Abstract
PURPOSE
The essence of guideline recommendations often is intertwined in large texts. This impedes clinical implementation and evaluation and delays timely modular revisions needed to deal with an ever-growing amount of knowledge and application of personalized medicine. The aim of this project was to model guideline recommendations as data-driven clinical decision trees (CDTs) that are clinically interpretable and suitable for implementation in decision support systems.
METHODS
All recommendations of the Dutch national breast cancer guideline for nonmetastatic breast cancer were translated into CDTs. CDTs were constructed by nodes, branches, and leaves that represent data items (patient and tumor characteristics [eg, T stage]), data item values (eg, T2 or less), and recommendations (eg, chemotherapy), respectively. For all data items, source of origin was identified (eg, pathology), and where applicable, data item values were defined on the basis of existing classification and coding systems (eg, TNM, Breast Imaging Reporting and Data System, Systematized Nomenclature of Medicine). All unique routes through all CDTs were counted to measure the degree of data-based personalization of recommendations.
RESULTS
In total, 60 CDTs were necessary to cover the whole guideline and were driven by 114 data items. Data items originated from pathology (49%), radiology (27%), clinical (12%), and multidisciplinary team (12%) reports. Of all data items, 101 (89%) could be classified by existing classification and coding systems. All 60 CDTs could be integrated in an interactive decision support app that contained 376 unique patient subpopulations.
CONCLUSION
By defining data items unambiguously and unequivocally and coding them to an international coding system, it was possible to present a complex guideline as systematically constructed modular data-driven CDTs that are clinically interpretable and accessible in a decision support app.
INTRODUCTION
The National Academy of Medicine defines clinical practice guidelines as “statements that include recommendations, intended to optimize patient care, that are informed by a systematic review of evidence and an assessment of the benefits and harms of alternative care options.”1(p4) It has been shown that their implementation reduces unwanted variability in clinical practice and improves outcome, therefore improving the quality of care.2
Quick and continuous revision and subsequent implementation and evaluation of guidelines in clinical practice are essential but challenging for several reasons.3 First, guideline development is time consuming and modular revision (meant to accelerate the revision process) cumbersome because modules often are intertwined in the entire guideline text. Second, as cancer treatment is getting more personalized and based on (biomarker) data, guideline recommendations need to be defined for ever-smaller and more-specific patient populations, which makes them more complex. Finally, routine, explicit guideline utilization for each patient is cumbersome because of ambiguity in guideline texts attributable to the use of equivocal terms.
Methods for transforming guidelines into computer-interpretable formats are well studied and have been successfully used for relatively simple guidelines, such as medication alerts to warn for potential contraindications.4-6 However, these methods often are aimed at how to describe guidelines in formal computer languages and not as much at the actual translation of free-text guidelines into such formats. Moreover, such descriptions are difficult to grasp and interpret by physicians involved in guideline committees. Therefore, the application of such an approach to complex multidisciplinary (oncology) guidelines has remained challenging. Occasionally, guidelines, such as those of the National Comprehensive Cancer Network (NCCN), are presented in widely used, compact flowcharts. However, these flowcharts are not fully data modulated, and a strict relationship between all possible (combinations of) patient/disease characteristics and guideline recommendations is not always present.
CONTEXT SUMMARY
Key Objective
To develop a scalable method for representing textual guideline recommendations as systematically designed, modular, data-driven clinical decision trees (CDTs) and to apply this method on a complex guideline. We tested this method using the Dutch national breast cancer guideline.
Knowledge Generated
The rules that comprise CDTs can be systematically derived from guideline recommendations. At each point in the care path, the CDT describes the most appropriate new interventions on the basis of the accumulating patient data available up to that point. We demonstrate the feasibility of applying the CDT method on a complex guideline. Data items that comprise the CDT were defined unequivocally and unambiguously on the basis of international classification and coding systems. In this way, interoperability with electronic health records and implementation of CDTs in decision support systems can be facilitated.
Relevance
The modular character of CDTs could provide a means for quick and clear implementation and accessibility of dynamic guidelines. Moreover, fast-growing knowledge could be taken into account more rapidly and easily by modular updating of CDTs, which supports implementation of data-driven personalized health care.
To anticipate these challenges and complement the NCCN flowcharts, we hypothesized that the transformation of guideline text into data-driven clinical decision trees (CDTs) can facilitate the continuous cycle of guideline development, implementation, evaluation, revision, and maintenance.7 We therefore set up a project that transformed the Dutch national breast cancer guideline into CDTs.8 Our aim was to model systematically all national breast cancer guideline recommendations for nonmetastatic breast cancer as data-driven CDTs on the basis of existing classification systems. When we succeed, we will try to develop an app that clinicians can use directly in daily practice, that complies with prerequisites for integration into the electronic health record (EHR), and that facilitates continuous learning from real-time data.
METHODS
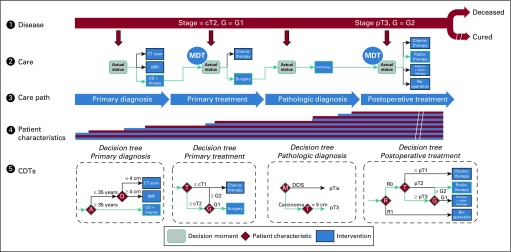
To represent text-based guideline recommendations in CDTs, we used a repetitive data collection approach for each step in a nominal patient-centric care pathway (Fig 1). Our method is based on a generic model for patient disease state that regresses, remains stable, or progresses either spontaneously or as a result of care interventions. During care, data that describe the disease state accumulate. The care pathway is decomposed into interventions for measuring the disease state (eg, diagnoses) that result in new data, which add to prior knowledge, and into interventions that influence the disease state. At each point, the CDTs describe the most appropriate new intervention on the basis of data available up to that point. The rules that comprise the CDT are derived from guideline recommendations. All CDTs can be used independently from one another (eg, a CDT for postoperative treatment can be used independently from a CDT for preoperative treatment and its outcome). By connecting CDTs head to tail, the actual care pathway can be reconstructed. The model is scalable across diseases because at no point are assumptions made on care process or type of data.
FIG 1.
Conceptual and simplified reflection of the breast cancer care pathway and related clinical decision trees (CDTs). See the Methods section for a detailed description. A, age; CT, computed tomography; D, tumor diameter; DCIS, ductal carcinoma in situ; G, tumor grade; M, tumor morphology; MDT, multidisciplinary team; MRI, magnetic resonance imaging; R, residual tumor; T, tumor stage; US, ultrasound.
Guideline text recommendations were mapped according to this approach and subsequently for each step modeled into data-driven CDTs. The method was applied to the 2012 version of the Dutch national breast cancer guideline. CDTs were developed together with researchers of the Netherlands Comprehensive Cancer Organization assisted by a multidisciplinary panel of breast cancer specialists (including surgeons, medical and radiation oncologists, radiologists, and pathologists) and supervised by the members of the Dutch national guideline working group to ensure accuracy and clinical interpretability.
CDTs
CDTs consist of nodes, branches, and leaves. For modeling and visualization of a CDT, the following concepts were used: The trunk of the tree represents the step in the care pathway to which the recommendation applies (eg, postoperative treatment tree). The nodes represent patient or tumor characteristics (eg, T stage) formulated as data items (eg, T2 or less). Data items are derived from medical history, physical examination, or diagnostic tests and can be independent (eg, tumor diameter) or dependent (eg, a data item that is classified as a category on the basis of another data item, such as T stage derived from tumor diameter). The branches represent cutoff points. The leaves represent patient-specific recommendations (eg, treatment recommendations, advice to perform a diagnostic test). It is optional to present the level of evidence that underlies a specific recommendation.
In each CDT, the patient or population, intervention, comparison, and outcome (PICO) system is represented. Leaves represent recommended interventions, and the collection of nodes and cutoff points that lead to a leaf represent the patient or population to whom the recommendation applies. Information such as background literature, studies that compare outcomes of various diagnostic or treatment strategies, and level of evidence is provided as meta-information that underlies the leaves (recommendations).9 This PICO strategy is supported by the Grading of Recommendations Assessment, Development and Evaluation (GRADE) system to judge and grade the quality of scientific publications that indicate levels of evidence.10,11 CDTs were constructed manually by systematically applying this approach to translate each guideline recommendation. All CDTs were checked and formally approved by the Dutch national guideline working group.
Implementation in Decision Support Systems and Interoperability
To establish the source of data that drives guideline recommendations, we identified the source record (eg, pathology or radiology report) for each data item. Data items analyzed included source of data origin (eg, pathology, radiology) and relation to classification systems (eg, TNM, Breast Imaging Reporting and Data System [BIRADS]).
To quantify linguistic unity (or lack thereof), we kept track of the number of different terms used in the free-text guidelines for each data item. For the purpose of linguistic unity, data items in nodes and interventions in leaves were described as much as possible using the most accepted, to our knowledge, international classification and coding systems (eg, TNM; BIRADS; International Classification of Diseases for Oncology, Third Edition; Systematized Nomenclature of Medicine Clinical Terms [SNOMED CT (SNOMED International 2018 version 1.37.3)]). Prerequisites for implementing CDTs in decision support systems and interoperability with EHRs are the systematic construction of CDTs as described herein and the unequivocal and unambiguous definition of data items on the basis of internationally acknowledged classification and coding systems. The reason for using international classification or coding systems is interoperability. Technically, our method allows for associating a single data item with codes from one or more coding or classification systems (one-to-many relationship) that express the same (eg, as can be the case in SNOMED CT and Logical Observation Identifiers Names and Codes). In this way, interoperability challenges faced during implementation can be solved pragmatically, depending on the choices made in source systems such as an EHR. Furthermore, the choice of the appropriate classification systems is included, in most cases, in the guideline itself (eg, TNM for cancer staging, BIRADS for radiology outcomes, New York Heart Association classification for heart failure, Eastern Cooperative Oncology Group or WHO for performance score).
Personalization
To express the complexity and degree to which guideline recommendations are personalized, all unique patient routes through all CDTs, which reflect patient subpopulations, were counted.
Decision Support
The CDTs were developed using Gaston (Medical Decision Support Systems BV, Eindhoven, the Netherlands).12 CDTs were created by simple drag-and-drop actions, using the data items as building blocks. Data items themselves were defined and coded (eg, SNOMED CT) in ART-DECOR (Advanced Tooling Requirements-Data Elements Codes, Object Identifiers and Rules), which is an international open source tool used by the Dutch National Institute for Information and Communication Technology (https://www.nictiz.nl/standaardisatie/art-decor) for development, maintenance, and publication of information standards for interoperability. Accordingly, the CDTs are interoperable per design, and a direct relation between information derived from different sources (eg, pathology or radiology reports) is modeled in such a way to enable (digital) information exchange and interoperability among the various actors in the care pathway.
CDTs were implemented in an interactive decision support application, Oncoguide (www.oncoguide.nl), which is also available for tablet computers. The application is designed to be used as a stand-alone app for manual data entry and to connect to EHRs for automatic electronic data exchange. For the latter, Oncoguide is accessible through RESTful Web Services (an application programming interface) and thereby follows the latest development on the Fast Health Interoperability Resources infrastructure of the international HL7 community (http://www.hl7.org/Special/committees/fiwg/index.cfm).
RESULTS
CDTs
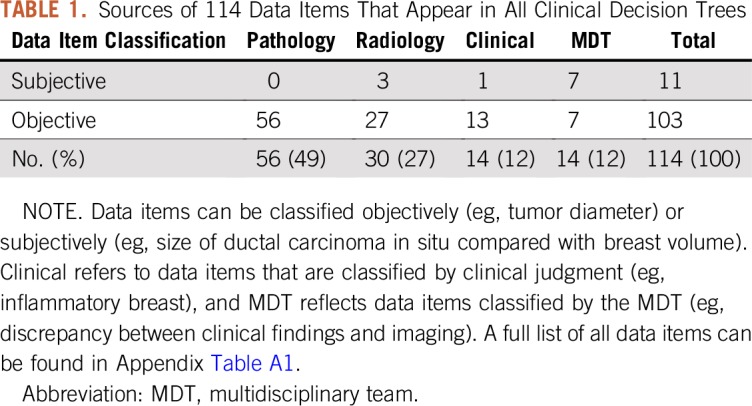
We translated the recommendations of the Dutch national breast cancer guideline (199 pages, A4 text format, 9,920 line numbers, 100,564 words, 13 chapters, and seven appendices) into 60 CDTs driven by 98 independent and 16 dependent data items. Figure 2 shows an example of a CDT. Table 1 lists a classification of data items with respect to their record source. Most objective data items originated from pathology reports (56 of 114; 49%), followed by radiology reports (30 of 114; 27%), clinical patient characteristics (14 of 114; 12%), and multidisciplinary team interpretation/validation (14 of 114; 12%). A list of all data items can be found in Appendix Table A1.
FIG 2.
Example of a clinical decision tree (CDT). (A) The top rectangle reflects the trunk of the CDT postoperative treatment. The rhombuses reflect the nodes and represent the data items. The branches define the cutoff values, which lead to additional nodes (rhombuses) or guideline recommendations (bottom rectangles; a delineated recommendation [rectangle with a curly bottom] means referral to another CDT, such as locoregional treatment after breast-conserving surgery[BCS]). (B) Note the double-delineated rhombus margin status, which can be unfolded to define the value of margin status. In contrast to other countries, the Dutch national breast cancer guideline does not recommend re-excision for focally positive margins after BCS in invasive tumor and recommends whole-breast irradiation, including boost.13
TABLE 1.
Sources of 114 Data Items That Appear in All Clinical Decision Trees
Implementation in Decision Support and Interoperability
Of all data items, 89% could be classified by existing classification and/or coding systems. On the basis of existing classification systems only, 75 (65%) of 114 data items were classified (66 to TNM, eight to BIRADS, and one to Response Evaluation Criteria in Solid Tumors [RECIST]). On the basis of the coding system SNOMED CT, only 90 (79%) of 114 data items could be classified. Ten data items could be qualified as too ambiguous to quantify (eg, size of ductal carcinoma in situ compared with breast volume; Appendix Table A1).
Twenty-two of 60 CDTs concerned recommendations for diagnostics, whereas 33 of 60 CDTs involved treatment recommendations. Five (8%) of 60 CDTs lacked recommendations because guideline recommendations were not available (eg, evaluation of surgical margin after treatment, recurrent disease).
By constructing CDTs systematically, all possible outcome values of each data item should lead to a recommendation. Four situations were identified for which the guideline represents no recommendation (as yet). For example, when a left ventricular ejection fraction of less than 50% is found in human epidermal growth factor receptor 2–positive breast cancer, the guideline lacks recommendations. As an example of ambiguity of data items within the guideline, Table 2 lists the many definitions of the data item margin status used in different classification systems.
TABLE 2.
Definitions of Margin Status Used in the Dutch National Breast Cancer Guideline and Other National and International Scientific Authoritative Sources
Personalization
In each CDT, there were one or more possible routes to reach one of the available recommendations, each of them defining a specific patient subpopulation. The total possible number of unique patient routes through all CDTs that led to one or more recommendations was 376 (see Appendix Fig A1). The mean number of possible patient routes per CDT was equal for CDTs that lead to treatment recommendations (6.4; median, four; range, one to 24) compared with CDTs that lead to diagnostic recommendations (6.3; median, three; range, one to 18).
Decision Support
All CDTs were successfully integrated in the interactive decision support app Oncoguide and are accessible free of charge (see Methods section). In the app, patient data are projected on the CDTs and show the path to the automatically generated patient-specific recommendation.
DISCUSSION
We show that it is feasible to transform a complex text-based guideline, such as the Dutch national breast cancer guideline, into data-driven CDTs. Although the concept of decision trees is not new, the clinical application of data-driven, moderated CDTs on a complex medical multidisciplinary guideline in such a way that they are both clinically interpretable and suitable for implementation in decision support systems has not been described earlier.14 By defining the data items needed for the CDTs, it was mostly possible to adhere to international classification and coding standards, although 21% of the data items needed to cover the complete guideline was not available in SNOMED CT. Although this does not limit the possibility to model recommendations as CDTs, closing this gap is important because different international guidelines (NCCN, European Society of Medical Oncology) can only give complementary recommendations if there is consensus about the definitions of all data items that determine diagnostic or treatment recommendations. For items currently not covered in SNOMED CT, we have put forward change requests for their inclusion at SNOMED International.
Because of the vagueness of recommendations, guidelines are sometimes criticized for not being helpful in practice.15,16 Different definitions of certain data values (Table 2) were encountered in the different chapters of the guideline, which automatically show up while designing CDTs. In this way, the CDT method is also a quality control instrument because it needs consequent and equivocal definitions. In contrast with text-based guidelines, our method can help the guideline updating process because it is based on compact guideline modules, and one can focus on the modules that need revision.
Clinical decision making is more and more personalized, and this is reflected in 376 unique subpopulations already described in the Dutch national breast cancer guideline. It is likely that this number will increase substantially because available pathology and genomic data will affect guideline recommendations soon. Moreover, in 2016, ASCO recommended the integration of higher-quality genomic data into clinical practice.17 These data can be modularly incorporated in CDTs.
In contrast to NCCN guidelines and flowcharts, our method of systematically constructed CDTs is fully data driven and delivers unambiguously and unequivocally defined and coded data items that relate all possible (combinations of) patient/disease characteristics to subpopulation-specific guideline recommendations. To our knowledge, only a few guideline-based clinical decision support systems have been routinely used for breast cancer management in the hospital setting.18-21 However, these local initiatives use decision rules and do not cover full national guidelines.
In 2012, ASCO started with the development of CancerLinQ, a rapid learning system to improve the quality and efficiency of cancer care by generating new knowledge on the basis of aggregated real-world patient data extracted from EHRs.22,23 Decision support systems, such as the Oncoguide app, can help to bring this goal a step closer because this tool is able to register patient subpopulation treatment choices and reasons for guideline deviations. The application of Oncoguide for decision support has been evaluated in comparison with Watson (IBM, Armonk, NY) on the basis of synthetic patient cases.24 However, additional research is needed to evaluate the value of guidelines-based decision support in daily clinical practice.
All CDTs together cover the whole care pathway for nonmetastatic breast cancer and follow all diagnostic and therapeutic options on the basis of the guideline. From our systematic method of constructing CDTs emerged five CDTs that lack recommendations, which pinpoints guideline gaps or, in other words, patient subpopulations that are not fully discussed in the guideline. These gaps can be addressed in future guideline updates.
Protocols, standardized reporting, and decision trees are sometimes put aside as cookbook medicine that ignores the fundamentally uncertain nature of medicine.25 However, the method described in this article does not reduce the level of evidence and secondary strength of recommendations in CDTs where evidence is weak or lacking. Although CDTs lack large text documents, no information is lost. Classification of recommendations by international grading systems, such as GRADE, is maintained.11 Likewise, as with text-based guidelines, it is up to the physicians to adhere to or deviate from the recommendations mentioned in the CDTs.7
A strength of this study is that we tested the method of CDTs by applying it to a highly complex multidisciplinary guideline while strictly adhering to international classification and coding systems. Moreover, our method is generalizable for guidelines in other disease areas where recommendations are based on the PICO system. The method already has been applied successfully to guidelines for other types of cancer, including to NCCN guidelines and Dutch nursing guidelines for pain management and wound care (all accessible online in Oncoguide).26 Furthermore, because no assumptions are needed for how the care process is organized, this model is scalable toward the future when more data become available. Similarly, this model is scalable across diseases, provided that CDTs are constructed systematically as described in the Methods section.
In this study, we tested the feasibility of modeling a complex textual guideline into systematically designed, modular, data-driven CDTs as the basis for a decision support system that can be used as a stand-alone application and has the ability to connect to EHRs through modern Web interfaces using international standards for electronic patient data exchange. Data on the practical application of CDTs in clinical practice is needed, however. Therefore, we are currently working with Dutch hospitals and EHR vendors to implement and evaluate the application of CDT-based decision support in routine clinical practice. Application of CDTs in practice and compliance of EHR documentation with the associated information standard offer to facilitate decision making and continuously evaluate and improve guidelines by comparison with real-live data. In addition to guideline recommendations, other types of knowledge can be represented as CDTs, such as clinical trial or genomic testing (MammaPrint [Agendia, Amsterdam, the Netherlands] and Oncotype DX [Genomic Health, Redwood City, CA]) indications. As an example of the latter, Figure 3 shows a CDT with recommendations for genomic testing in the Oncoguide application.
FIG 3.
Screenshot that shows a part of the clinical decision tree (CDT) for indication genomic testing in Oncoguide (translated into English). The green path through the CDT highlights the data provided in the data panel on the left side projected onto the CDT, which in this case leads to the recommendation that genomic testing is indicated. The full tree (in Dutch) is also accessible in an interactive format through https://oncoguide.nl/#!/projects/7/guideline/17/tree/153/10494. ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; neg, negative; pos, positive.
In conclusion, it is possible to present the complex Dutch national breast cancer guideline as clinically interpretable, modular, data-driven CDTs by using a set of 114 data items, 89% of which are defined by existing international classification and coding systems. The modular character of CDTs could provide a means for quick and clear implementation and accessibility of dynamic guidelines. Moreover, fast-growing knowledge could more rapidly and easily be taken into account by modularly updating CDTs, which supports implementation of data-driven personalized health care.
To demonstrate the potential application of CDTs as decision support, all CDTs were successfully implemented in an interactive decision support app, Oncoguide. Oncoguide provides a framework to register unique patient subpopulations and has the potency to report on physician, and potentially patient, motivation for guideline adherence or nonadherence in daily practice, which facilitates collaborative learning and improves the quality of care.27 Connection of Oncoguide to the EHR will be an essential next step to enable routine use of decision support in daily practice. The unequivocal and unambiguous definition of data items is an essential prerequisite for implementation of CDTs in decision support systems, and reaching consensus internationally on these definitions is a challenge for all national and international guideline working groups.
APPENDIX
FIG A1.
A unique patient route within a clinical decision tree that is based on the Dutch breast cancer guideline 2012. ALND, axillary lymph node dissection; BCS, breast-conserving surgery; LCIS, lobular carcinoma in situ; mi, micrometastasis; MDT, multidisciplinary team; paget, Paget’s disease; sn, sentinel node; SNP, sentinel node procedure.
TABLE A1.
All 114 Unique Data Items
Footnotes
Presented at the National Comprehensive Cancer Network 23rd Annual Conference, Orlando, FL, March 22-24, 2018.
AUTHOR CONTRIBUTIONS
Conception and design: Mathijs P. Hendriks, Xander A.A.M. Verbeek, Thijs van Vegchel, Maurice J.C. van der Sangen, Luc J.A. Strobbe, Jos W.S. Merkus, Sabine Siesling
Administrative support: Thijs van Vegchel
Provision of study material or patients: All authors
Collection and assembly of data: Mathijs P. Hendriks, Xander A.A.M. Verbeek, Thijs van Vegchel
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Mathijs P. Hendriks
Consulting or Advisory Role: MSD
Carolien H. Smorenburg
Travel, Accommodations, Expenses: Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1. Graham R, Mancher M, Miller Wolman D, et al (eds): Clinical Practice Guidelines We Can Trust. Washington, DC, National Academies Press, 2011. [PubMed] [Google Scholar]
- 2.Panella M, Marchisio S, Di Stanislao F. Reducing clinical variations with clinical pathways: Do pathways work? Int J Qual Health Care. 2003;15:509–521. doi: 10.1093/intqhc/mzg057. [DOI] [PubMed] [Google Scholar]
- 3.Martínez García L, Sanabria AJ, García Alvarez E, et al. The validity of recommendations from clinical guidelines: A survival analysis. CMAJ. 2014;186:1211–1219. doi: 10.1503/cmaj.140547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clercq P, Kaiser K, Hasman A. Computer-interpretable guideline formalisms. Stud Health Technol Inform. 2008;139:22–43. [PMC free article] [PubMed] [Google Scholar]
- 5.Peleg M. Computer-interpretable clinical guidelines: A methodological review. J Biomed Inform. 2013;46:744–763. doi: 10.1016/j.jbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Robertson J, Walkom E, Pearson SA, et al. The impact of pharmacy computerised clinical decision support on prescribing, clinical and patient outcomes: A systematic review of the literature. Int J Pharm Pract. 2010;18:69–87. [PubMed] [Google Scholar]
- 7. Quality of Care Directorate: Directive for Guidelines: 20 Criteria for Developing and Implementing a Clinical Guideline [in Dutch]. The Hague, the Netherlands, Quality of Care Directorate, 2012.
- 8. NABON: Breast Cancer: Dutch Guideline, Version 2.0, 2012. https://www.oncoline.nl/uploaded/docs/mammacarcinoom/Dutch%20Breast%20Cancer%20Guideline%202012.pdf.
- 9.Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annu Symp Proc. 2006:359–363. [PMC free article] [PubMed] [Google Scholar]
- 10. Burgers JS, van Everdingen JJ: Evidence-based guideline development in the Netherlands: The EBRO platform [in Dutch]. Ned Tijdschr Geneeskd 148:2057-2059, 2004. [PubMed] [Google Scholar]
- 11.Hultcrantz M, Rind D, Akl EA, et al. The GRADE Working Group clarifies the construct of certainty of evidence. J Clin Epidemiol. 2017;87:4–13. doi: 10.1016/j.jclinepi.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Clercq PA, Hasman A, Blom JA, et al. Design and implementation of a framework to support the development of clinical guidelines. Int J Med Inform. 2001;64:285–318. doi: 10.1016/s1386-5056(01)00189-7. [DOI] [PubMed] [Google Scholar]
- 13.Vos EL, Siesling S, Baaijens MHA, et al. Omitting re-excision for focally positive margins after breast-conserving surgery does not impair disease-free and overall survival. Breast Cancer Res Treat. 2017;164:157–167. doi: 10.1007/s10549-017-4232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiffman RN, Greenes RA. Improving clinical guidelines with logic and decision-table techniques: Application to hepatitis immunization recommendations. Med Decis Making. 1994;14:245–254. doi: 10.1177/0272989X9401400306. [DOI] [PubMed] [Google Scholar]
- 15.Merkow RP, Korenstein D, Yeahia R, et al. Quality of cancer surveillance clinical practice guidelines: Specificity and consistency of recommendations. JAMA Intern Med. 2017;177:701–709. doi: 10.1001/jamainternmed.2017.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codish S, Shiffman RN. A model of ambiguity and vagueness in clinical practice guideline recommendations. AMIA Annu Symp Proc. 2005:146–150. [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes KS, Ambinder EP, Hess GP, et al. Identifying health information technology needs of oncologists to facilitate the adoption of genomic medicine: Recommendations from the 2016 American Society of Clinical Oncology Omics and Precision Oncology Workshop. J Clin Oncol. 2017;35:3153–3159. doi: 10.1200/JCO.2017.74.1744. [DOI] [PubMed] [Google Scholar]
- 18.Patkar V, Acosta D, Davidson T, et al. Using computerised decision support to improve compliance of cancer multidisciplinary meetings with evidence-based guidance. BMJ Open. 2012;2:e000439. doi: 10.1136/bmjopen-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eccher C, Seyfang A, Ferro A. Implementation and evaluation of an Asbru-based decision support system for adjuvant treatment in breast cancer. Comput Methods Programs Biomed. 2014;117:308–321. doi: 10.1016/j.cmpb.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Bouaud J, Spano JP, Lefranc JP, et al. Physicians’ attitudes towards the advice of a guideline-based decision support system: A case study with OncoDoc2 in the management of breast cancer patients. Stud Health Technol Inform. 2015;216:264–269. [PubMed] [Google Scholar]
- 21.Séroussi B, Guézennec G, Lamy JB, et al. Reconciliation of multiple guidelines for decision support: A case study on the multidisciplinary management of breast cancer within the DESIREE project. AMIA Annu Symp Proc. 2018;2017:1527–1536. [PMC free article] [PubMed] [Google Scholar]
- 22.Sledge GW, Jr, Miller RS, Hauser R. CancerLinQ and the future of cancer care. Am Soc Clin Oncol Educ Book. 2013;33:430–434. doi: 10.14694/EdBook_AM.2013.33.430. [DOI] [PubMed] [Google Scholar]
- 23.Schilsky RL, Michels DL, Kearbey AH, et al. Building a rapid learning health care system for oncology: The regulatory framework of CancerLinQ. J Clin Oncol. 2014;32:2373–2379. doi: 10.1200/JCO.2014.56.2124. [DOI] [PubMed] [Google Scholar]
- 24. Keikes L, Medlock S, van de Berg DJ, et al: The first steps in the evaluation of a “black-box” decision support tool: A protocol and feasibility study for the evaluation of Watson for oncology. J Clin Transl Res 3:411-423, 2018 (suppl 3) [PMC free article] [PubMed] [Google Scholar]
- 25. Morris AH: Computerized protocols and bedside decision support. Crit Care Clin 15:523-545, 1999. [DOI] [PubMed]
- 26. Ebben K, Lamb P, van der Werf J, et al: A Method for structured comparison of oncologic clinical practice guidelines. National Comprehensive Cancer Network Annual Conference, Orlando, FL, March 22-24, 2018.
- 27.Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]