nat Protoc. 2014;9(12):2755–2770. doi: 10.1038/nprot.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strain MC, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4):e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhummakupt A, et al. The latent human immunodeficiency virus (HIV) reservoir resides primarily in CD32-CD4+ T cells in perinatally HIV-infected adolescents with long-term virologic suppression. J Infect Dis. 2019;219(1):80–88. doi: 10.1093/infdis/jiy461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.