Abstract
Background
High-grade serous ovarian cancer (HGSOC) is the most malignant gynecologic tumor. This study reveals biomarkers related to HGSOC incidence and progression using the bioinformatics method.
Material/Methods
Five gene expression profiles were downloaded from GEO. Differentially-expressed genes (DEGs) in HGSOC and normal ovarian tissue samples were screened using limma and the function of DEGs was annotated by KEGG and GO analysis using clusterProfiler. A co-expression network utilizing the WGCNA package was established to define several hub genes from the key module. Furthermore, survival analysis was performed, followed by expression validation with datasets from TCGA and GTEx. Finally, we used single-gene GSEA to detect the function of prognostic hub genes.
Results
Out of the 1874 DEGs detected from 114 HGSOC versus 49 normal tissue samples, 956 were upregulated and 919 were downregulated. The functional annotation indicated that upregulated DEGs were mostly enriched in cell cycle, whereas the downregulated DEGs were enriched in the MAPK or Ras signaling pathway. Two modules significantly associated with HGSOC were excavated through WGCNA. After survival analysis and expression validation of hub genes, we found that 2 upregulated genes (MAD2L1 and PKD2) and 3 downregulated genes (DOCK5, FANCD2 and TBRG1) were positively correlated with HGSOC prognosis. GSEA for single-hub genes revealed that MAD2L1 and PKD2 were associated with proliferation, while DOCK5, FANCD2, and TBRG1 were associated with immune response.
Conclusions
We found that FANCD2, PKD2, TBRG1, and DOCK5 had prognostic value and could be used as potential biomarkers for HGSOC treatment.
MeSH Keywords: Computational Biology, Ovarian Neoplasms, Survival Analysis
Background
Ovarian cancer has a high mortality rate, which ranks first among gynecologic malignant tumors. Most deaths (70%) of patients presented with advanced-staged, high-grade serous ovarian cancer (HGSOC) due to the lack of specific symptoms at the early stage [1]. Therefore, it is of great significance to study the potential prognostic biomarkers related to the development of HGSOC.
In recent years, bioinformatics-assisted analyses of expression profile have been widely used to detect the biomarkers of human diseases [2]. Weighted gene co-expression network analysis (WGCNA) is a biological approach to determine highly synergistic gene sets and to identify the association between gene modules and phenotype of samples [3]. WGCNA has been comprehensively utilized in multiple cancer-associated studies to determine hub genes that could be associated with respective traits, such as pancreatic carcinoma [4], colon cancer [5], and ovarian cancer [6–10]. However, few previous studies have focused on HGSOC.
To identify potential biomarkers for specific diagnosis and therapy targets in HGSOC, WGCNA was performed to discover the hub genes that play an essential role in the development of HGSOC.
Material and Methods
Data collection
Five gene expression profiles were downloaded from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Datasets GSE18520 [11], GSE27651 [12], GSE54388 [13], GSE10971 [14] and GSE14001 [15] are listed in Supplementary Table 1, with a sample size of 114 for HGSOC and 49 normal tissue samples. All samples were processed using the Affymetrix human genome U133 plus 2.0 array. Genomic and clinical data were obtained from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) and GTEx (https://gtexportal.org/home/) [16] using the TCGAbiolinks package (Version 2.14.0; https://github.com/BioinformaticsFMRP/TCGAbiolinks) [17]. The RNA expression profiles were sampled from 363 high-grade serous ovarian cancer and 108 normal tissues.
Research design and data preprocessing
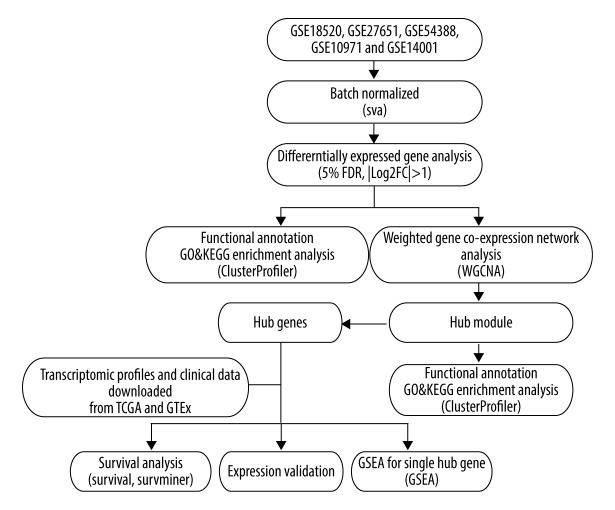
The research design is shown in a flowchart (Figure 1). The raw data from 5 datasets were chosen for integrated analysis using the affy package (Version 3.8; http://bioconductor.org/packages/release/bioc/html/affy.html) [18]. The batch effect of datasets was removed using the SVA package (Version 3.8; http://bioconductor.org/packages/release/bioc/html/sva.html) with its combat function (Supplementary Figure 1) [19].
Figure 1.
Flow diagram of this study.
Differential gene expression analysis
We detected the DEGs between HGSOC and normal ovarian tissue samples using the limma package (Version 3.30.0; http://www.bioconductor.org/packages/release/bioc/html/limma.html) [20]. A false discovery rate (FDR) <0.05 and |log2FC|>1 were set as the criteria value. The expression intensity and direction of DEGs were represented using the pheatmap package (Version 1.0.12, https://cran.r-project.org/web/packages/pheatmap).
Function enrichment analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of DEGs were conducted using the clusterProfiler package (Version 6.8; http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) [21] to predict their underlying molecular functions. A p value of <0.05 was considered statistically significant.
Weighted gene co-expression network analysis (WGCNA)
We utilized the WGCNA package (Version 1.67; https://cran.r-project.org/web/packages/WGCNA/index.html) to construct a co-expression network for DEGs. To identify key modules, soft-thresholding power was set as β=9 (scale-free R2=0.84), and cut height was set as 0.25 (Supplementary Figure 2). We then explored the biological function of the modules that had the highest correlation with traits through GO and KEGG pathway analyses, and hub genes in a module were selected with |MM|>0.85 and |GS|>0.3.
Survival analysis and expression validation of hub genes
Survival analysis was performed for hub genes using the survival package (Version 2.43.3; https://cran.r-project.org/package=survival) and survminer package (Version 0.4.3; https://cran.r-project.org/package=survminer). The Kaplan-Meier curves were plotted by the expression profiles from TCGA, which were divided into 2 groups based on a certain gene’s cutoff value as determined by survminer. The hub gene expression levels between HGSOC and normal tissue samples were also validated.
Gene set enrichment analysis (GSEA)
GSEA analysis of each hub gene with the TCGA-OV dataset was performed. The HGSOC samples (n=363) were divided into 2 groups according to the median expression value of each hub gene (high vs. low). A p value of <0.05 was considered as statistically significant. The “h.all.v6.2.entrez.gmt” were selected as reference gene sets, which were downloaded from the Molecular Signature Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb).
Results
Differential expression analysis
We screened 1874 DEGs, including 919 downregulated and 956 upregulated genes, between HGSOC samples and normal tissue samples. The expression changes of DEGs were represented by a heatmap (Supplementary Figure 3), which showed that the samples were divided into 2 clusters.
Function enrichment for DEGs
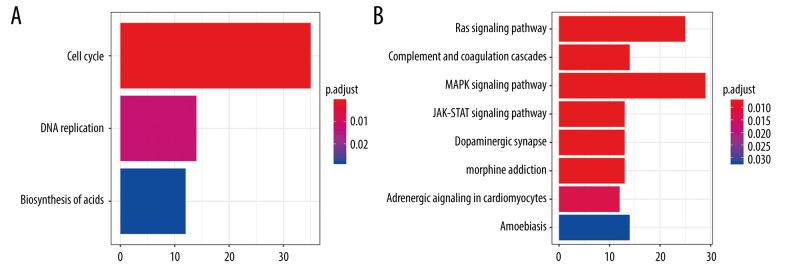
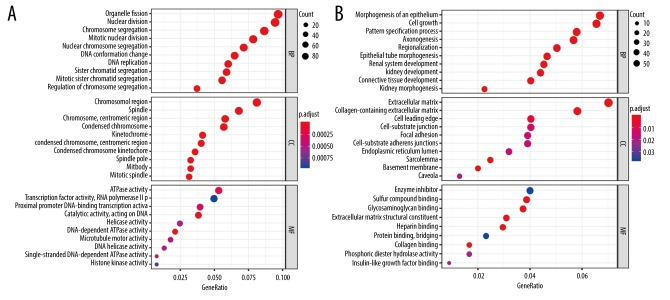
The potential biological functions of the upregulated and downregulated differentially-expressed genes were annotated by clusterProfiler R package. The KEGG pathway analysis revealed that the upregulated genes were mainly involved in regulation of cell cycle, DNA replication, and biosynthesis of amino acids, while the downregulated genes were mainly linked to the Ras signaling pathway, complement and coagulation cascades, and the MAPK and JAK-STAT signaling pathways (Figure 2 and Supplementary Table 2). Furthermore, we performed GO enrichment analysis, the biological progress of which was in line with the KEGG enrichment results. Chromosome segregation and mitotic nuclear division were indicated for the upregulated genes, while morphogenesis of an epithelium and collagen-containing extracellular matrix were indicated for the downregulated genes (Figure 3 and Supplementary Table 3).
Figure 2.
The KEGG pathway enrichment analysis of differently-expressed genes. (A) KEGG pathways enrichment for upregulated genes. (B) KEGG pathways enrichment for downregulated genes.
Figure 3.
The GO enrichment analysis of differently-expressed genes. (A) GO enrichment analysis of upregulated genes. (B) GO enrichment analysis of downregulated genes.
Co-expression modules construction
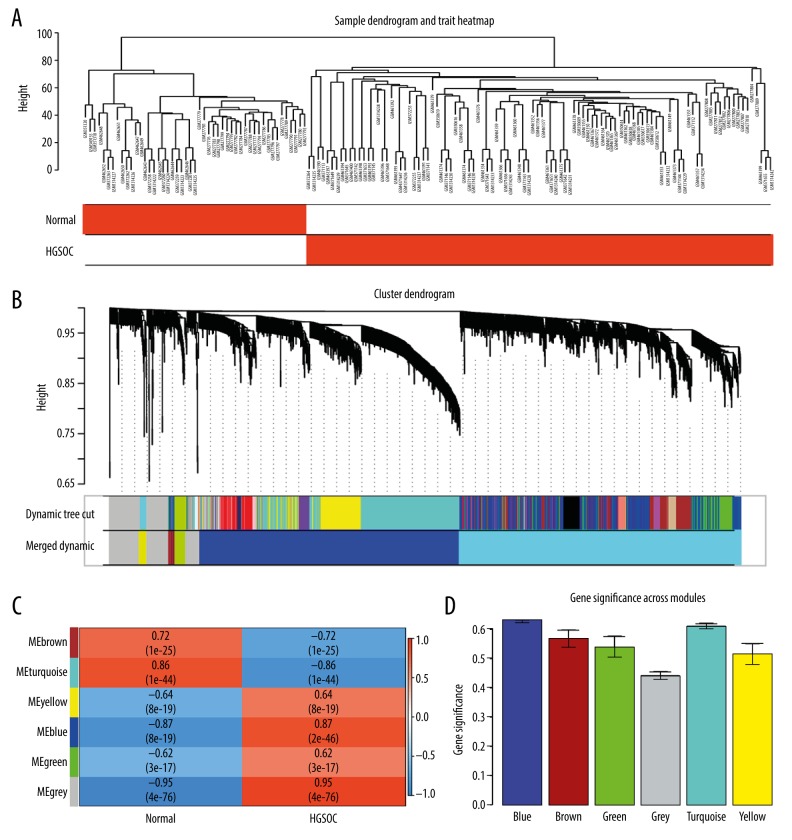
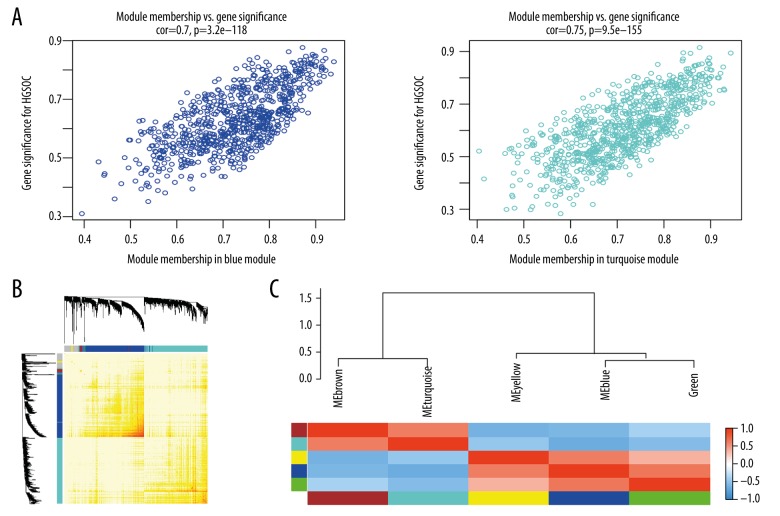
To construct co-expression modules and find the key modules related to HGSOC, the expression profiles of 1874 DEGs were assessed with the WGCNA package. Hierarchical clustering analysis is presented in Figure 4A. Then, the highly related genes were put into modules. The MED threshold was set as 0.25, and 6 modules were excavated (Figure 4B). The genes that did not belong to any module were collected in the gray module, and were not used in any subsequent analysis. The other 5 modules are shown in blue, turquoise, yellow, brown, and green, respectively (Figure 4C). Among the 5 modules, the turquoise and blue modules had remarkable relevance for tumor progression (Figure 4C, 4D).
Figure 4.
Co-expression modules construction and selection. (A) Samples clustering and trait heatmap of datasets from GEO according to the DEGs expression between HGSOC and normal tissue samples. (B) Dendrogram of all DEGs were clustered with dissimilarity according to topological overlap (1-TOM). (C) associations between modules and traits. In each cell, the upper number is the correlation coefficient of the module in the trait, and the lower number is the p value. Among them, the turquoise and blue modules were the most correlative with normal and cancer traits. (D) Distribution of average gene significance in the modules correlated with HGSOC. TOM – topological overlap matrix.
Moreover, intramodular analysis for GS and MM resulted in the identification of genes in the turquoise module, which were negatively correlated with HGSOC (correlation=0.75 and p<9.5e–155) and genes in the blue module revealed a highly positive correlation with HGSOC (correlation=0.7 and p<3.2e–118), as shown in Figure 5A.
Figure 5.
Select hub genes in significant co-expression modules. (A) The scatter plot of gene significance (GS) versus module membership (MM) in the blue module and turquoise module. (B) The heatmap presents the TOM among all genes. Colors beneath and right of the dendrograms explain the color-coding for each module. The more saturated yellow and red indicates a high co-expression inter-connectedness in the heatmap (C). Clustering of module eigengenes and the heatmap of the adjacencies.
The heatmap was plotted to show all genes (Figure 5B). To quantify co-expression similarity of the 5 modules, we calculated the connectivity of eigengenes. Positively correlated eigengenes were grouped together, with 2 of 5 modules were classified into one cluster and 3 into another. The heatmap of the adjacencies is also presented (Figure 5C).
There were 76 hub genes from the turquoise module and 76 hub genes from the blue module selected, with a threshold module membership (MM) >0.85 and gene significance (GS) >0.3 (Supplementary Table 4).
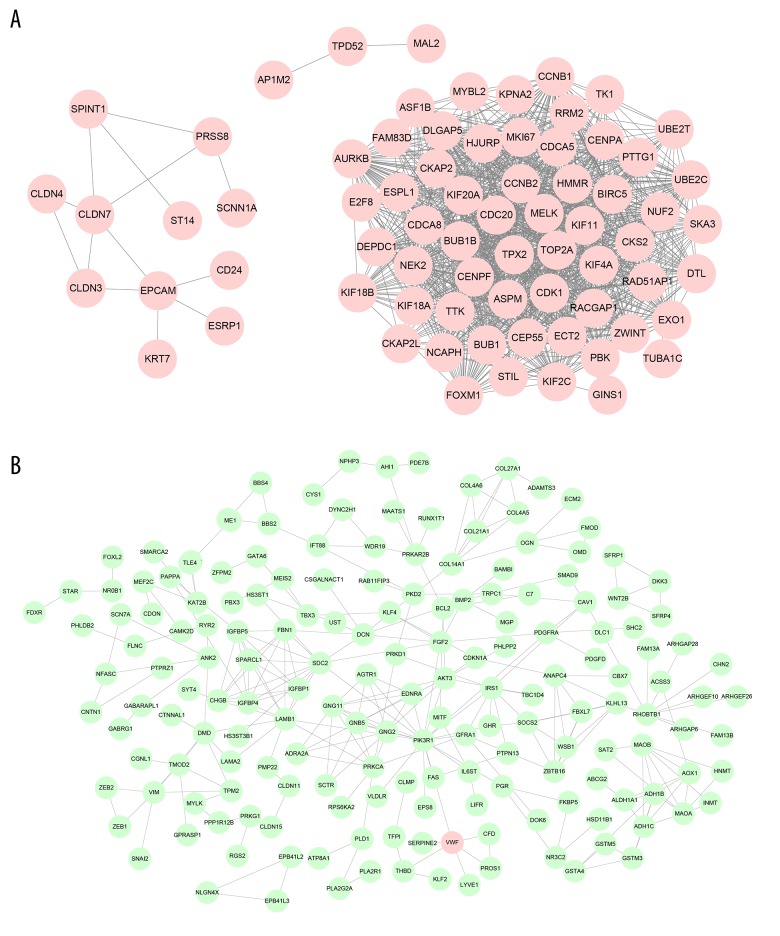
The turquoise and blue modules were analyzed by STRING database, with a combined score >0.7 and were visualized by Cytoscape software (Figure 6).
Figure 6.
Protein–Protein Interaction (PPI) network of genes in 2 modules. (A) The genes in blue module. (B) The genes in turquoise module. The color presents the fold change (upregulated genes are red, downregulated genes are green).
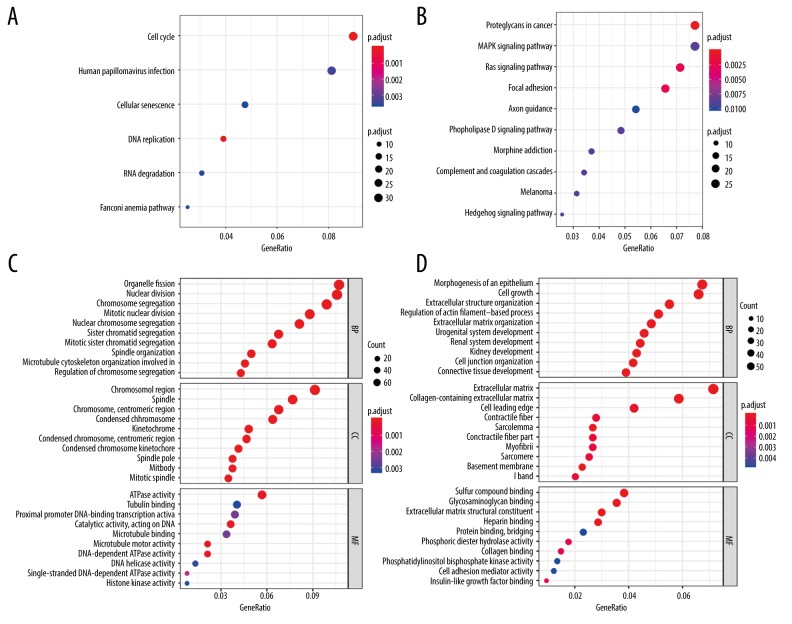
To investigate the potential functions of the genes within the 2 modules (turquoise and blue), we performed GO and KEGG pathway analyses, and showed the most significant GO terms and KEGG pathways in Figure 7. This analysis revealed that genes in the blue module were mainly enriched in cell cycle and DNA replication, while genes in the turquoise module played their roles in different signal pathways.
Figure 7.
GO and KEGG pathway analysis of the 2 modules. (A) KEGG pathway analysis of blue module; (B) KEGG pathway analysis of turquoise module; (C) GO analysis of blue module; (D) GO analysis of turquoise module. GO analysis includes biological process (BP), cellular component (CC), and molecular function (MF). The count represents the number of genes in each pathway and dot size corresponds to “count”.
Validation of hub genes
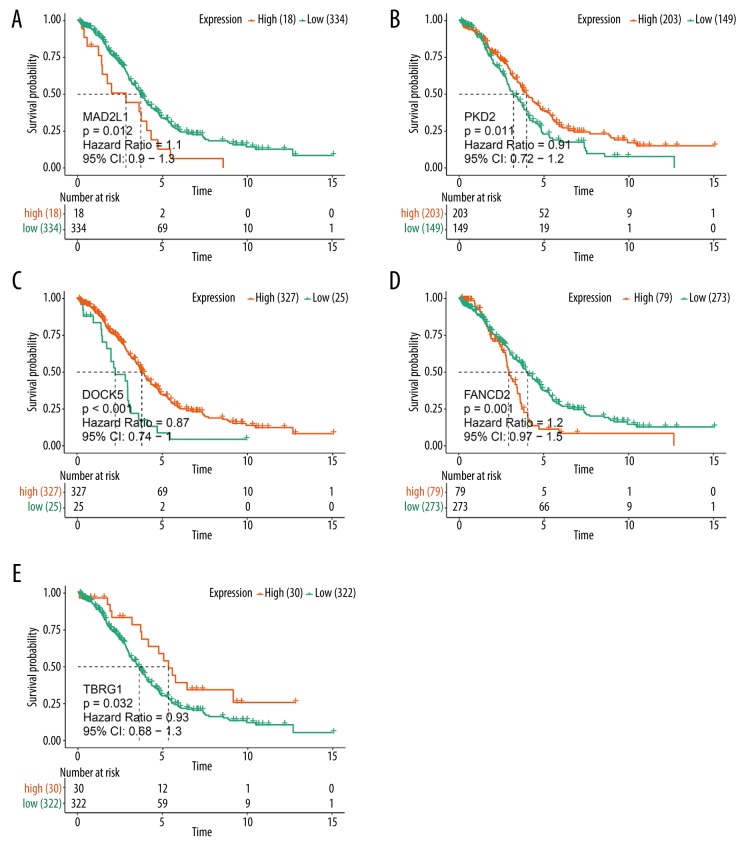
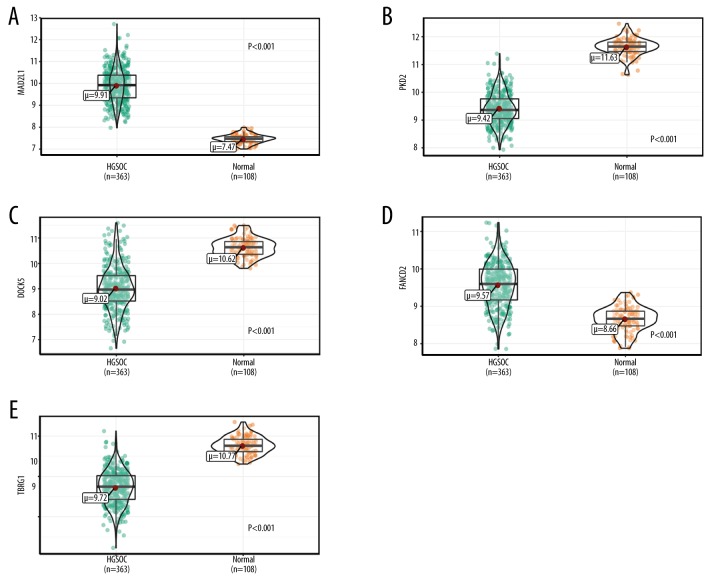
Analyzing the results of WGCNA, we found that the turquoise and blue modules had the highest association with HGSOC. Accordingly, we hypothesized that the genes in the turquoise module might act as tumor suppressors and genes in the blue module might act as tumor promoters. Survival analyses were performed among the 152 hub genes selected from the 2 modules. We found that MAD2L1 and FANCD2 in the blue module and PKD2, TBRG1, and DOCK5 in the turquoise module were consistent with our speculation. Survival curves showed that higher expression of MAD2L1 and FANCD2 was significantly associated with poor prognosis of patients, as was the lower expression of PKD2, TBRG1, and DOCK5 (Figure 8). Finally, we used gene profiles downloaded from TCGA and GTEx to validate the expression of these genes, and the results were similar to the expression exhibited by GEO (Figure 9).
Figure 8.
Kaplan-Meier analysis of (A) MAD2L1, (B) PKD2, (C) DOCK5, (D) FANCD2, and (E) TBRG1 by comparing the higher (red) and lower (green) expressions with overall survival outcomes for patients with HGSOC.
Figure 9.
Validation of hub gene expressions in the TCGA and GTEx datasets. (A) MAD2L1, (B) PKD2, (C) DOCK5, (D) FANCD2, and (E) TBRG1 gene expression differences between HGSOC and normal tissues.
Potential function of hub genes through GSEA
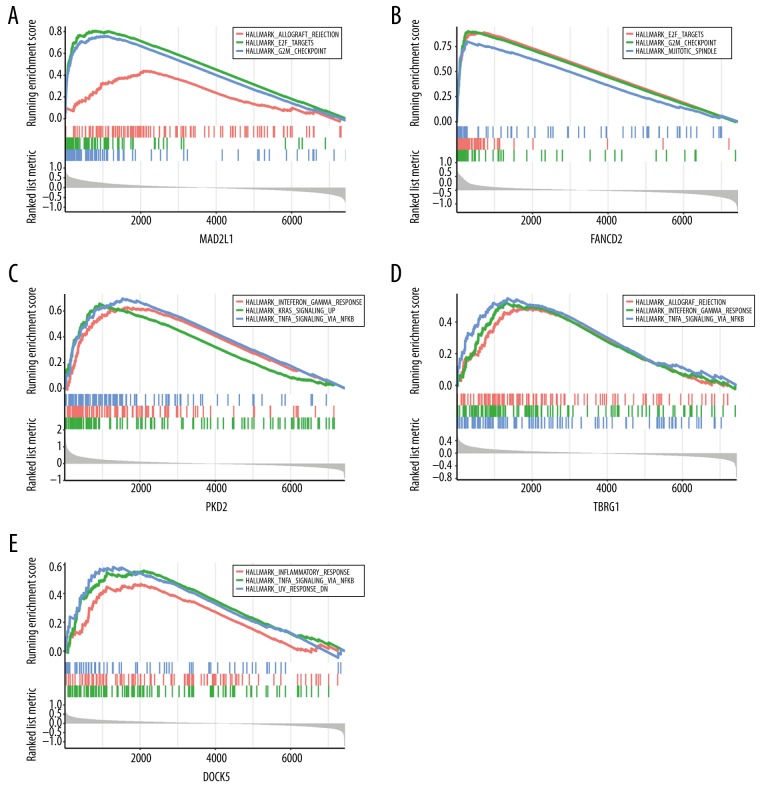
To better understand the potential biological functions of MAD2L1, FANCD2, PKD2, TBRG1, and DOCK5 in HGSOC, we performed GSEA based on the TCGA-OV dataset. As shown in Figure 10, genes in higher-expression groups of MAD2L1 and FANCD2 were all involved in “E2F TARGETS” and “G2M CHECKPOINT” of the cell cycle, which indicated that these 2 upregulated genes are closely associated with tumor proliferation, whereas “TNFA SIGNALING VIA NFKB”, “interferon gamma RESPONSE” and “inflammatory response” were enriched in the PKD2, TBRG1, and DOCK5 high-expression groups, which indicated these downregulated genes are involved in immune response (Supplementary Table 5).
Figure 10.
Gene set enrichment analysis (GSEA) of hub genes in the TCGA-OV dataset. Three gene sets enriched in the high-expressed group of single-hub genes. (A) MAD2L1, (B) FANCD2, (C) PKD2, (D) TBRG1, and (E) DOCK5.
Discussion
With the purpose of identifying the molecular mechanism of HGSOC and to investigate potential biomarkers for better detection and therapy, we integrated the gene expression profiles of GSE54388, GSE27651, GSE10971, GSE18520, and GSE14001, which contained 114 samples of HGSOC tissue and 49 samples of normal tissue.
We identified 1874 DEGs that were correlated with HGSOC, and the cutoff criteria were p<0.05 and |logFC|≥1. In KEGG analysis, the upregulated genes were predominantly involved in cell cycle and DNA replication, while the downregulated genes were highly involved in Ras signaling, complement and coagulation cascades, and MAPK signaling pathways. The GO analysis supported the previous enrichment analysis, which both help to understand the role of DEGs in HGSOC.
WGCNA analysis was used to select co-expression modules related to the development of HGSOC, and 2 modules (blue and turquoise) were found to have the highest correlation with HGSOC. We showed the Protein–Protein Interactions (PPI) network and also performed GO and KEGG analyses for genes in the 2 modules. The results indicated that genes in the blue module were enriched in cell cycle and DNA replication, while genes in the turquoise module were involved in different signaling pathways. After filtering with MM and GS value, we detected 152 hub genes from the 2 modules. Five genes – MAD2L1 and FANCD2 in the blue module and PKD2, TBRG1, and DOCK5 in the turquoise module – were excavated after survival analysis and expression validation with datasets downloaded from TCGA, and were found to have prognostic value for HGSOC. Among these 5 hub genes, MAD2L1 and FANCD2 are associated with ovarian cancer.
As a component of the mitotic checkpoint, high levels of MAD2L1 are related to increased cellular proliferation, migration, and metastasis, which can lead to shorter survival in various cancers [22–26]. However, in ovarian cancer, the role of MAD2L1 did not agree with previous findings that patients with lower MAD2L1 levels were less sensitive to paclitaxel and had shorter progression-free survival (PFS) and overall survival (OS) [27,28]. This discrepancy might have been caused by our analysis, ignoring the mutations of p53 and BRCA1, which are known regulators of MAD2L1 and are commonly mutated in HGSOC [29,30].
High FANCD2 levels have been shown to be associated with poor prognosis in many types of cancer [31–35], as well as in ovarian cancer [36]. FANCD2 overexpression can stabilize the replication fork, and create BRCA1/2 mutant tumor resistance towards PARP1/2 inhibitor treatments [37]. The results indicated that FANCD2 expression can influence cancer sensitivity to PARP1/2 inhibitors and thus could be used as a potential target of therapy.
To further explore the biological functions of the 5 selected hub genes, we conducted single-gene GSEA. “E2F TARGETS” and “G2M CHECKPOINT” were enriched in the high-expression groups of MAD2L1 and FANCD2, indicating their contribution to HGSOC proliferation. In the high-expression groups of PKD2, TBRG1, and DOCK5, immune-related signals, such as “TNFA SIGNALING VIA NFKB”, “INTERFERON GAMMA RESPONSE” and “INFLAMMATORY RESPONSE” were enriched, indicating the activity of immune response.
Conclusions
We identified several DEGs and meaningful gene modules in HGSOC. Four valuable hub genes (FANCD2, PKD2, TBRG1, and DOCK5) were strongly dysregulated in HGSOC tissues. GSEA further suggested that FANCD2 is associated with tumor proliferation, while PKD2, TBRG1, and DOCK5 influence immune response. More work is needed to fully reveal their individual contributions towards the pathogenesis of HGSOC and to validate their value as prognostic biomarkers.
Limitations of this study include the lack of analysis for detailed clinical classification of HGSOC, such as grade, stage, lymph node metastasis, and prognosis. In future research, we will explore hub genes and their potential function based on this clinical information in detail.
Supplementary Data
Supplementary Table 1.
Characteristics of the included datasets.
Samples clustering of 5 datasets after removing the batch effects.
Soft-thresholding power determination in WGCNA. (A) Analysis of the scale-free fit index for different soft-thresholding powers. (B) Mean connectivity for various soft-thresholding powers. (C) Histogram of connectivity distribution when β=9. (D) Check scale-free topology when β=9.
Heatmap of the top 200 DEGs based on the value of |logFC|. High or low expression is shown as a red or blue strip, respectively. The experimental group was labelled HGSOC, while the control group was named Nor.
Supplementary Table 2.
The KEGG enrichment analysis of genes.
| ID | Description | p. adjust | Count | Regulation |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 0.000633 | 35 | up |
| hsa03030 | DNA replication | 0.012093 | 14 | up |
| hsa01230 | Biosynthesis of amino acids | 0.028478 | 12 | up |
| hsa04014 | Ras signaling pathway | 0.007503 | 25 | down |
| hsa04610 | Complement and coagulation cascades | 0.007503 | 14 | down |
| hsa04010 | MAPK signaling pathway | 0.007503 | 29 | down |
| hsa04630 | JAK-STAT signaling pathway | 0.007503 | 13 | down |
| hsa04728 | Dopaminergic synapse | 0.007503 | 13 | down |
| hsa05032 | Morphine addiction | 0.007503 | 13 | down |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 0.013251 | 12 | down |
| hsa05146 | Amoebiasis | 0.03213 | 14 | down |
Supplementary Table 3.
The GO enrichment analysis of genes.
| Ontology | ID | Description | p. adjust | Count | Regulation |
|---|---|---|---|---|---|
| BP | GO: 0007059 | Chromosome segregation | 7.51E-30 | 76 | up |
| BP | GO: 0140014 | Mitotic nuclear division | 8.88E-28 | 69 | up |
| BP | GO: 0000280 | Nuclear division | 1.11E-26 | 83 | up |
| BP | GO: 0048285 | Organelle fission | 3.71E-25 | 85 | up |
| BP | GO: 0098813 | Nuclear chromosome segregation | 5.13E-25 | 63 | up |
| CC | GO: 0098687 | Chromosomal region | 1.26E-26 | 74 | up |
| CC | GO: 0000775 | Chromosome, centromeric region | 2.73E-24 | 53 | up |
| CC | GO: 0000793 | Condensed chromosome | 1.10E-20 | 52 | up |
| CC | GO: 0000779 | Condensed chromosome, centromeric region | 2.30E-19 | 37 | up |
| CC | GO: 0005819 | Spindle | 2.30E-19 | 62 | up |
| MF | GO: 0140097 | Catalytic activity, acting on DNA | 2.32E-08 | 34 | up |
| MF | GO: 0008094 | DNA-dependent ATPase activity | 1.57E-05 | 19 | up |
| MF | GO: 0016887 | ATPase activity | 0.000226 | 47 | up |
| MF | GO: 0043142 | Single-stranded DNA-dependent ATPase activity | 0.000325 | 7 | up |
| MF | GO: 0001077 | Proximal promoter DNA-binding transcription activator activity, RNA polymerase II-specific | 0.000325 | 35 | up |
| BP | GO: 0002009 | Morphogenesis of an epithelium | 9.98E-07 | 53 | down |
| BP | GO: 0016049 | Cell growth | 2.29E-06 | 52 | down |
| BP | GO: 0001822 | Kidney development | 6.68E-06 | 35 | down |
| BP | GO: 0072001 | Renal system development | 6.68E-06 | 36 | down |
| BP | GO: 0003002 | Regionalization | 6.68E-06 | 40 | down |
| CC | GO: 0062023 | Collagen-containing extracellular matrix | 1.22E-11 | 49 | down |
| CC | GO: 0031012 | Extracellular matrix | 1.29E-10 | 59 | down |
| CC | GO: 0042383 | Sarcolemma | 3.36E-05 | 21 | down |
| CC | GO: 0005604 | Basement membrane | 3.64E-05 | 17 | down |
| CC | GO: 0031252 | Cell leading edge | 0.006422 | 34 | down |
| MF | GO: 0005201 | Extracellular matrix structural constituent | 4.99E-05 | 24 | down |
| MF | GO: 0005539 | Glycosaminoglycan binding | 5.13E-05 | 29 | down |
| MF | GO: 1901681 | Sulfur compound binding | 6.67E-05 | 30 | down |
| MF | GO: 0008201 | Heparin binding | 0.000129 | 23 | down |
| MF | GO: 0005518 | Collagen binding | 0.000836 | 13 | down |
Supplementary Table 4.
Hub genes in blue and turquoise module (|MM|>0.85 and |GS|>0.3).
| Blue module | Turquoise module | ||||
|---|---|---|---|---|---|
| Gene | MM | GS | Gene | MM | GS |
| CSE1L | 0.885182 | 0.654936 | LRRN4 | 0.863969 | −0.76695 |
| PCNA | 0.853268 | 0.634531 | LINC01105 | 0.85413 | −0.83749 |
| HNRNPAB | 0.859002 | 0.665006 | DAB2 | 0.928442 | −0.84279 |
| TOP2A | 0.891679 | 0.822887 | CELF2 | 0.888927 | −0.77641 |
| SMC4 | 0.917093 | 0.806692 | LAMA4 | 0.850672 | −0.68463 |
| MTHFD2 | 0.877458 | 0.806098 | SPOCK1 | 0.867854 | −0.80181 |
| PSRC1 | 0.871528 | 0.788497 | PAPSS2 | 0.879786 | −0.72486 |
| CKS1B | 0.934063 | 0.787083 | DAPK1 | 0.899065 | −0.7982 |
| MCM2 | 0.922548 | 0.841694 | PROCR | 0.898493 | −0.7677 |
| PCLAF | 0.899258 | 0.817579 | PKD2 | 0.929768 | −0.75857 |
| CRABP2 | 0.87272 | 0.877783 | GSDME | 0.91672 | −0.80166 |
| CCNB2 | 0.897672 | 0.848503 | IGFBP6 | 0.892364 | −0.79082 |
| LSM4 | 0.871655 | 0.664114 | THBD | 0.869457 | −0.80323 |
| CDC20 | 0.893728 | 0.758763 | KDR | 0.884189 | −0.74272 |
| UBE2C | 0.876499 | 0.834623 | FRY | 0.899738 | −0.80017 |
| CDK1 | 0.876549 | 0.8119 | GNG11 | 0.930805 | −0.75466 |
| EZH2 | 0.862178 | 0.795034 | ABCA8 | 0.944703 | −0.89654 |
| MAD2L1 | 0.889283 | 0.76929 | GPRASP1 | 0.899009 | −0.87775 |
| PTTG1 | 0.870574 | 0.824798 | GFPT2 | 0.866683 | −0.7817 |
| BUB1B | 0.892962 | 0.819369 | RNASE4 | 0.903103 | −0.80729 |
| DLGAP5 | 0.877017 | 0.82571 | CALB2 | 0.897513 | −0.85688 |
| ZWINT | 0.922712 | 0.79987 | BCHE | 0.922752 | −0.81917 |
| TRIP13 | 0.884367 | 0.77344 | NPY1R | 0.882911 | −0.85536 |
| RAD51AP1 | 0.858395 | 0.802612 | GHR | 0.888245 | −0.71151 |
| NDC80 | 0.86773 | 0.721595 | ECM2 | 0.850415 | −0.66158 |
| CKS2 | 0.94052 | 0.829763 | ARHGAP6 | 0.869659 | −0.73929 |
| KIF11 | 0.885283 | 0.822206 | WNT2B | 0.877108 | −0.80996 |
| NEK2 | 0.89035 | 0.839637 | PTGIS | 0.883089 | −0.79366 |
| KIF23 | 0.853223 | 0.7629 | LGALS2 | 0.85777 | −0.75854 |
| FEN1 | 0.909533 | 0.760884 | MAF | 0.898868 | −0.73781 |
| TTK | 0.894859 | 0.855031 | SYNE1 | 0.854181 | −0.8207 |
| MELK | 0.900554 | 0.813158 | PLPP1 | 0.920489 | −0.7545 |
| STIL | 0.867957 | 0.835405 | TCEAL2 | 0.877183 | −0.81491 |
| SAC3D1 | 0.874805 | 0.712282 | TBC1D2B | 0.862424 | −0.70643 |
| HMGA1 | 0.869641 | 0.806441 | PDE8B | 0.878248 | −0.91687 |
| GINS1 | 0.893705 | 0.7861 | ATP10D | 0.88824 | −0.74775 |
| CENPF | 0.880011 | 0.84842 | TFPI | 0.86169 | −0.75802 |
| AURKA | 0.914224 | 0.801243 | CHN2 | 0.855714 | −0.81706 |
| EIF4G1 | 0.869979 | 0.737708 | BICC1 | 0.864204 | −0.82881 |
| NR2F6 | 0.887103 | 0.752212 | DIXDC1 | 0.858203 | −0.8161 |
| BUB1 | 0.913764 | 0.809106 | DIRAS3 | 0.875531 | −0.8524 |
| PUF60 | 0.866461 | 0.739985 | OLFML1 | 0.886786 | −0.70603 |
| TPX2 | 0.862716 | 0.819783 | CSGALNACT1 | 0.90082 | −0.83324 |
| RPL39L | 0.857416 | 0.742065 | PDGFD | 0.866505 | −0.68393 |
| EIF6 | 0.86057 | 0.671087 | RADX | 0.868631 | −0.84742 |
| XPOT | 0.857586 | 0.684634 | KLF2 | 0.894365 | −0.78132 |
| SCRIB | 0.867393 | 0.771012 | SMPD3 | 0.867973 | −0.78767 |
| CCNA2 | 0.868501 | 0.7858 | PPP1R3B | 0.85511 | −0.72434 |
| CCNB1 | 0.904717 | 0.755995 | OGN | 0.88218 | −0.80725 |
| PRC1 | 0.886914 | 0.802267 | ABI3BP | 0.872052 | −0.78816 |
| MRPL15 | 0.853847 | 0.663766 | ITLN1 | 0.884383 | −0.81818 |
| NUSAP1 | 0.867073 | 0.795277 | MGARP | 0.903375 | −0.8306 |
| SLC52A2 | 0.868392 | 0.803613 | ARHGAP18 | 0.918993 | −0.80114 |
| TACC3 | 0.875177 | 0.752928 | DDR2 | 0.852443 | −0.67497 |
| KIF4A | 0.871167 | 0.797892 | ANTXR2 | 0.858928 | −0.72284 |
| CEP55 | 0.850744 | 0.827425 | LIX1L | 0.878142 | −0.68937 |
| DTL | 0.865255 | 0.780689 | MCC | 0.870811 | −0.71797 |
| KIF20A | 0.888407 | 0.82511 | TBRG1 | 0.85824 | −0.79791 |
| CENPU | 0.863996 | 0.732426 | PTPN21 | 0.863405 | −0.7201 |
| KIF15 | 0.882733 | 0.786989 | CNRIP1 | 0.889837 | −0.80871 |
| ECT2 | 0.916732 | 0.831958 | PPM1K | 0.895422 | −0.86713 |
| CDCA8 | 0.85916 | 0.797452 | MEDAG | 0.866729 | −0.84797 |
| MCM4 | 0.914084 | 0.813176 | LINC01279 | 0.857322 | −0.66728 |
| RACGAP1 | 0.918941 | 0.779139 | PLEKHH2 | 0.865982 | −0.76522 |
| PSAT1 | 0.854015 | 0.826163 | SLC30A4 | 0.904509 | −0.70542 |
| UBE2T | 0.850996 | 0.725215 | TCEAL3 | 0.856911 | −0.7193 |
| SLC25A33 | 0.868987 | 0.6974 | CDON | 0.869831 | −0.69303 |
| CDCA3 | 0.857304 | 0.797015 | TCEAL7 | 0.870405 | −0.84944 |
| NUF2 | 0.91985 | 0.868254 | ERN1 | 0.880253 | −0.77167 |
| RCC2 | 0.909521 | 0.783731 | MUM1L1 | 0.899449 | −0.85402 |
| FAM83D | 0.901393 | 0.849648 | RNASEL | 0.872061 | −0.72795 |
| POC1A | 0.856044 | 0.816551 | DOCK5 | 0.893588 | −0.83622 |
| DEPDC1B | 0.854264 | 0.764695 | RBMS3 | 0.850965 | −0.82463 |
| CENPL | 0.857368 | 0.768059 | HAND2-AS1 | 0.858159 | −0.89488 |
| KIF14 | 0.902918 | 0.830047 | DTWD1 | 0.879321 | −0.75556 |
| FANCD2 | 0.890071 | 0.736399 | IFFO1 | 0.857298 | −0.73892 |
MM – module membership; GS – gene significance.
Supplementary Table 5.
The Gene Set Enrichment Analysis (GSEA) of hub genes.
| Description | setSize | enrichmentScore | NES | p.adjust | core_enrichment | |
|---|---|---|---|---|---|---|
| MAD2L1 | ALLOGRAFT REJECTION | 120 | 0.438738 | 1.761702 | 0.007716 | CDKN2A/NME1/GZMB/MMP9/CXCL9/CCL5/CXCL13/IL15/CCL11/EIF5A/TAP1/CCL13/GZMA/SRGN/IL2RG/CCL2/UBE2N/CCL7/HLA-DOB/CTSS/CCL4/B2M/CD3D/PRF1/CD2/LTB/TNF/SIT1/IL2RA/CD7/HLA-G/CD8A/CD3E/ST8SIA4/CD86/FCGR2B/IFNG/IL12A/CXCR3/LY86/CD8B/RIPK2/UBE2D1/TPD52/HLA-DQA1/MRPL3/CD80/WARS/CD79A/CCR1/LCK/HDAC9/IGSF6/BCL10/TRAT1/CAPG/CD3G/CD96/IL11/IL2RB/MAP4K1/KRT1 |
| E2F TARGETS | 105 | 0.805846 | 3.155588 | 0.007716 | MAD2L1/CDKN2A/BIRC5/CKS2/CKS1B/CCNE1/TK1/UBE2S/PTTG1/UBE2T/MYBL2/NME1/CCNB2/AURKB/PLK1/DEPDC1/KPNA2/CDC20/RRM2/CENPM/CDKN3/CDK1/PLK4/AURKA/PCNA/SNRPB/KIF2C/SPC25/TRIP13/JPT1/ASF1B/ORC6/H2AFX/TOP2A/MELK/RNASEH2A/TACC3/CDCA8/DLGAP5/KIF4A/DCTPP1/SPC24/RFC3/CENPE/HMMR/RAD51AP1/DIAPH3/STMN1/POP7/BUB1B/DCK/MTHFD2/RPA3/GINS1/SPAG5/RACGAP1/KIF22/GINS4/ DDX39A/DSCC1/CDC25A/KIF18B/RAN/E2F8/RFC2/TUBG1/SLBP/BRCA2/HMGB3/SUV39H1/CHEK1/PRIM2/GINS3/ESPL1/SMC4/MXD3 | |
| G2M CHECKPOINT | 104 | 0.759634 | 2.975861 | 0.007716 | MAD2L1/CCNA2/UBE2C/BIRC5/CKS2/CKS1B/UBE2S/PTTG1/MYBL2/PBK/CCNB2/AURKB/PLK1/KPNA2/CDC20/CENPA/CDKN3/TTK/CDK1/PLK4/NEK2/AURKA/GINS2/KIF2C/JPT1/ORC6/H2AFX/CDC45/TOP2A/TROAP/TACC3/CDC6/SNRPD1/TPX2/KIF4A/NUSAP1/CENPE/HMMR/NDC80/STMN1/BUB1/EXO1/DTYMK/KIF23/TRAIP/PRC1/RACGAP1/KIF22/E2F1/DDX39A/CDC25A/POLQ/KIF15/FBXO5/RAD54L/KNL1/KIF11/BRCA2/HMGB3/E2F2/SUV39H1/CHEK1/CENPF/PRIM2/ESPL1/SMC4/ODC1/CCNF/STIL/SMC2/CDC7/MCM6/HIST1H2BK/EZH2/MCM2 | |
| FANCD2 | G2M CHECKPOINT | 104 | 0.901015 | 3.369604 | 0.006028 | MYBL2/KIF15/TPX2/TOP2A/KIF2C/UBE2C/BIRC5/ESPL1/MAD2L1/HMMR/PBK/KIF4A/PLK1/TROAP/TTK/BUB1/CDC20/POLQ/ NUSAP1/RACGAP1/CCNB2/AURKB/CENPA/MKI67/CCNA2/KNL1/CDK1/TACC3/TRAIP/ PLK4/E2F2/CENPF/AURKA/KIF23/KIF11/ BRCA2/NEK2/CDC45/NDC80/EXO1/CDC25A/E2F1/CKS1B/CDC6/UBE2S/PRC1/KPNA2/RAD54L/CKS2/CENPE/SMC2/STIL/CCNF/LMNB1/CDKN3/PTTG1/STMN1/EZH2/ORC6/GINS2/CDC7/FBXO5/MCM2/ODC1/NSD2/H2AFX/KIF22/MCM6/INCENP/SMC4/CHEK1/DDX39A/KIF20B/BARD1/DTYMK/CHAF1A/SUV39H1 |
| E2F TARGETS | 105 | 0.889041 | 3.34287 | 0.006028 | MYBL2/CDKN2A/DEPDC1/MELK/CCNE1/ASF1B/TOP2A/KIF2C/TRIP13/BIRC5/ESPL1/BUB1B/MAD2L1/HMMR/KIF4A/PLK1/CDC20/CIT/CDCA8/SPAG5/SPC24/RACGAP1/CCNB2/AURKB/RRM2/TK1/MKI67/KIF18B/DLGAP5/CDK1/TACC3/PLK4/GINS4/AURKA/BRCA2/E2F8/RFC3/DIAPH3/SPC25/CDC25A/CKS1B/TIMELESS/UBE2S/RAD51AP1/KPNA2/CKS2/CENPE/GINS1/LMNB1/CDKN3/PTTG1/STMN1/UBE2T/CENPM/EZH2/ORC6/ATAD2/MCM2/MCM4/NCAPD2/HELLS/RNASEH2A/PCNA/H2AFX/KIF22/MCM6/SMC4/CHEK1/DDX39A/BARD1/DSCC1/GINS3/TCF19/SUV39H1/RFC2/CSE1L/UNG/MSH2/SNRPB/PRIM2/HMGB3/RAN/DCLRE1B/JPT1/NME1/TUBG1/DCK/MTHFD2/DCTPP1/TUBB/PAICS/DEK/PA2G4/DONSON/SLBP | |
| MITOTIC SPINDLE | 80 | 0.798921 | 2.888574 | 0.006028 | KIF15/TPX2/TOP2A/KIF2C/BIRC5/ESPL1/ KIF4A/PLK1/TTK/BUB1/ANLN/NUSAP1/RACGAP1/CCNB2/ECT2/DLGAP5/CDK1/CENPF/AURKA/KIF23/KIF11/BRCA2/NEK2/NDC80/PRC1/PIF1/CENPE/LMNB1/FBXO5/KIF22/INCENP/SMC4/KIF20B/CENPJ/SASS6 | |
| PKD2 | KRAS SIGNALING UP | 118 | 0.656511 | 2.270659 | 0.004293 | MMP11/PRRX1/PLAU/TMEM158/ETV1/CFH/GFPT2/LIF/PLAT/SPARCL1/ADGRA2/ TMEM176A/MMP9/LAPTM5/ITGB2/PCSK1N/TMEM176B/RGS16/EPB41L3/ENG/NRP1/TNFAIP3/IL2RG/APOD/MALL/EPHB2/IKZF1/PLAUR/WNT7A/MAFB/TFPI/AKAP12/TRIB2/KLF4/CXCL10/SPP1/BMP2/C3AR1/SPON1/ ETV5/ADAMDEC1/LCP1/FCER1G/FLT4/GYPC/G0S2/TRAF1/DUSP6/CTSS/ADAM8/SOX9/PPP1R15A/MMD/IRF8 |
| PKD2 | TNFA SIGNALING VIA NFKB | 109 | 0.696773 | 2.395892 | 0.004293 | SERPINE1/PLAU/FOSB/KLF2/ICAM1/GFPT2/LIF/EGR1/SLC2A3/FOS/ZFP36/DUSP1/EGR2/TNFAIP6/NR4A1/GEM/OLR1/CCL5/NR4A3/EGR3/TNFAIP3/LDLR/TNFAIP2/GADD45B/PLAUR/PLEK/NFAT5/CDKN1A/CCL2/KDM6B/KLF4/CXCL1/CXCL10/BMP2/SIK1/IL6ST/ DUSP4/FOSL2/CCL4/CXCL11/IER3/G0S2/ TRAF1/JUNB/F3/CD44/PPP1R15A/SERPINB2/RHOB/NR4A2/KLF9/SGK1/PTGER4/IFIT2/B4GALT5/MAFF/IER5/CXCL6/ETS2/PER1/BCL6/TAP1/TNFRSF9/SMAD3/ID2/PLPP3/IL1B/PTX3/SLC2A6/RNF19B/BIRC3/IFIH1 |
| INTERFERON GAMMA RESPONSE | 107 | 0.628308 | 2.160028 | 0.004293 | C1S/CFH/CXCL9/ICAM1/C1R/XAF1/TNFAIP6/OAS2/IL2RB/LATS2/CCL5/CSF2RB/LCP2/ IFI44L/OAS3/HLA-DQA1/RSAD2/TNFAIP3/HLA-B/TNFAIP2/MX1/HELZ2/SLAMF7/ CDKN1A/CCL2/STAT1/CXCL10/FAS/EPSTI1/IFIT3/CD38/PIM1/TAPBP/CXCL11/SELP/CD74/WARS/ST8SIA4/IRF8/ST3GAL5/IFI44/LY6E/CD86/LGALS3BP/IFIT2/FCGR1A/OASL/EIF2AK2/MYD88/IFI30/CFB/TAP1/IFIT1/CMPK2/B2M/HLA-DRB1/PML/IFIH1/TXNIP/IFI27/HLA-G/ JAK2/TRIM14 | |
| TBRG1 | ALLOGRAFT REJECTION | 120 | 0.489708 | 1.924057 | 0.008729 | IL18/THY1/LIF/CD74/HLA-DOA/HLA-DMA/HLA-DQA1/C2/HLA-DRA/LTB/IL2RG/FAS/ELF4/PRKCB/CD47/PRKCG/B2M/CD3E/LY75/ICAM1/INHBB/TAP1/TAPBP/IL2RB/HDAC9/CD2/IL16/CCL5/GZMA/FYB1/CD96/CD4/JAK2/CXCL9/IL15/STAB1/CD7/CCL4/ITGAL/HLA-DOB/IGSF6/IKBKB/HLA-G/ITGB2/LYN/TNF/IL12A/SPI1/PTPRC/CRTAM/CD8A/PRF1/CCL22/WAS/LCP2/CTSS/CD3D/FASLG/CXCR3 |
| TNFA SIGNALING VIA NFKB | 109 | 0.549517 | 2.128181 | 0.008729 | BIRC3/IL18/CCND1/FOS/FOSB/LIF/CCL20/GADD45B/EGR1/CEBPD/EDN1/JUNB/SGK1/CCNL1/NR4A1/NFAT5/ZFP36/F3/IRF1/KLF2/TNFAIP2/IFIT2/CLCF1/SMAD3/ETS2/DUSP1/ICAM1/TAP1/LAMB3/MAFF/SERPINB2/PLAU/TRIB1/EGR3/BTG2/CCL5/TRAF1/IL6ST/CCL4/BTG3/TRIP10/TNFAIP3/IER3/TIPARP/EGR2/BMP2/TNF | |
| INTERFERON GAMMA RESPONSE | 107 | 0.524759 | 2.03116 | 0.008729 | CFB/XAF1/CD74/HLA-DMA/HLA-DQA1/IFITM3/HLA-DRB1/MX2/RTP4/PSMB8/IFI27/PSMB9/IRF1/IDO1/IFIT3/IFIT1/LY6E/FAS/TNFAIP2/ IFIT2/EPSTI1/B2M/ZBP1/TXNIP/ICAM1/TAP1/TAPBP/IL2RB/PML/TNFSF10/ITGB7/HLA-B/CCL5/CASP8/GZMA/SLC25A28/JAK2/C1R/CXCL9/IL15/NMI/SECTM1/MX1/HLA-G/TNFAIP3/UBE2L6/C1S/PARP12 | |
| DOCK5 | TNFA SIGNALING VIA NFKB | 109 | 0.558475 | 2.319462 | 0.007567 | CD44/CCND1/FOSB/FOS/BIRC3/LAMB3/TNFAIP2/IL18/NFAT5/LDLR/EGR3/KLF2/EGR1/ZFP36/KLF9/BCL6/SIK1/SMAD3/DUSP1/ NR4A1/ETS2/IL6ST/SGK1/BTG2/CEBPD/GADD45B/DUSP4/PER1/KLF4/IRF1/EDN1/TRIP10/ICAM1/NR4A2/F3/TRAF1/SLC2A3/RHOB/FOSL2/IFIT2/STAT5A/CDKN1A/OLR1/KYNU/PLAU/LIF/TNFAIP3/CXCL1/MAFF/EGR2/JUNB/GFPT2/RIPK2/IL1B/RNF19B/F2RL1/ CXCL6/G0S2/PPP1R15A/PLEK/IER5/ICOSLG/TNFAIP8/TRIB1/MAP2K3 |
| INFLAMMATORY RESPONSE | 104 | 0.465585 | 1.916731 | 0.007567 | SLC7A2/CD82/GPR132/STAB1/IL18/LDLR/TNFSF15/TAPBP/P2RX7/CYBB/PTAFR/BTG2/CLEC5A/TPBG/SLC7A1/MET/AHR/RASGRP1/IL2RB/IRF1/SGMS2/EDN1/LYN/ICAM1/ GABBR1/F3/TNFSF10/ITGB8/C3AR1/APLNR/LCP2/CDKN1A/OLR1/AQP9/LIF/RGS16/CCL22/RGS1/SELE/RTP4/RIPK2/IL1B/ITGA5/CXCL6/PCDH7/CD14/CCR7/SLC11A2/ICOSLG | |
| UV RESPONSE DN | 62 | 0.584505 | 2.210784 | 0.007567 | CELF2/MGLL/RUNX1/IRS1/DLC1/RBPMS/LDLR/MT1E/SYNE1/SMAD3/PTPN21/DUSP1/GCNT1/PTPRM/VLDLR/SIPA1L1/CAV1/SLC7A1/MET/FHL2/PDGFRB/RND3/EFEMP1/F3/NRP1/ ANXA2/APBB2/PRDM2/PPARG |
Footnotes
Conflict of interest
None.
Source of support: This work was supported by a grant from the National Natural Science Foundation of China (grant number 81472761 to GL)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Servant N, Roméjon J, Gestraud P, et al. Bioinformatics for precision medicine in oncology: Principles and application to the SHIVA clinical trial. Front Genet. 2014;5:152. doi: 10.3389/fgene.2014.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Z, Cheng Y, Jiang Y, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci. 2018;14:124–36. doi: 10.7150/ijbs.22619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou XG, Huang XL, Liang SY, et al. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. OncoTargets Ther. 2018;11:2815–30. doi: 10.2147/OTT.S163891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li N, Zhan X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J. 2019;10:273–90. doi: 10.1007/s13167-019-00175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Q, Fan C. A novel risk score system for assessment of ovarian cancer based on co-expression network analysis and expression level of five lncRNAs. BMC Med Genet. 2019;20:103. doi: 10.1186/s12881-019-0832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Bi F, An Y, et al. Identification of pathological grade and prognosis-associated lncRNA for ovarian cancer. J Cell Biochem. 2019;120:14444–54. doi: 10.1002/jcb.28704. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Bi F, An Y, et al. Coexpression network analysis identified Kruppel-like factor 6 (KLF6) association with chemosensitivity in ovarian cancer. J Cell Biochem. 2018 doi: 10.1002/jcb.27567. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Li S, Liang J, et al. ITLNI identified by comprehensive bioinformatic analysis as a hub candidate biological target in human epithelial ovarian cancer. Cancer Manag Res. 2019;11:2379–92. doi: 10.2147/CMAR.S189784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mok SC, Bonome T, Vathipadiekal V, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: Microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–32. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King ER, Tung CS, Tsang YT, et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am J Surg Pathol. 2011;35:904–12. doi: 10.1097/PAS.0b013e318212ae22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeung TL, Leung CS, Wong KK, et al. ELF3 is a negative regulator of epithelial-mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:16951–63. doi: 10.18632/oncotarget.15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tone AA, Virtanen C, Shaw PA, et al. Decreased progesterone receptor isoform expression in luteal phase fallopian tube epithelium and high-grade serous carcinoma. Endocr Relat Cancer. 2011;18:221–34. doi: 10.1530/ERC-10-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tung CS, Mok SC, Tsang YT, et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod Pathol. 2009;22:1243–50. doi: 10.1038/modpathol.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carithers LJ, Ardlie K, Barcus M, et al. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015;13:311–19. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mounir M, Lucchetta M, Silva TC, et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol. 2019;15:e1006701. doi: 10.1371/journal.pcbi.1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier L, Cope L, Bolstad BM, et al. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 19.Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–83. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Wang LG, Han Y, et al. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–87. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladhaug IP, Westgaard A, Schjolberg AR, et al. Spindle proteins in resected pancreatic head adenocarcinomas: BubR1 is an independent prognostic factor in pancreatobiliary-type tumours. Histopathology. 2010;56:345–55. doi: 10.1111/j.1365-2559.2010.03489.x. [DOI] [PubMed] [Google Scholar]
- 23.Genga KR, Filho FD, Ferreira FV, et al. Proteins of the mitotic checkpoint and spindle are related to chromosomal instability and unfavourable prognosis in patients with myelodysplastic syndrome. J Clin Pathol. 2015;68:381–87. doi: 10.1136/jclinpath-2014-202728. [DOI] [PubMed] [Google Scholar]
- 24.Choi JW, Kim Y, Lee JH, et al. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch. 2013;463:681–87. doi: 10.1007/s00428-013-1473-6. [DOI] [PubMed] [Google Scholar]
- 25.Nascimento AV, Singh A, Bousbaa H, et al. Mad2 checkpoint gene silencing using epidermal growth factor receptor-targeted chitosan nanoparticles in non-small cell lung cancer model. Mol Pharm. 2014;11:3515–27. doi: 10.1021/mp5002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Xu DB, Zhao XL, et al. Combination analysis of Bub1 and Mad2 expression in endometrial cancer: Act as a prognostic factor in endometrial cancer. Arch Gynecol Obstet. 2013;288:155–65. doi: 10.1007/s00404-012-2706-7. [DOI] [PubMed] [Google Scholar]
- 27.Furlong F, Fitzpatrick P, O’Toole S, et al. Low MAD2 expression levels associate with reduced progression-free survival in patients with high-grade serous epithelial ovarian cancer. J Pathol. 2012;226:746–55. doi: 10.1002/path.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano Y, Sumi T, Teramae M, et al. Expression of the mitotic-arrest deficiency 2 is associated with chemotherapy resistance in ovarian serous adenocarcinoma. Oncol Rep. 2012;28:1200–4. doi: 10.3892/or.2012.1907. [DOI] [PubMed] [Google Scholar]
- 29.Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci U S A. 2004;101:17108–13. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torlakovic EE, Riddell R, Banerjee D, et al. Canadian Association of Pathologists-Association canadienne des pathologistes National Standards Committee/Immunohistochemistry: Best practice recommendations for standardization of immunohistochemistry tests. Am J Clin Pathol. 2010;133:354–65. doi: 10.1309/AJCPDYZ1XMF4HJWK. [DOI] [PubMed] [Google Scholar]
- 31.Balacescu O, Balacescu L, Tudoran O, et al. Gene expression profiling reveals activation of the FA/BRCA pathway in advanced squamous cervical cancer with intrinsic resistance and therapy failure. BMC Cancer. 2014;14:246. doi: 10.1186/1471-2407-14-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Groep P, Hoelzel M, Buerger H, et al. Loss of expression of FANCD2 protein in sporadic and hereditary breast cancer. Breast Cancer Res Treat. 2008;107:41–47. doi: 10.1007/s10549-007-9534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozawa H, Iwatsuki M, Mimori K, et al. FANCD2 mRNA overexpression is a bona fide indicator of lymph node metastasis in human colorectal cancer. Ann Surg Oncol. 2010;17:2341–48. doi: 10.1245/s10434-010-1002-7. [DOI] [PubMed] [Google Scholar]
- 34.Han SS, Tompkins VS, Son DJ, et al. CDKN1A and FANCD2 are potential oncotargets in Burkitt lymphoma and multiple myeloma. Exp Hematol Oncol. 2015;4:9. doi: 10.1186/s40164-015-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu H, Masuda T, Iguchi T, et al. Clinical significance of FANCD2 gene expression and its association with tumor progression in hepatocellular carcinoma. Anticancer Res. 2017;37:1083–90. doi: 10.21873/anticanres.11420. [DOI] [PubMed] [Google Scholar]
- 36.Moes-Sosnowska J, Rzepecka IK, Chodzynska J, et al. Clinical importance of FANCD2, BRIP1, BRCA1, BRCA2 and FANCF expression in ovarian carcinomas. Cancer Biol Ther. 2019;20:843–54. doi: 10.1080/15384047.2019.1579955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kais Z, Rondinelli B, Holmes A, et al. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15:2488–99. doi: 10.1016/j.celrep.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Characteristics of the included datasets.
Samples clustering of 5 datasets after removing the batch effects.
Soft-thresholding power determination in WGCNA. (A) Analysis of the scale-free fit index for different soft-thresholding powers. (B) Mean connectivity for various soft-thresholding powers. (C) Histogram of connectivity distribution when β=9. (D) Check scale-free topology when β=9.
Heatmap of the top 200 DEGs based on the value of |logFC|. High or low expression is shown as a red or blue strip, respectively. The experimental group was labelled HGSOC, while the control group was named Nor.
Supplementary Table 2.
The KEGG enrichment analysis of genes.
| ID | Description | p. adjust | Count | Regulation |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 0.000633 | 35 | up |
| hsa03030 | DNA replication | 0.012093 | 14 | up |
| hsa01230 | Biosynthesis of amino acids | 0.028478 | 12 | up |
| hsa04014 | Ras signaling pathway | 0.007503 | 25 | down |
| hsa04610 | Complement and coagulation cascades | 0.007503 | 14 | down |
| hsa04010 | MAPK signaling pathway | 0.007503 | 29 | down |
| hsa04630 | JAK-STAT signaling pathway | 0.007503 | 13 | down |
| hsa04728 | Dopaminergic synapse | 0.007503 | 13 | down |
| hsa05032 | Morphine addiction | 0.007503 | 13 | down |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 0.013251 | 12 | down |
| hsa05146 | Amoebiasis | 0.03213 | 14 | down |
Supplementary Table 3.
The GO enrichment analysis of genes.
| Ontology | ID | Description | p. adjust | Count | Regulation |
|---|---|---|---|---|---|
| BP | GO: 0007059 | Chromosome segregation | 7.51E-30 | 76 | up |
| BP | GO: 0140014 | Mitotic nuclear division | 8.88E-28 | 69 | up |
| BP | GO: 0000280 | Nuclear division | 1.11E-26 | 83 | up |
| BP | GO: 0048285 | Organelle fission | 3.71E-25 | 85 | up |
| BP | GO: 0098813 | Nuclear chromosome segregation | 5.13E-25 | 63 | up |
| CC | GO: 0098687 | Chromosomal region | 1.26E-26 | 74 | up |
| CC | GO: 0000775 | Chromosome, centromeric region | 2.73E-24 | 53 | up |
| CC | GO: 0000793 | Condensed chromosome | 1.10E-20 | 52 | up |
| CC | GO: 0000779 | Condensed chromosome, centromeric region | 2.30E-19 | 37 | up |
| CC | GO: 0005819 | Spindle | 2.30E-19 | 62 | up |
| MF | GO: 0140097 | Catalytic activity, acting on DNA | 2.32E-08 | 34 | up |
| MF | GO: 0008094 | DNA-dependent ATPase activity | 1.57E-05 | 19 | up |
| MF | GO: 0016887 | ATPase activity | 0.000226 | 47 | up |
| MF | GO: 0043142 | Single-stranded DNA-dependent ATPase activity | 0.000325 | 7 | up |
| MF | GO: 0001077 | Proximal promoter DNA-binding transcription activator activity, RNA polymerase II-specific | 0.000325 | 35 | up |
| BP | GO: 0002009 | Morphogenesis of an epithelium | 9.98E-07 | 53 | down |
| BP | GO: 0016049 | Cell growth | 2.29E-06 | 52 | down |
| BP | GO: 0001822 | Kidney development | 6.68E-06 | 35 | down |
| BP | GO: 0072001 | Renal system development | 6.68E-06 | 36 | down |
| BP | GO: 0003002 | Regionalization | 6.68E-06 | 40 | down |
| CC | GO: 0062023 | Collagen-containing extracellular matrix | 1.22E-11 | 49 | down |
| CC | GO: 0031012 | Extracellular matrix | 1.29E-10 | 59 | down |
| CC | GO: 0042383 | Sarcolemma | 3.36E-05 | 21 | down |
| CC | GO: 0005604 | Basement membrane | 3.64E-05 | 17 | down |
| CC | GO: 0031252 | Cell leading edge | 0.006422 | 34 | down |
| MF | GO: 0005201 | Extracellular matrix structural constituent | 4.99E-05 | 24 | down |
| MF | GO: 0005539 | Glycosaminoglycan binding | 5.13E-05 | 29 | down |
| MF | GO: 1901681 | Sulfur compound binding | 6.67E-05 | 30 | down |
| MF | GO: 0008201 | Heparin binding | 0.000129 | 23 | down |
| MF | GO: 0005518 | Collagen binding | 0.000836 | 13 | down |
Supplementary Table 4.
Hub genes in blue and turquoise module (|MM|>0.85 and |GS|>0.3).
| Blue module | Turquoise module | ||||
|---|---|---|---|---|---|
| Gene | MM | GS | Gene | MM | GS |
| CSE1L | 0.885182 | 0.654936 | LRRN4 | 0.863969 | −0.76695 |
| PCNA | 0.853268 | 0.634531 | LINC01105 | 0.85413 | −0.83749 |
| HNRNPAB | 0.859002 | 0.665006 | DAB2 | 0.928442 | −0.84279 |
| TOP2A | 0.891679 | 0.822887 | CELF2 | 0.888927 | −0.77641 |
| SMC4 | 0.917093 | 0.806692 | LAMA4 | 0.850672 | −0.68463 |
| MTHFD2 | 0.877458 | 0.806098 | SPOCK1 | 0.867854 | −0.80181 |
| PSRC1 | 0.871528 | 0.788497 | PAPSS2 | 0.879786 | −0.72486 |
| CKS1B | 0.934063 | 0.787083 | DAPK1 | 0.899065 | −0.7982 |
| MCM2 | 0.922548 | 0.841694 | PROCR | 0.898493 | −0.7677 |
| PCLAF | 0.899258 | 0.817579 | PKD2 | 0.929768 | −0.75857 |
| CRABP2 | 0.87272 | 0.877783 | GSDME | 0.91672 | −0.80166 |
| CCNB2 | 0.897672 | 0.848503 | IGFBP6 | 0.892364 | −0.79082 |
| LSM4 | 0.871655 | 0.664114 | THBD | 0.869457 | −0.80323 |
| CDC20 | 0.893728 | 0.758763 | KDR | 0.884189 | −0.74272 |
| UBE2C | 0.876499 | 0.834623 | FRY | 0.899738 | −0.80017 |
| CDK1 | 0.876549 | 0.8119 | GNG11 | 0.930805 | −0.75466 |
| EZH2 | 0.862178 | 0.795034 | ABCA8 | 0.944703 | −0.89654 |
| MAD2L1 | 0.889283 | 0.76929 | GPRASP1 | 0.899009 | −0.87775 |
| PTTG1 | 0.870574 | 0.824798 | GFPT2 | 0.866683 | −0.7817 |
| BUB1B | 0.892962 | 0.819369 | RNASE4 | 0.903103 | −0.80729 |
| DLGAP5 | 0.877017 | 0.82571 | CALB2 | 0.897513 | −0.85688 |
| ZWINT | 0.922712 | 0.79987 | BCHE | 0.922752 | −0.81917 |
| TRIP13 | 0.884367 | 0.77344 | NPY1R | 0.882911 | −0.85536 |
| RAD51AP1 | 0.858395 | 0.802612 | GHR | 0.888245 | −0.71151 |
| NDC80 | 0.86773 | 0.721595 | ECM2 | 0.850415 | −0.66158 |
| CKS2 | 0.94052 | 0.829763 | ARHGAP6 | 0.869659 | −0.73929 |
| KIF11 | 0.885283 | 0.822206 | WNT2B | 0.877108 | −0.80996 |
| NEK2 | 0.89035 | 0.839637 | PTGIS | 0.883089 | −0.79366 |
| KIF23 | 0.853223 | 0.7629 | LGALS2 | 0.85777 | −0.75854 |
| FEN1 | 0.909533 | 0.760884 | MAF | 0.898868 | −0.73781 |
| TTK | 0.894859 | 0.855031 | SYNE1 | 0.854181 | −0.8207 |
| MELK | 0.900554 | 0.813158 | PLPP1 | 0.920489 | −0.7545 |
| STIL | 0.867957 | 0.835405 | TCEAL2 | 0.877183 | −0.81491 |
| SAC3D1 | 0.874805 | 0.712282 | TBC1D2B | 0.862424 | −0.70643 |
| HMGA1 | 0.869641 | 0.806441 | PDE8B | 0.878248 | −0.91687 |
| GINS1 | 0.893705 | 0.7861 | ATP10D | 0.88824 | −0.74775 |
| CENPF | 0.880011 | 0.84842 | TFPI | 0.86169 | −0.75802 |
| AURKA | 0.914224 | 0.801243 | CHN2 | 0.855714 | −0.81706 |
| EIF4G1 | 0.869979 | 0.737708 | BICC1 | 0.864204 | −0.82881 |
| NR2F6 | 0.887103 | 0.752212 | DIXDC1 | 0.858203 | −0.8161 |
| BUB1 | 0.913764 | 0.809106 | DIRAS3 | 0.875531 | −0.8524 |
| PUF60 | 0.866461 | 0.739985 | OLFML1 | 0.886786 | −0.70603 |
| TPX2 | 0.862716 | 0.819783 | CSGALNACT1 | 0.90082 | −0.83324 |
| RPL39L | 0.857416 | 0.742065 | PDGFD | 0.866505 | −0.68393 |
| EIF6 | 0.86057 | 0.671087 | RADX | 0.868631 | −0.84742 |
| XPOT | 0.857586 | 0.684634 | KLF2 | 0.894365 | −0.78132 |
| SCRIB | 0.867393 | 0.771012 | SMPD3 | 0.867973 | −0.78767 |
| CCNA2 | 0.868501 | 0.7858 | PPP1R3B | 0.85511 | −0.72434 |
| CCNB1 | 0.904717 | 0.755995 | OGN | 0.88218 | −0.80725 |
| PRC1 | 0.886914 | 0.802267 | ABI3BP | 0.872052 | −0.78816 |
| MRPL15 | 0.853847 | 0.663766 | ITLN1 | 0.884383 | −0.81818 |
| NUSAP1 | 0.867073 | 0.795277 | MGARP | 0.903375 | −0.8306 |
| SLC52A2 | 0.868392 | 0.803613 | ARHGAP18 | 0.918993 | −0.80114 |
| TACC3 | 0.875177 | 0.752928 | DDR2 | 0.852443 | −0.67497 |
| KIF4A | 0.871167 | 0.797892 | ANTXR2 | 0.858928 | −0.72284 |
| CEP55 | 0.850744 | 0.827425 | LIX1L | 0.878142 | −0.68937 |
| DTL | 0.865255 | 0.780689 | MCC | 0.870811 | −0.71797 |
| KIF20A | 0.888407 | 0.82511 | TBRG1 | 0.85824 | −0.79791 |
| CENPU | 0.863996 | 0.732426 | PTPN21 | 0.863405 | −0.7201 |
| KIF15 | 0.882733 | 0.786989 | CNRIP1 | 0.889837 | −0.80871 |
| ECT2 | 0.916732 | 0.831958 | PPM1K | 0.895422 | −0.86713 |
| CDCA8 | 0.85916 | 0.797452 | MEDAG | 0.866729 | −0.84797 |
| MCM4 | 0.914084 | 0.813176 | LINC01279 | 0.857322 | −0.66728 |
| RACGAP1 | 0.918941 | 0.779139 | PLEKHH2 | 0.865982 | −0.76522 |
| PSAT1 | 0.854015 | 0.826163 | SLC30A4 | 0.904509 | −0.70542 |
| UBE2T | 0.850996 | 0.725215 | TCEAL3 | 0.856911 | −0.7193 |
| SLC25A33 | 0.868987 | 0.6974 | CDON | 0.869831 | −0.69303 |
| CDCA3 | 0.857304 | 0.797015 | TCEAL7 | 0.870405 | −0.84944 |
| NUF2 | 0.91985 | 0.868254 | ERN1 | 0.880253 | −0.77167 |
| RCC2 | 0.909521 | 0.783731 | MUM1L1 | 0.899449 | −0.85402 |
| FAM83D | 0.901393 | 0.849648 | RNASEL | 0.872061 | −0.72795 |
| POC1A | 0.856044 | 0.816551 | DOCK5 | 0.893588 | −0.83622 |
| DEPDC1B | 0.854264 | 0.764695 | RBMS3 | 0.850965 | −0.82463 |
| CENPL | 0.857368 | 0.768059 | HAND2-AS1 | 0.858159 | −0.89488 |
| KIF14 | 0.902918 | 0.830047 | DTWD1 | 0.879321 | −0.75556 |
| FANCD2 | 0.890071 | 0.736399 | IFFO1 | 0.857298 | −0.73892 |
MM – module membership; GS – gene significance.
Supplementary Table 5.
The Gene Set Enrichment Analysis (GSEA) of hub genes.
| Description | setSize | enrichmentScore | NES | p.adjust | core_enrichment | |
|---|---|---|---|---|---|---|
| MAD2L1 | ALLOGRAFT REJECTION | 120 | 0.438738 | 1.761702 | 0.007716 | CDKN2A/NME1/GZMB/MMP9/CXCL9/CCL5/CXCL13/IL15/CCL11/EIF5A/TAP1/CCL13/GZMA/SRGN/IL2RG/CCL2/UBE2N/CCL7/HLA-DOB/CTSS/CCL4/B2M/CD3D/PRF1/CD2/LTB/TNF/SIT1/IL2RA/CD7/HLA-G/CD8A/CD3E/ST8SIA4/CD86/FCGR2B/IFNG/IL12A/CXCR3/LY86/CD8B/RIPK2/UBE2D1/TPD52/HLA-DQA1/MRPL3/CD80/WARS/CD79A/CCR1/LCK/HDAC9/IGSF6/BCL10/TRAT1/CAPG/CD3G/CD96/IL11/IL2RB/MAP4K1/KRT1 |
| E2F TARGETS | 105 | 0.805846 | 3.155588 | 0.007716 | MAD2L1/CDKN2A/BIRC5/CKS2/CKS1B/CCNE1/TK1/UBE2S/PTTG1/UBE2T/MYBL2/NME1/CCNB2/AURKB/PLK1/DEPDC1/KPNA2/CDC20/RRM2/CENPM/CDKN3/CDK1/PLK4/AURKA/PCNA/SNRPB/KIF2C/SPC25/TRIP13/JPT1/ASF1B/ORC6/H2AFX/TOP2A/MELK/RNASEH2A/TACC3/CDCA8/DLGAP5/KIF4A/DCTPP1/SPC24/RFC3/CENPE/HMMR/RAD51AP1/DIAPH3/STMN1/POP7/BUB1B/DCK/MTHFD2/RPA3/GINS1/SPAG5/RACGAP1/KIF22/GINS4/ DDX39A/DSCC1/CDC25A/KIF18B/RAN/E2F8/RFC2/TUBG1/SLBP/BRCA2/HMGB3/SUV39H1/CHEK1/PRIM2/GINS3/ESPL1/SMC4/MXD3 | |
| G2M CHECKPOINT | 104 | 0.759634 | 2.975861 | 0.007716 | MAD2L1/CCNA2/UBE2C/BIRC5/CKS2/CKS1B/UBE2S/PTTG1/MYBL2/PBK/CCNB2/AURKB/PLK1/KPNA2/CDC20/CENPA/CDKN3/TTK/CDK1/PLK4/NEK2/AURKA/GINS2/KIF2C/JPT1/ORC6/H2AFX/CDC45/TOP2A/TROAP/TACC3/CDC6/SNRPD1/TPX2/KIF4A/NUSAP1/CENPE/HMMR/NDC80/STMN1/BUB1/EXO1/DTYMK/KIF23/TRAIP/PRC1/RACGAP1/KIF22/E2F1/DDX39A/CDC25A/POLQ/KIF15/FBXO5/RAD54L/KNL1/KIF11/BRCA2/HMGB3/E2F2/SUV39H1/CHEK1/CENPF/PRIM2/ESPL1/SMC4/ODC1/CCNF/STIL/SMC2/CDC7/MCM6/HIST1H2BK/EZH2/MCM2 | |
| FANCD2 | G2M CHECKPOINT | 104 | 0.901015 | 3.369604 | 0.006028 | MYBL2/KIF15/TPX2/TOP2A/KIF2C/UBE2C/BIRC5/ESPL1/MAD2L1/HMMR/PBK/KIF4A/PLK1/TROAP/TTK/BUB1/CDC20/POLQ/ NUSAP1/RACGAP1/CCNB2/AURKB/CENPA/MKI67/CCNA2/KNL1/CDK1/TACC3/TRAIP/ PLK4/E2F2/CENPF/AURKA/KIF23/KIF11/ BRCA2/NEK2/CDC45/NDC80/EXO1/CDC25A/E2F1/CKS1B/CDC6/UBE2S/PRC1/KPNA2/RAD54L/CKS2/CENPE/SMC2/STIL/CCNF/LMNB1/CDKN3/PTTG1/STMN1/EZH2/ORC6/GINS2/CDC7/FBXO5/MCM2/ODC1/NSD2/H2AFX/KIF22/MCM6/INCENP/SMC4/CHEK1/DDX39A/KIF20B/BARD1/DTYMK/CHAF1A/SUV39H1 |
| E2F TARGETS | 105 | 0.889041 | 3.34287 | 0.006028 | MYBL2/CDKN2A/DEPDC1/MELK/CCNE1/ASF1B/TOP2A/KIF2C/TRIP13/BIRC5/ESPL1/BUB1B/MAD2L1/HMMR/KIF4A/PLK1/CDC20/CIT/CDCA8/SPAG5/SPC24/RACGAP1/CCNB2/AURKB/RRM2/TK1/MKI67/KIF18B/DLGAP5/CDK1/TACC3/PLK4/GINS4/AURKA/BRCA2/E2F8/RFC3/DIAPH3/SPC25/CDC25A/CKS1B/TIMELESS/UBE2S/RAD51AP1/KPNA2/CKS2/CENPE/GINS1/LMNB1/CDKN3/PTTG1/STMN1/UBE2T/CENPM/EZH2/ORC6/ATAD2/MCM2/MCM4/NCAPD2/HELLS/RNASEH2A/PCNA/H2AFX/KIF22/MCM6/SMC4/CHEK1/DDX39A/BARD1/DSCC1/GINS3/TCF19/SUV39H1/RFC2/CSE1L/UNG/MSH2/SNRPB/PRIM2/HMGB3/RAN/DCLRE1B/JPT1/NME1/TUBG1/DCK/MTHFD2/DCTPP1/TUBB/PAICS/DEK/PA2G4/DONSON/SLBP | |
| MITOTIC SPINDLE | 80 | 0.798921 | 2.888574 | 0.006028 | KIF15/TPX2/TOP2A/KIF2C/BIRC5/ESPL1/ KIF4A/PLK1/TTK/BUB1/ANLN/NUSAP1/RACGAP1/CCNB2/ECT2/DLGAP5/CDK1/CENPF/AURKA/KIF23/KIF11/BRCA2/NEK2/NDC80/PRC1/PIF1/CENPE/LMNB1/FBXO5/KIF22/INCENP/SMC4/KIF20B/CENPJ/SASS6 | |
| PKD2 | KRAS SIGNALING UP | 118 | 0.656511 | 2.270659 | 0.004293 | MMP11/PRRX1/PLAU/TMEM158/ETV1/CFH/GFPT2/LIF/PLAT/SPARCL1/ADGRA2/ TMEM176A/MMP9/LAPTM5/ITGB2/PCSK1N/TMEM176B/RGS16/EPB41L3/ENG/NRP1/TNFAIP3/IL2RG/APOD/MALL/EPHB2/IKZF1/PLAUR/WNT7A/MAFB/TFPI/AKAP12/TRIB2/KLF4/CXCL10/SPP1/BMP2/C3AR1/SPON1/ ETV5/ADAMDEC1/LCP1/FCER1G/FLT4/GYPC/G0S2/TRAF1/DUSP6/CTSS/ADAM8/SOX9/PPP1R15A/MMD/IRF8 |
| PKD2 | TNFA SIGNALING VIA NFKB | 109 | 0.696773 | 2.395892 | 0.004293 | SERPINE1/PLAU/FOSB/KLF2/ICAM1/GFPT2/LIF/EGR1/SLC2A3/FOS/ZFP36/DUSP1/EGR2/TNFAIP6/NR4A1/GEM/OLR1/CCL5/NR4A3/EGR3/TNFAIP3/LDLR/TNFAIP2/GADD45B/PLAUR/PLEK/NFAT5/CDKN1A/CCL2/KDM6B/KLF4/CXCL1/CXCL10/BMP2/SIK1/IL6ST/ DUSP4/FOSL2/CCL4/CXCL11/IER3/G0S2/ TRAF1/JUNB/F3/CD44/PPP1R15A/SERPINB2/RHOB/NR4A2/KLF9/SGK1/PTGER4/IFIT2/B4GALT5/MAFF/IER5/CXCL6/ETS2/PER1/BCL6/TAP1/TNFRSF9/SMAD3/ID2/PLPP3/IL1B/PTX3/SLC2A6/RNF19B/BIRC3/IFIH1 |
| INTERFERON GAMMA RESPONSE | 107 | 0.628308 | 2.160028 | 0.004293 | C1S/CFH/CXCL9/ICAM1/C1R/XAF1/TNFAIP6/OAS2/IL2RB/LATS2/CCL5/CSF2RB/LCP2/ IFI44L/OAS3/HLA-DQA1/RSAD2/TNFAIP3/HLA-B/TNFAIP2/MX1/HELZ2/SLAMF7/ CDKN1A/CCL2/STAT1/CXCL10/FAS/EPSTI1/IFIT3/CD38/PIM1/TAPBP/CXCL11/SELP/CD74/WARS/ST8SIA4/IRF8/ST3GAL5/IFI44/LY6E/CD86/LGALS3BP/IFIT2/FCGR1A/OASL/EIF2AK2/MYD88/IFI30/CFB/TAP1/IFIT1/CMPK2/B2M/HLA-DRB1/PML/IFIH1/TXNIP/IFI27/HLA-G/ JAK2/TRIM14 | |
| TBRG1 | ALLOGRAFT REJECTION | 120 | 0.489708 | 1.924057 | 0.008729 | IL18/THY1/LIF/CD74/HLA-DOA/HLA-DMA/HLA-DQA1/C2/HLA-DRA/LTB/IL2RG/FAS/ELF4/PRKCB/CD47/PRKCG/B2M/CD3E/LY75/ICAM1/INHBB/TAP1/TAPBP/IL2RB/HDAC9/CD2/IL16/CCL5/GZMA/FYB1/CD96/CD4/JAK2/CXCL9/IL15/STAB1/CD7/CCL4/ITGAL/HLA-DOB/IGSF6/IKBKB/HLA-G/ITGB2/LYN/TNF/IL12A/SPI1/PTPRC/CRTAM/CD8A/PRF1/CCL22/WAS/LCP2/CTSS/CD3D/FASLG/CXCR3 |
| TNFA SIGNALING VIA NFKB | 109 | 0.549517 | 2.128181 | 0.008729 | BIRC3/IL18/CCND1/FOS/FOSB/LIF/CCL20/GADD45B/EGR1/CEBPD/EDN1/JUNB/SGK1/CCNL1/NR4A1/NFAT5/ZFP36/F3/IRF1/KLF2/TNFAIP2/IFIT2/CLCF1/SMAD3/ETS2/DUSP1/ICAM1/TAP1/LAMB3/MAFF/SERPINB2/PLAU/TRIB1/EGR3/BTG2/CCL5/TRAF1/IL6ST/CCL4/BTG3/TRIP10/TNFAIP3/IER3/TIPARP/EGR2/BMP2/TNF | |
| INTERFERON GAMMA RESPONSE | 107 | 0.524759 | 2.03116 | 0.008729 | CFB/XAF1/CD74/HLA-DMA/HLA-DQA1/IFITM3/HLA-DRB1/MX2/RTP4/PSMB8/IFI27/PSMB9/IRF1/IDO1/IFIT3/IFIT1/LY6E/FAS/TNFAIP2/ IFIT2/EPSTI1/B2M/ZBP1/TXNIP/ICAM1/TAP1/TAPBP/IL2RB/PML/TNFSF10/ITGB7/HLA-B/CCL5/CASP8/GZMA/SLC25A28/JAK2/C1R/CXCL9/IL15/NMI/SECTM1/MX1/HLA-G/TNFAIP3/UBE2L6/C1S/PARP12 | |
| DOCK5 | TNFA SIGNALING VIA NFKB | 109 | 0.558475 | 2.319462 | 0.007567 | CD44/CCND1/FOSB/FOS/BIRC3/LAMB3/TNFAIP2/IL18/NFAT5/LDLR/EGR3/KLF2/EGR1/ZFP36/KLF9/BCL6/SIK1/SMAD3/DUSP1/ NR4A1/ETS2/IL6ST/SGK1/BTG2/CEBPD/GADD45B/DUSP4/PER1/KLF4/IRF1/EDN1/TRIP10/ICAM1/NR4A2/F3/TRAF1/SLC2A3/RHOB/FOSL2/IFIT2/STAT5A/CDKN1A/OLR1/KYNU/PLAU/LIF/TNFAIP3/CXCL1/MAFF/EGR2/JUNB/GFPT2/RIPK2/IL1B/RNF19B/F2RL1/ CXCL6/G0S2/PPP1R15A/PLEK/IER5/ICOSLG/TNFAIP8/TRIB1/MAP2K3 |
| INFLAMMATORY RESPONSE | 104 | 0.465585 | 1.916731 | 0.007567 | SLC7A2/CD82/GPR132/STAB1/IL18/LDLR/TNFSF15/TAPBP/P2RX7/CYBB/PTAFR/BTG2/CLEC5A/TPBG/SLC7A1/MET/AHR/RASGRP1/IL2RB/IRF1/SGMS2/EDN1/LYN/ICAM1/ GABBR1/F3/TNFSF10/ITGB8/C3AR1/APLNR/LCP2/CDKN1A/OLR1/AQP9/LIF/RGS16/CCL22/RGS1/SELE/RTP4/RIPK2/IL1B/ITGA5/CXCL6/PCDH7/CD14/CCR7/SLC11A2/ICOSLG | |
| UV RESPONSE DN | 62 | 0.584505 | 2.210784 | 0.007567 | CELF2/MGLL/RUNX1/IRS1/DLC1/RBPMS/LDLR/MT1E/SYNE1/SMAD3/PTPN21/DUSP1/GCNT1/PTPRM/VLDLR/SIPA1L1/CAV1/SLC7A1/MET/FHL2/PDGFRB/RND3/EFEMP1/F3/NRP1/ ANXA2/APBB2/PRDM2/PPARG |