Abstract
Coxiella burnetii is an intracellular bacterium that causes acute and chronic Q fever. This unique pathogen has been historically challenging to study due to obstacles in genetically manipulating the organism and the inability of small animal models to fully mimic human Q fever. Here, we review the current state of C. burnetii research, highlighting new approaches that allow the mechanistic study of infection in disease relevant settings.
Keywords: Coxiella burnetii, Q fever, Intracellular, Macrophage
1. Introduction
1.1. Q fever and the discovery of Coxiella burnetii
In 1935, Australian researchers Drs. Macfarlane Burnet (University of Melbourne) and Edward Derrick (Queensland Health Department) were in the midst of investigating a disease of unknown origin prevalent among slaughterhouse workers in Queensland [1]. This mystifying disease, initially called abattoir fever, was renamed Query (Q) fever due to unknown cause. The disease was characterized by an incapacitating high fever lasting up to two weeks, often presenting with pulmonary and influenza-like complications. The only consistently observed trait of this fever was prevalence among individuals working with animal material. Concurrently, over 7,500 miles away in the United States at Rocky Mountain Laboratories, Drs. Herald Cox and Gordon Davis were attempting to isolate the causative agent of Rocky Mountain Spotted Fever, Rickettsia rickettsii, from ticks in the Nine Mile region of the Bitterroot Valley in Montana. During their studies, they serendipitously discovered a new agent that induced fever in guinea pigs [2]. Through a series of pioneering studies, and demonstration of human transmission by Dr. Rolla Dyer [3], Cox and Davis discovered they were handling the same agent as the Australian researchers. Interestingly, Dr. Dyer inadvertently demonstrated transmission of the pathogen after working with Dr. Cox, becoming infected himself while handling contaminated samples, and developing symptoms of this mysterious disease. By documenting his own symptomatic time course, Dr. Dyer provided the first detailed glimpse of pathogenic transmission to humans. These seminal studies demonstrated disease development in guinea pigs inoculated with Dr. Dyer’s samples, confirming the Nine Mile agent and the Australian Q fever agent as the same organism. The pathogen underwent several name changes, being referred to initially as a filter-passing virus, briefly Rickettsia diaporica, then Rickettsia burnetii [4], before settling on the genus and species that will forever honor two of the early pioneers, Coxiella burnetii. Over 80 years later, C. burnetii continues to intrigue researchers with many “Queries” as one of the most highly infectious and unique intracellular pathogens in the microbial world. The discoveries highlighted in this review include development of axenic cultivation, genetic manipulation, secreted protein characterization, and novel in vivo and ex vivo mammalian infection models that have led to an explosion of new knowledge in the past two decades. These new areas are paving the way to improved vaccines and treatments, and highlight the substantial impact of novel approaches on obligate intracellular pathogen research.
C. burnetii is an environmentally-stable bacterium and has the lowest bacterial infectious dose known to man, with fewer than 10 bacteria capable of causing disease [5, 6]. Infection ensues following inhalation by a susceptible host, and this aerosol route of exposure, combined with environmental stability and a low infectious dose, have resulted in classification of the organism as a category B select agent with the potential for bioweapon use [7]. The biological warfare aspect of C. burnetii holds historical significance, with multiple nations, including the United States, assessing potential weaponization characteristics of the pathogen in the mid-1900s. In the United States, C. burnetii was among a group of agents that were field tested in human and animal trials conducted at the Dugway Proving Ground in Utah, USA. Using aerosolized natural infection, researchers were able to observe the course of Q fever progression and assess infectious doses, disease symptoms, and treatment strategies [8]. Although the Dugway studies were ultimately ended, surveys of the area isolated C. burnetii Dugway strains from infected rodents, confirming transmission to mammals in the region and enhancing our understanding of C. burnetii infection [8]. These strains are still studied today due to genomic variation compared to the Nine Mile reference isolate and a severely attenuated phenotype in animal models of infection.
1.2. Prevalence, statistics, and global Q fever outbreaks
Global Q fever incidence and prevalence rates are poorly defined due to varying symptoms, misdiagnoses, and vast geographical locations of occurrence. A healthy immune system can combat acute Q fever, resulting in decreased diagnoses and incidence reports [9]. Approximately 50 individuals out of 100,000 persons per year obtain acute Q fever, whereas 1 individual out of 1,000,000 develops chronic Q fever [9]. The risk of exposure to C. burnetii increases in warmer climates, during summer months, and while working on farms, in slaughterhouses, or in research laboratories studying the pathogen [9–11]. Increased exposure rates often correlate with increased potential for outbreaks in the affected region.
C. burnetii outbreaks occur across the globe, with incident rates approaching thousands of infected individuals in some cases. In 1937, the first documented outbreak occurred in Australia when Macfarlane Burnet contracted Q fever, leading to several additional cases within the Melbourne facility where he conducted research [12]. Another small outbreak occurred in the United States in 1940 at the National Institutes of Health (NIH). After initiating studies on Q fever, 15 infected individuals were identified [12]. Following this small outbreak, 47 individuals at the NIH developed Q fever in 1946 [12]. A common theme among these outbreaks was that disease not only affected laboratory personnel working directly with C. burnetii, but also non-Q fever researchers. The most likely reason for these additional cases was incorrect handling procedures that allowed release of the pathogen into facility air [12].
In 1955, Q fever emerged in Africa, the third continent to document the disease, where nine countries reported varying numbers of infected individuals. In the same year, Q fever spread to South America, with the first documented case in Cayenne, New Guinea [13]. Small outbreaks have continued in South America over the years, culminating in an all-time high rate of incidence in 2005, with 150 Q fever cases per 100,000 individuals [14]. In 1984, 18 cases of Q fever were identified in Idaho, USA following exposure to infected sheep at an outdoor research facility [15, 16]. In 2002, in South Wales, United Kingdom, another Q fever outbreak developed, with 95 people of varying occupations infected during building renovations, suggesting exposure was due to C. burnetii aerosolization [17].
The largest Q fever outbreak on record occurred in the Netherlands between 2007 and 2010 on dairy farms that reported a high prevalence of spontaneous abortions in goats and sheep [16]. In 2007, numerous people were diagnosed with the disease, with over 4,000 documented cases and 25 deaths by 2016. In addition, over 150 individuals developed chronic Q fever. It is suggested that over 40,000 people were infected with C. burnetii based on seroprevalence studies in the area. Not all infected individuals in the area worked directly with animals, but were located downwind of dairy farms, implicating an aerosol mode of transmission [18, 16, 19]. Unfortunately, the long-term impact of chronic disease in this area is difficult to predict, posing health issues for many years to come.
1.3. Acute vs. chronic Q fever
Due to an aerosol mode of exposure, C. burnetii infection first presents as an acute, pulmonary disease characterized by pneumonia and flu-like symptoms [19]. Acute infection is accompanied by prolonged high fever and is often mis-diagnosed due to non-descript symptoms. If diagnosed properly, acute Q fever patients are treated with a 1–2 week regimen of doxycycline to alleviate symptoms [20]. However, healthy individuals can recover from acute disease without medical intervention, further contributing to under diagnosis. In fact, while fewer than 200 cases of acute Q fever are diagnosed annually in the U.S., 3–5% of the population are seropositive for C. burnetii antigens, indicative of previous exposure. This statistic suggests the infection rate is much higher than the number of diagnosed cases [21].
By an undefined mechanism, C. burnetii can establish a persistent infection that results in chronic Q fever. Chronic disease can manifest as hepatitis [22], osteomyelitis [23], fibrosis [24], and chronic fatigue syndrome [25], but the most commonly observed presentation is endocarditis following hematogenous spread from the lungs [26]. C. burnetii is responsible for the majority of non-cultivatable infectious endocarditis reported in the clinic [26]. During chronic infection, C. burnetii replicates within heart cells, promoting vegetative outgrowth on heart valves; however, the cellular niche in this organ has not been determined. Valve replacement is often needed to eradicate the infection and, in contrast to acute disease, chronic infection is extremely difficult to treat. The most effective current regimen is doxycycline combined with hydroxychloroquine for up to 1.5 years [27]. Dual antibiotic treatment is necessary due to the acidic nature of the pathogen’s intracellular niche (described in section 5.1). Combinatorial treatment elicits a bactericidal effect and decreases patient relapse [19]. This regimen is clearly not an optimal treatment course or prognosis, highlighting the need for improved therapeutic options. Moreover, a doxycycline-resistant strain was isolated in 2005 [28, 29], further underscoring the need for novel, improved treatment options.
1.4. Current vaccines
Q-Vax, created from an inactivated whole cell phase I strain of C. burnetii (Henzerling RSA334), is the only current commercially available vaccination against Q fever, and provides lifelong immunity to C. burnetii, making it extremely effective [30]. However, Australia is currently the only country using Q-Vax. Though Q-Vax has proven to be an important tool to control Q fever outbreaks in Australia, caveats to the vaccine remain. One of the largest obstacles to widespread use of Q-Vax is that pre-screening must be completed for each patient prior to clearance for vaccination because previous exposure to C. burnetii can elicit severe localized and systemic reactions. The pre-screening process involves skin and serological tests, and individuals who test positive are not cleared to receive the vaccine [31, 32]. Logistically, this process is not plausible for any globally-used vaccine strategy. Additionally, Q-Vax failure primarily occurs due to exposure to C. burnetii within 15 days after vaccination. Rare vaccine failure can also occur following exposure to C. burnetii after this 15-day period [33]. Due to the obstacles of using Q-Vax, further research is needed to develop a widely-accepted vaccine to combat Q fever.
2. Genomic features
2.1. Chromosome
Numerous C. burnetii isolates have been described, and each contains a chromosome that varies between 1,989,565 and 2,214,254 base pairs (bp) [34, 35]. Extensive genome comparison studies have allowed grouping of C. burnetii isolates according to genetic composition [36, 37]. Eight genomic groups (I-VIII) have been described that differ substantially at the genomic level, yet have not necessitated Coxiella speciation. Of particular note are substantial rearrangements and genome reduction in certain isolates that collectively influence type IV secretion system (T4SS) effector gene repertoires. For an extensive discussion on this topic, please refer to Larson et al [38]. C. burnetii isolates encode genes for biosynthesis of many amino acids, but is auxotrophic for some amino acids, including cysteine. In contrast to other obligate intracellular bacteria, presence of many biosynthetic genes allows C. burnetii to generate many necessary macromolecules apart from a host cell. This feature, along with C. burnetii-specific parameters such as acidic pH, allowed development of an axenic growth medium, termed acidified citrate cysteine medium (ACCM), that has revolutionized the C. burnetii field [39]. The original ACCM formulation has been further refined for robust bacterial growth apart from host cells [40, 41], which has allowed development of genetic manipulation methods, such as transposon mutagenesis and gene-specific knockouts [42–44]. These tools are being used to satisfy molecular Koch’s postulates for C. burnetii virulence factors.
2.2. Plasmids
In addition to the chromosome, all C. burnetii isolates maintain plasmid sequences, with most harboring a large plasmid between 37,000 and 55,000 bp, and a small number of isolates incorporate plasmid genes into the chromosome. Plasmid sequences vary substantially between isolates and 5 plasmids have been described: QpH1, QpRS, QpDV, QpDG, and an unnamed plasmid from a strain (CBQY) isolated in China [45–49, 35, 50–52]. Early work by pioneers in the field suggested that plasmid content determines whether a given isolate causes acute or chronic disease [53]. However, analysis of many additional isolates suggests this distinction is not absolute [54]. Although multiple secreted proteins (described below) are encoded on C. burnetii plasmids, these extrachromosomal elements remain largely uncharacterized and their requirement for virulence has not been proven.
2.3. Phase variation
Early infection studies showed that C. burnetii virulence differs depending on the structure of the isolate’s lipopolysaccharide (LPS) [55, 5, 56, 57]. LPS is still the most well characterized and recognized determinant of C. burnetii virulence and has been the focus of many vaccine design studies [58]. Full length LPS is produced by most virulent C. burnetii isolates (bacteria in phase I; typified by the Nine Mile I RSA493 reference isolate) and is predicted to shield the organism from recognition by dendritic cells [59]. Through repeated passaging in eukaryotic cells or axenic medium, LPS is increasingly truncated until C. burnetii loses the ability to cause disease and is cleared by immunocompetent hosts. These avirulent bacteria are referred to as being in phase II (typified by the avirulent Nine Mile II RSA439 isolate). The genetic basis for LPS variation was recently defined by Beare et al [60], who used whole genome comparison data and targeted mutagenesis to identify six LPS biosynthesis genes (cbu0533, cbu0678, cbu0839, cbu0845, cbu1655, and cbu1657) needed for construction of full-length LPS. These exciting results will pave the way for enhanced understanding of a major virulence determinant and improve vaccine design.
3. Modeling Q fever
3.1. Animal models of C. burnetii infection
Multiple small animal models, including mice, hamsters, and guinea pigs, have been used to advance understanding of the host response to C. burnetii. Unfortunately, mice are inherently resistant to C. burnetii, with many strains efficiently clearing the organism shortly after infection [61]. However, severe combined immunodeficient (SCID) mice have been used to effectively model distinct aspects of the immune response to the pathogen. These studies implicate a major role for interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), T cells, and B cells in the anti-C. burnetii response [62]. Additionally, SCID mice have been used to assess virulence potential of strains deficient in individual T4SS effectors [63]. In contrast to mice, guinea pigs replicate the fever seen in C. burnetii-infected humans, but lack genetic tools available in mice [64–67]. A recent study using the guinea pig model characterized infection dynamics with multiple C. burnetii isolates [68]. This study showed the most severe form of Q fever is caused by group I isolates while no virulence is evident in guinea pigs infected with group VI isolates that were isolated from the Dugway studies described in section 1.1.
Alternative infection platforms, such as non-mammalian insect and non-human primate models, are also used to probe distinct aspects of C. burnetii pathogenesis. Galleria mellonella (wax moth) are susceptible to phase I and phase II C. burnetii, which requires a T4SS for replication in these insects, and can be treated with doxycycline to antagonize infection [69]. Drosophila melanogaster are also susceptible to C. burnetii infection in a T4SS-dependent manner, and elicit an innate immune response to the pathogen [70]. These models are important in the field due to lower animal costs and ease of genetic manipulation. Clinical symptoms, serological analyses, and clinical chemistry analyses determined the primate model resembles human acute Q fever [61]. Thus, primates are used as an important model for identification of potential improved vaccines for Q fever. Collectively, these studies highlight the importance of current animal models of infection, but also underscore the need for new models that recapitulate all features of human disease.
3.2. Cellular models of C. burnetii infection
While animal models have provided a substantial amount of information about the host response to C. burnetii, several ex vivo infection systems have been used to model host-pathogen interactions that occur at the tissue and cellular level during Q fever. Immortalized eukaryotic cell lines have provided reliable systems for characterizing C. burnetii manipulation of mammalian host cells, and several host species have been exploited. In vitro, C. burnetii replicates in most cell types regardless of species or tissue origin [71]. Cell lines used to date include epithelial, fibroblast, trophoblast, macrophage, and endothelial cells from mice, guinea pigs, non-human primates, and humans. These cell lines have proven crucial for modeling host cell responses to infection and allowing characterization of C. burnetii replication vacuole formation. However, recent work using human pulmonary cell lines demonstrated that C. burnetii does not replicate as efficiently in alveolar and airway epithelial cells compared to alveolar macrophages, highlighting the need to better understand the pathogen’s interaction with multiple cell types and define cell intrinsic antibacterial responses. In addition to immortalized cell lines, primary cells from mice, guinea pigs, non-human primates, and humans have been used in recent years to more closely mimic in vivo conditions [72, 73]. Finally, a novel infection system was recently developed using an ex vivo human lung platform to study C. burnetii interaction with primary human lung tissue and cells [72, 74]. This disease-relevant platform (discussed in section 5.3) is being used to model the human pulmonary innate immune response to C. burnetii.
4. Dot/Icm type IV secretion system
4.1. Structural features
One critical virulence determinant encoded on the C. burnetii chromosome is termed the defect in organelle trafficking/intracellular multiplication (Dot/Icm) T4SS [75]. This system is related to the Dot/Icm apparatus in Legionella pneumophila and directly injects effectors, proteins that control numerous infection events, into the cytoplasm of infected host cells. The Dot/Icm T4SS is composed of 26 different components [34] that assemble into an injector evolutionarily related to conjugation pili. Inner membrane-associated components interact with cytoplasmic chaperones, including IcmS and IcmW [76], that deliver substrates to the secretion channel formed by DotF, DotG, and DotH. This channel allows effectors to transit through the periplasmic space and across the outer membrane of C. burnetii. The secretion system then penetrates the replication vacuole membrane, serving as a conduit for entry of the effector into the host cytosol where the protein controls distinct infection events. Over the past decade, numerous T4SS-dependent events have been characterized during C. burnetii infection, including parasitophorous vacuole (PV) formation, interaction with autophagosomes, prevention of host cell death, and interplay with the macrophage inflammatory response [77]. Importantly, the T4SS is required for virulence in D. melanogaster and G. mellonella, two recently developed models of the innate immune response to C. burnetii (refer to section 3.1) [78, 79, 69, 70]. T4SS-defective bacteria now await detailed testing in animal models to study the innate and adaptive immune responses to secreted effectors. Nonetheless, the absolute requirement of a functional T4SS to generate the vacuole needed for replication indicates this virulence determinant is vital for disease progression.
4.2. Effector identification
Early effector identification studies used L. pneumophila as a surrogate model of secretion and well-established readouts borrowed from the type III secretion field. These readouts of effector injection into the host cytosol include the widely-used adenylate cyclase (CyaA) and b-lactamase (BlaM) systems [80–82]. Using these assays to monitor secretion of tagged proteins produced by L. pneumophila and C. burnetii, over 130 T4SS substrates have been identified. However, most of these substrates await confirmation during natural C. burnetii infection of host cells to be established as bona fide T4SS-secreted effectors. In addition, a limited subset of effectors has been shown to be absolutely required for host cell infection, PV formation, intracellular replication, and disease in a competent host (refer to section 5.1). As a result, this area of the C. burnetii research field is ripe for exploration.
A major advantage in the C. burnetii field compared to other obligate intracellular pathogens is the ability to culture the organism axenically [40, 39]. This advance allowed generation of strains that replicate efficiently in axenic medium, yet contain mutations in genes required for intracellular growth. This work is not yet possible in other obligate intracellular pathogen fields, preventing satisfaction of molecular Koch’s postulates when studying possible virulence factors. In addition, the continually evolving area of C. burnetii genetic manipulation has provided numerous approaches for altering specific genetic content [38]. Combining axenic growth with new genetic methods has allowed generation of several transposon mutant libraries and detailed mechanistic studies of immunomodulatory LPS biosynthesis [60, 83, 84]. These techniques will continue to advance future studies of T4SS effector activity during infection.
Genome sequence comparisons of multiple isolates indicate altered effector repertoires exist, with some effector-encoding genes specific to an individual isolate or group of isolates. As mentioned in section 2.1, Beare et al showed substantial genomic arrangements among distinct genomic groups that influence the effector repertoire available during infection [36]. This work was followed by the identification of multiple isolate-specific T4SS effectors containing eukaryotic ankyrin repeat domains [85, 86]. These, and other, eukaryotic domain-containing proteins suggest horizontal gene exchange has occurred in C. burnetii and these proteins likely control isolate-specific events that contribute to differential disease presentation. In addition, recent reports identified a large repertoire of conserved and isolate-specific plasmid-encoded T4SS effectors [87, 81], implicating a role for these extrachromosomal elements in C. burnetii infection.
Note: Individual effectors will be described in the sections below in relation to impacted infection events.
5. Establishment of infection and intracellular replication
5.1. Intracellular replication
C. burnetii has a biphasic developmental cycle, consisting of small cell variant (SCV) and large cell variant (LCV) morphological forms, which are both infectious in vitro. The SCV is metabolically repressed and resistant to many environmental stressors including high temperature and nutrient limitation. The SCV is likely the form of the pathogen that initiates natural infections. The LCV is larger, metabolically active, and replication proficient, multiplying to high numbers within eukaryotic cells [71, 88]. Proteomic analysis and protein characterization studies suggest SCV abundant proteins are involved in immune evasion, whereas LCV-abundant proteins are related to cell division and intracellular survival [89].
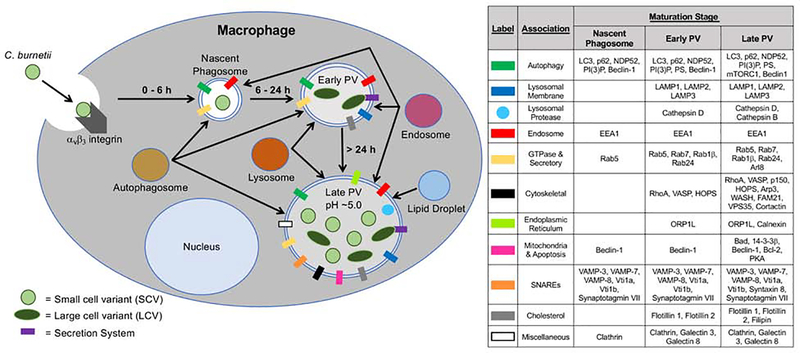
After binding to, and uptake by, a susceptible host cell, phagosome-harboring phase I and phase II C. burnetii SCVs transit through the phagolysosomal maturation pathway (Fig 1, left). At each maturation stage, distinct host markers with specific functions are present on C. burnetii-containing compartments (Fig. 1, right). Autophagosomes and endosomes quickly fuse with the phagosome to form the nascent phagosome. From host cell entry to ~ 6 h post-infection, endosomes, autophagosomes, and lysosomes containing acid phosphatase fuse with the nascent phagosome, forming the early PV. In the early PV, C. burnetii transitions from the SCV to the LCV, is metabolically activated, and produces the T4SS to translocate effector proteins into the host cytoplasm. Continual fusion with multiple intracellular compartments results in formation of the acidic (pH ~5.0) late PV compartment. After roughly 6 days, C. burnetii transitions from LCV back to SCV. Continual effector secretion is necessary to maintain the expanded late PV that ultimately encompasses most of the infected cell [13, 71]. The promiscuous fusogenicity of the PV is unique among intracellular bacterial pathogens. Membrane markers, proteases, and lipid droplets (LD), which are involved in metabolism and membrane synthesis, are present within the PV. LD accumulation is T4SS-dependent and necessary for proper bacterial growth, although the reasons for this requirement are not yet understood [90].
Fig. 1. Formation of the C. burnetii PV.
(Left) Schematic representation of the PV maturation pathway. C. burnetii SCV bind to the αvβ3 integrin receptor and are engulfed via phagocytosis by the macrophage, occupying a nascent phagosome from 0–6 h post-infection. Through autophagosome, endosome, and lysosome fusion, the early PV is formed within 6–24 h. Here, the SCV differentiates into the metabolically active LCV, allowing T4SS effector secretion. Through continuous fusion of autophagosomes, endosomes, and low pH lysosomes, the PV expands from 24 h onward, with an acidic pH ~ 5.0. In the late PV, the LCV transitions back to the SCV, maintaining both morphological forms of C. burnetii at late times post-infection. Lipid droplets also associate with the PV to regulate intracellular growth. (Right) Host proteins that associate with the PV at each point in the maturation stage.
Several T4SS-translocated effectors modify the PV and promote proper vacuole development. Coxiella vacuolar protein B (CvpB; also known as Cig2) interacts with phosphatidylinositol 3-phosphate and phosphatidylserine, altering host metabolism to promote proper PV formation [78]. CvpB is necessary for microtubule-associated protein 1A/1 B-light chain 3 (LC3) localization to the PV. CvpA activity is necessary for PV biogenesis via engagement of clathrin-mediated transport [91]. Other effectors that promote PV development include CvpC, which also engages autophagy, CvpD, and CvpE [92]. Cig57 is also required for optimal PV formation and intracellular replication via manipulation of vesicular trafficking [93]. Coxiella effector for intracellular replication A (CirA) aids PV formation via interactions with Rho GTPase and is necessary for bacterial virulence [94]. These studies demonstrate that C. burnetii requires secretion of multiple effectors to form and maintain the PV throughout infection.
5.2. Engagement of autophagy
Autophagy is a eukaryotic process of recycling damaged cellular components that removes unwanted materials, cytoplasmic components, and damaged organelles via trafficking to double-membrane autophagosomes. Autophagosomes ultimately fuse with lysosomes containing hydrolytic enzymes that degrade unwanted material into amino acids and peptides to recycle nutrients, allowing the cell to maintain proper functionality [95–98]. Many intracellular pathogens engage the autophagy system, either to obtain nutrients or prevent destruction in lysosomes. A functional autophagy system is necessary for efficient C. burnetii PV expansion and replication. When autophagy-related components are inhibited, a significant reduction in C. burnetii replication, number of infected cells, and overall PV development occurs [84, 99, 97]. Autophagosomes are recruited to the PV in a T4SS-dependent manner and LC3-II protein levels increase in infected cells [100], indicating active engagement of this host process by C. burnetii [101]. C. burnetii-infected cells also contain stable levels of the autophagy-related cargo protein sequestosome 1 (SQSTM1/p62), and recruit this protein to the PV in a T4SS-dependent manner, further underscoring the importance of autophagy during C. burnetii infection [102, 100]. Importantly, p62 localizes to PVs of both virulent phase I and avirulent phase II C. burnetii, and p62 protein levels increase during virulent isolate infection, suggesting isolate-specific variation in manipulating p62 [95]. p62 remains stable during induction of autophagy-dependent degradation in C. burnetii-infected cells, implying the necessity of p62 for intracellular growth [103]. Mammalian target of rapamycin complex 1 (mTORCI) is an autophagy-inhibiting protein located on lysosomal membranes, and activity of this protein is inhibited via the T4SS, promoting proper PV expansion [100]. Overall, these results indicate C. burnetii infection requires autophagic processes for proper PV development and bacterial survival.
5.3. Alveolar macrophages and the innate immune response to C. burnetii
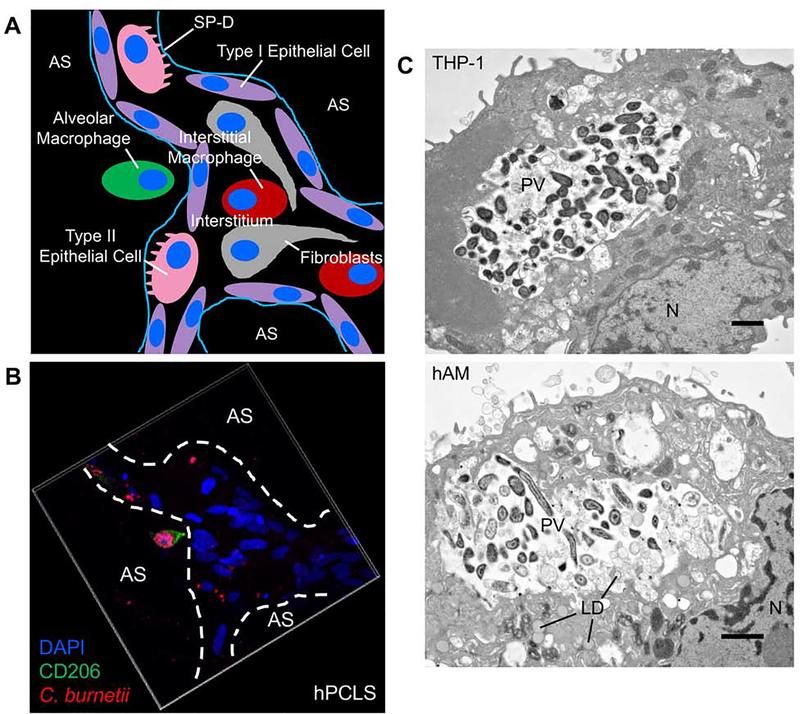
The human lung environment contains multiple cell types including neutrophils, fibroblasts, epithelial cells, endothelial cells, interstitial macrophages, and alveolar macrophages (AMs) [104]. Although numerous intracellular residence possibilities exist (Fig. 2A), the AM remains the preferential replication niche of C. burnetii [74, 72]. Recent establishment of an ex vivo human lung infection system allowed further assessment of this specificity. This system uses bronchioalveolar lavage fluid obtained from human lungs post mortem to isolate primary human AMs (hAMs) for cellular studies [74, 72]. To analyze all cells within the lung environment, lung tissue is processed and sectioned to obtain human precision-cut lung slices (hPCLS) [74, 72]. In hPCLS, C. burnetii replicates to high numbers within a PV in hAMs (Fig. 2B–C), but can also be found in interstitial macrophages, epithelial cells, and fibroblasts [74, 72]. However, only AMs support development of C. burnetii-filled, expanded PV. Further in vitro studies using human alveolar and airway epithelial cells, primary human lung fibroblast (HLFs), and human macrophage-like cells showed that epithelial cell lines do not support efficient C. burnetii replication, whereas macrophage-like and fibroblast cells allow robust growth. To investigate potential differences between isolated HLFs and fibroblasts in hPCLS, surfactant protein D (SP-D), an immunomodulatory protein in the pulmonary environment produced by type II alveolar epithelial cells, was introduced to isolated HLFs. A dose-dependent decrease in PV expansion and replication occurs in the presence of SP-D, suggesting a potential mechanism for suppression of bacterial growth in fibroblasts in the human lung [72]. Valuable information has been gained from the ex vivo hPCLS system; however, a few caveats remain. These include the absence of an adaptive immune response and neutrophil influx, although neutrophil attractant signaling can be assessed [72]. Nonetheless, the hPCLS model provides a novel glimpse into the innate interactions between C. burnetii and its human host. Collectively, these findings underscore the importance of closely mimicking the human lung environment using the ex vivo hPCLS infection platform and primary hAMs.
Fig. 2. C. burnetii targets alveolar macrophages in the human lung.
(A) Schematic representation of the alveolar environment and components present. SP-D = surfactant protein D. AS = alveolar space. (B) hPCLS were infected with mCherry-expressing C. burnetii (red) for 72 h, then processed for confocal immunofluorescence microscopy using CD206 antibody (green) to probe for alveolar macrophages and DAPI to stain nuclei (blue). Alveolar macrophages harbor expanded PV containing replicating C. burnetii while other cell types contain very few bacteria and no enlarged PV. (C) THP-1 cells (top) or hAMs (bottom) were infected with C. burnetii for 72 h, then processed for electron microscopy. N = nucleus. LD = lipid droplet. PV = parasitophorous vacuole. Bar, 20 μm. Both macrophage cell types support expanded PV development and robust bacterial growth.
AMs are present within lung airways and alveolar spaces, surveying the area for foreign material and invading pathogens [105]. Stimulation of AMs by external triggers can promote two major polarization phenotypes: M1 and M2. Macrophages polarized to a M1 state produce pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α, to initiate a robust immune response. M1-polarized macrophages also produce nitric oxide (NO) and reactive oxygen species (ROS) to combat intracellular bacteria. In contrast, M2-polarized macrophages have an anti-inflammatory phenotype, and are associated with wound healing and Th2 immune responses. M2 macrophages produce IL-10 and exhibit decreased production of NO and ROS. Interestingly, macrophages are never permanently differentiated into one phenotype, but can switch depending upon local environmental stimuli [106]. During infection, C. burnetii triggers a phenotypic switch from a M1 to a M2 macrophage as shown by an increase in M2 polarization genes such as il10, tgfβ1, and arg1 in monocyte-derived macrophages [107]. CXCL10-positive M1 cells also decrease during infection, with a corresponding increase in TGM2-positive M2 cells, and sustained production of IL-10 [74, 72, 107]. These findings suggest the pathogen attempts to convert AMs to a M2 phenotype, a more hospitable environment, to avoid immune system detection and allow optimal time for intracellular replication.
During initial interactions with macrophages, phase I C. burnetii is recognized by extracellular Toll-like receptor 4 (TLR4) and the αvβ3 integrin receptor, while phase II C. burnetii is recognized by TLR4, TLR2, complement receptor 3, and the αvβ3 integrin receptor [108, 109]. Following engagement of these receptors, phagocytosis and pinocytosis have both been suggested to play a role during uptake [38, 110]. Importantly, C. burnetii Outer membrane protein A (OmpA) was the first invasin identified as necessary for internalization in non-phagocytic cells. OmpA is also involved in bacterial replication in phagocytic and non-phagocytic cells [111]. These findings and limited studies on uptake highlight the need for additional research to expand our understanding of C. burnetii entry into host cells.
6. Alteration of host cell signaling
6.1. Apoptosis
C. burnetii replicates throughout a prolonged infectious cycle within eukaryotic cells, often growing within the same cell for 1–2 weeks before exiting by an undefined mechanism. This prolonged growth cycle necessitates preventing death of the heavily infected host cell. To preserve host cell integrity, C. burnetii potently prevents intrinsic and extrinsic apoptosis [112–121]. Early studies showed that C. burnetii antagonizes staurosporine-induced intrinsic apoptosis and TNF-α-induced extrinsic apoptosis of human macrophage-like cells and primate alveolar macrophages. The pathogen inhibits processing of executioner caspase-3 and the DNA repair enzyme poly (ADP-ribose) polymerase (PARP) [121], while preventing mitochondrial cytochrome c release [120]. C. burnetii further promotes a pro-survival response by activating multiple host signaling cascades, including Erk1/2, Akt, and PKA [112, 113, 118]. In addition, the pathogen engages the B-cell lymphoma (Bcl) family of mitochondrial proteins, recruiting anti-apoptotic Bcl-2 to the PV, inactivating pro-apoptotic Bcl-2-associated agonist of cell death (Bad) [113, 116], and triggering increased expression of myeloid cell leukemia 1 (Mcl-1) in neutrophils [112]. Subversion of these host signaling cascades results in efficient prevention of multiple modes of apoptosis.
In addition to engaging host signaling proteins, a subset of T4SS-secreted effectors antagonizes distinct aspects of the apoptotic response. Initial anti-apoptotic effector studies were performed using L. pneumophila, which is readily amenable to genetic manipulation and secretes C. burnetii T4SS effectors into the host cytosol. L. pneumophila normally triggers apoptosis of host cells [117]. However, when L. pneumophila expresses the C. burnetii T4SS effector AnkG, the organism is no longer able to kill host cells. Ensuing studies showed that AnkG binds to host p32, a versatile protein that interacts with components of the complement cascade, allowing the effector to enter the nucleus and prevent apoptosis by an undefined mechanism [122]. In addition to AnkG, Coxiella antiapoptosis effector A (CaeA) and CaeB prevent mitochondrial-dependent intrinsic apoptosis, albeit by distinct undefined mechanisms [114]. These studies have significantly advanced understanding of C. burnetii prevention of host cell death; however, there is still much to learn about effector mechanisms of action and the identity of additional anti-apoptotic effectors.
6.2. p62 and the Nrf2-Keap1 pathway
Nuclear erythroid 2-related factor 2 (Nrf2) is a leucine zipper transcription factor that regulates antioxidant processes in response to oxidative stress. Nrf2 is typically bound to Kelch-like ECH-associated protein 1 (Keap1) in the cytoplasm, promoting constitutive proteasome-dependent degradation of Nrf2 following ubiquitination. p62 is a cargo receptor described in section 5.2 that localizes to the PV and autophagosomes, targeting cellular proteins for degradation in a proteasome-independent fashion. p62 phosphorylated at S349 binds to Keap1 during times of cellular stress, preventing the Nrf2-Keap1 interaction. Released Nrf2 translocates into the nucleus and binds to antioxidant response elements (AREs) on DNA to induce transcription of cytoprotective genes and antioxidant enzymes to control stress-induced death [95, 123, 124].
p62 levels are stabilized and S349-phosphorylated p62 levels increase throughout infection, suggesting an important role for the Nrf2-Keap1 pathway [95]. Limited studies show phosphorylated p62 binding to Keapl and consequent activation of Nrf2 occurs in infections caused by other bacteria, such as Salmonella spp., and this event targets the pathogen for destruction [125]. However, C. burnetii is not degraded in a PV decorated with T4SS-recruited p62 or in cells containing increased levels of S349-phosphorylated p62, suggesting a pro-bacterial role for the protein in this infection. This distinction between C. burnetii and other intracellular bacteria will allow future studies to define novel p62-related pathogenic studies and further understand the contribution of p62 to bacterial infection. Correlating with increased levels of S349-phosphorylated p62, C. burnetii infection increases and stabilizes levels of Nrf2, and Nrf2 translocates to the nucleus of infected cells [95]. These results indicate the Nrf2-Keapl pathway is activated during infection; however, downstream consequences are not defined. During Pseudomonas aeruginosa infection, Nrf2−/− mice display increased inflammation and innate cell infiltration in the lungs [126], suggesting Nrf2 is necessary for efficient infection. A similar Nrf2 activity may impact C. burnetii in the lung, allowing the pathogen to avoid a robust immune response.
6.3. Oxidative stress
ROS are chemically reactive oxygen molecules that constitute one of the most important cellular defense mechanisms against invading pathogens by generating superoxide and hydroxyl radicals [127, 128]. ROS are produced following formation of the nicotinamide adenine dinucleotide phosphate oxidase (NADPH oxidase) enzyme complex that contains cytosolic p67phox, p47phox, and p40phox, and flavocytochrome b558 (cyt b558) containing transmembrane p22phox and the catalytic subunit gp91phox [128]. Upon stimulation, such as bacterial infection, p47phox is phosphorylated, causing the cytosolic complex to translocate to cyt b558. Here, cytosolic and membrane components form an active NADPH oxidase complex. NADPH is then reduced to NADP+ + H+ in the cytosol while electrons are transferred to oxygen, creating ROS and harmful hydrogen peroxide (H2O2) that is released on the other side of the membrane.
C. burnetii is directly affected by NADPH oxidase activity and inducible nitric oxide synthase (iNOS), which produces free radical nitric oxide (NO). Macrophages defective in NADPH oxidase or iNOS activity, alongside addition of an iNOS inhibitor or H2O2-scavenging catalase, enhance C. burnetii replication, demonstrating the sensitivity of the pathogen to NO and ROS [129]. To combat the microbicidal oxidative stress response, C. burnetii decreases ROS production by preventing localization of the cytosolic NADPH oxidase complex to cyt b558. Using a human neutrophil model, Siemsen et al showed a significant decrease in p22phox and p47phox levels at the membrane of C. burnetii-infected cells [130]. This finding demonstrates a potential C. burnetii mechanism for antagonizing ROS production. Further investigation using an acid phosphatase (ACP) inhibitor showed that C. burnetii secretion of ACP inhibits oxidative stress, suggesting a potential mechanism for preventing NADPH oxidase complex formation [131]. In addition, C. burnetii superoxide dismutase and catalase activity combats free oxygen radical and H2O2 formation, respectively [132, 133]. Finally, transcriptional profiling reveals vast differentiation in SCV and LCV (refer to section 5.1) oxidative stress-related gene expression. SCV-upregulated genes include a peroxiredoxin (ahpC2) and peroxiredoxin reductase (ahpD) involved in H2O2 scavenging. cbu1677 and cbu0819, which are involved in free radical detoxification, are also upregulated in SCV along with cbu1678, an acetyltransferase that modifies and detoxifies molecules via addition of an acetyl group [134]. These results suggest C. burnetii employs an extensive panel of proteins to combat oxidative stress and promote intracellular bacterial survival.
6.4. Inflammatory pathways
As described above, C. burnetii specifically targets AMs for intracellular replication. Intracellular pathogens that exploit macrophages must confront the cellular inflammasome-directed innate immune response that promotes inflammation and recruits immune assistance to the site of infection. Recent studies have begun to address the importance of C. burnetii regulating macrophage inflammatory signaling. Avirulent C. burnetii triggers prolonged production of IL-1β that correlates with ASC-dependent inflammasome formation and non-canonical processing of human-specific caspase-4 and −5, which are functional homologs of murine-specific caspase-11 [74]. Virulent isolates do not trigger this response, suggesting either phase I C. burnetii actively suppresses the inflammasome response or does not efficiently trigger the response in human macrophages due to differences in LPS structure or secreted effectors. In contrast, Cunha et al showed that avirulent C. burnetii produces a T4SS effector termed Inhibition of caspase activation A (IcaA) that antagonizes inflammasome activation in murine bone marrow-derived macrophages [135]. The most obvious difference between these studies is the origin of host cells used. Mice produce innate immune components not found in humans, such as caspase-11 and immunity-related GTPases, and we do not currently understand the full impact of these differences on the macrophage response to intracellular pathogens. However, comparing human cell studies to murine studies will likely uncover major events defining the host-specific immune response to C. burnetii. This is still a major undefined area of the C. burnetii field, and differentiating responses to avirulent and virulent organisms will be key to understanding human disease.
7. Remaining questions and conclusions
How does C. burnetii escape the host cell?
One of the most intriguing, yet unexplained, aspects of C. burnetii infection is how the pathogen escapes an initial host cell following sufficient replication. C. burnetii inhibits apoptosis for an extended period of time, suggesting the cell does not release harbored bacteria through intentional activation of cell death [121]. Alternatively, it is quite possible that C. burnetii employs a lytic release pathway similar to Chlamydia. In this scenario, once the PV contains a vast number of bacteria, effector secretion or signaling stimulation would rupture the PV and plasma membranes, releasing bacteria [136]. A second possibility is that cell-to-cell contact is necessary for C. burnetii invasion of nearby cells. Macrophage interaction with other cells could allow rapid entry of C. burnetii into nearby cells, creating a new host environment while avoiding detection by extracellular sensors. This process could occur through an exocytosis program or budding of the PV from the host plasma membrane, transferring a pre-made vesicle containing the pathogen into a new cell. Lastly, it is possible that C. burnetii escapes by extrusion, a package-and-release process that would maintain bacterial and PV integrity. Phagocytosis of the extruded body into a nearby cell would effectively introduce C. burnetii to a new growth niche. In light of C. burnetii replicating for up to 2 weeks within the same cell, these possibilities await future testing using a prolonged experimental system [88].
How does C. burnetii migrate to the heart to cause endocarditis?
A major mystery in the Q fever field revolves around how the pathogen exits the lung, migrates to the heart, and persists to cause chronic disease. It is assumed that either free bacteria or infected myeloid cells spread through the bloodstream to the heart where C. burnetii replicates to high numbers in heart cells. It is speculated that the pathogen targets cardiac macrophages for growth, but this has never been shown. A major impediment to uncovering these answers is the lack of testable animal models of chronic Q fever. Attempts have been made to trigger cardiac injury before or after C. burnetii infection, as human endocarditis typically manifests following valvular injury [137]. Hackstadt induced cardiac lesions using catheters in rabbits that were subsequently infected; however, this model was unable to distinguish differences between acute and chronic disease isolates of C. burnetii [138]. La Scola et al used guinea pigs to model endocarditis but found that this model only mimicked acute endocarditis [65]. More recently, a mouse model of chronic endocarditis was developed by Meghari et al who used mice overexpressing IL-10 to elicit macrophages at the heart that are unable to clear C. burnetii, allowing chronic infection [139]. This model awaits further testing to identify additional determinants of chronic disease. Ultimately, an infection model is needed that allows establishment of acute disease followed by C. burnetii spread to other organs and development of endocarditis following valvular injury similar to human disease.
Have we identified all C. burnetii T4SS effectors?
Closely-related L. pneumophila secretes more than 300 effectors, while C. burnetii secretes over 130. Although a completely saturating screen has not yet been reported, it is likely that researchers have identified the majority of C. burnetii effectors. However, numerous critical effector-driven infection events are yet to be defined. These include effectors that regulate the macrophage inflammatory response, recruit autophagy-related proteins, activate pro-survival kinase cascades, and control transcriptional events that alter host gene expression. This is an expanding and exciting area of C. burnetii research that will not only allow characterization of bacterial virulence mechanisms but will also identify novel host cell activities and responses to intracellular pathogens.
Why does C. burnetii prefer AMs to other pulmonary cells?
As described above, the preferred replication niche for C. burnetii is the AM [140]. Multiple studies have demonstrated C. burnetii replication in murine macrophages, primary human macrophages, and macrophage-like cells [141, 72]. Moreover, C. burnetii replicates in non-macrophage cell lines, but replication is inhibited in these cells in the pulmonary environment, making AM preference striking [72]. A potential reason for this cellular tropism is that C. burnetii has evolved mechanisms to dismantle the bactericidal responses of the AM. For example, a phenotypic switch from M1 to M2 polarization occurs during hAM infection, creating a more permissive environment for prolonged bacterial growth [72]. Additionally, C. burnetii exploits specific host pathways (such as Nrf2-Keap1 signaling), represses the oxidative stress response, secretes effectors that manipulate the host cell, and subverts autophagy [95, 130, 38, 13]. Thus, the macrophage pathway of bacterial uptake and phagolysosomal maturation to an acidic vacuole is the perfect replicative niche for C. burnetii. This cell type is also the first responder to lung infection, providing sound rationale for why the pathogen would adapt to macrophages as the preferred niche for survival. Finally, AMs are not stationary cells, making them inviting for pathogenic invasion and systemic spread to other tissues [106].
In summary, the C. burnetii research field has exploded in the last 15 years, largely due to the advent of axenic growth, genetic manipulation methodology, and new infection models that allow mechanistic studies of host-pathogen interactions. These advances have promoted increased interest in the cell biology of infection using this unique pathogen as a tool to study disarmament of the cellular antibacterial response. Inevitably, future studies of novel virulence determinants, and mutant strains deficient in these components, will identify new host-pathogen interactions that can be exploited in other intracellular pathogen research. The last decade of research by several talented teams has proven that C. burnetii is quickly moving past the “Query” stage encountered by early rickettsial pioneers.
Acknowledgements
This work was supported by funding from the NIH/NIAID R21AI127931 (D.E.V.) and the NIH/NIGMS P20GM103625 (D.E.V.). We thank Tiffany Weinkopff, Katelynn Brann, Marissa Fullerton, and Catherine Lee for critical review of the manuscript.
Footnotes
Conflict of interest
There is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Derrick EH. “Q” fever, a new fever entity: clinical features, diagnosis, and laboratory investigation. Med J Aust 1937;2:281–99. [DOI] [PubMed] [Google Scholar]
- [2].Davis GE, Cox HR. A filter-passing infectious agent isolated from ticks. I. Isolation from Dermacentor andersonii, reactions in animals, and filtration. Public Health Rep 1938;53:2259–82.19315693 [Google Scholar]
- [3].Dyer RE. A filter-passing infectious agent isolated from ticks. Human infection. Pub. Health Rep 1938;53:2277–82. [Google Scholar]
- [4].Cox HR. Rickettsia Diaporica And American Q Fever. Am J Trop Med Hyd 1940;s1–20:463–9. [Google Scholar]
- [5].Moos A, Hackstadt T. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect. Immun 1987;55:1144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hackstadt T Biosafety concerns and Coxiella burnetii. Trends Microbiol 1996;4:341–2. [DOI] [PubMed] [Google Scholar]
- [7].Oyston PC, Davies C. Q fever: the neglected biothreat agent. J Med Microbiol 2011;60:9–21. [DOI] [PubMed] [Google Scholar]
- [8].Stoenner HG, Lackman DB. The biologic properties of Coxiella burnetii isolated from rodents collected in Utah. Am. J. Hyg 1960;71:45–51. [DOI] [PubMed] [Google Scholar]
- [9].Maurin M, Raoult D. Q fever. Clin. Microbiol. Rev 1999;12:518–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hartzell JD, Wood-Morris RN, Martinez LJ, Trotta RF. Q fever: epidemiology, diagnosis, and treatment. Mayo Clin Proc 2008;83:574–9. [DOI] [PubMed] [Google Scholar]
- [11].Kampschreur LM, Dekker S, Hagenaars JC, Lestrade PJ, Renders NH, de Jager-Leclercq MG, et al. Identification of risk factors for chronic Q fever, the Netherlands. Emerg Infect Dis 2012;18:563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hirschmann JV. The Discovery of Q Fever and Its Cause. Am J Med Sci 2019;358:3–10. [DOI] [PubMed] [Google Scholar]
- [13].Eldin C, Raoult D. Moving from Q fever to C. burnetii infection. Epidemiol Infect 2016;144:1163–4. [DOI] [PubMed] [Google Scholar]
- [14].Eldin C, Mahamat A, Demar M, Abboud P, Djossou F, Raoult D. Q fever in French Guiana. Am J Trop Med Hyg 2014;91:771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rauch AM, Tanner M, Pacer RE, Barrett MJ, Brokopp CD, Schonberger LB. Sheep-associated outbreak of Q fever, Idaho. Arch Intern Med 1987;147:341–4. [PubMed] [Google Scholar]
- [16].Oliveira RD, Mousel MR, Pabilonia KL, Highland MA, Taylor JB, Knowles DP, et al. Domestic sheep show average Coxiella burnetii seropositivity generations after a sheep-associated human Q fever outbreak and lack detectable shedding by placental, vaginal, and fecal routes. PLoS One 2017;12:e0188054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].van Woerden HC, Mason BW, Nehaul LK, Smith R, Salmon RL, Healy B, et al. Q fever outbreak in industrial setting. Emerg Infect Dis 2004;10:1282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kampschreur LM, Delsing CE, Groenwold RH, Wegdam-Blans MC, Bleeker-Rovers CP, de Jager-Leclercq MG, et al. Chronic Q fever in the Netherlands 5 years after the start of the Q fever epidemic: results from the Dutch chronic Q fever database. J Clin Microbiol 2014;52:1637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eldin C, Melenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q Fever to Coxiella burnetii Infection: a Paradigm Change. Clin Microbiol Rev 2017;30:115–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kersh GJ. Antimicrobial therapies for Q fever. Expert Rev Anti Infect Ther 2013;11:1207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anderson AD, Kruszon-Moran D, Loftis AD, McQuillan G, Nicholson WL, Priestley RA, et al. Seroprevalence of Q fever in the United States, 2003–2004. Am J Trop Med Hyg 2009;81:691–4. [DOI] [PubMed] [Google Scholar]
- [22].Gomes MM, Chaves A, Gouveia A, Santos L. Two rare manifestations of Q fever: splenic and hepatic abscesses and cerebral venous thrombosis, with literature review ma non troppo. BMJ Case Rep 2014;2014:bcr2013202843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Merhej V, Tattevin P, Revest M, Le Touvet B, Raoult D. Q fever osteomyelitis: a case report and literature review. Comp Immunol Microbiol Infect Dis 2012;35:169–72. [DOI] [PubMed] [Google Scholar]
- [24].Melenotte C, Izaaryene JJ, Gomez C, Delord M, Prudent E, Lepidi H, et al. Coxiella burnetii: A Hidden Pathogen in Interstitial Lung Disease? Clin Infect Dis 2018;67:1120–4. [DOI] [PubMed] [Google Scholar]
- [25].Roest HI, Bossers A, van Zijderveld FG, Rebel JM. Clinical microbiology of Coxiella burnetii and relevant aspects for the diagnosis and control of the zoonotic disease Q fever. Vet Q 2013;33:148–60. [DOI] [PubMed] [Google Scholar]
- [26].Aistleitner K, Jeske R, Wolfel R, Wiessner A, Kikhney J, Moter A, et al. Detection of Coxiella burnetii in heart valve sections by fluorescence in situ hybridization. J Med Microbiol 2018;67:537–42. [DOI] [PubMed] [Google Scholar]
- [27].Wiener-Well Y, Fink D, Schlesinger Y, Raveh D, Rudensky B, Yinnon AM. Q fever endocarditis; not always expected. Clin Microbiol Infect 2010;16:359–62. [DOI] [PubMed] [Google Scholar]
- [28].Rolain JM, Lambert F, Raoult D. Activity of telithromycin against thirteen new isolates of C. burnetii including three resistant to doxycycline. Ann N Y Acad Sci 2005;1063:252–6. [DOI] [PubMed] [Google Scholar]
- [29].Rouli L, Rolain JM, El Filali A, Robert C, Raoult D. Genome sequence of Coxiella burnetii 109, a doxycycline-resistant clinical isolate. J Bacteriol 2012;194:6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marmion BP, Ormsbee RA, Kyrkou M, Wright J, Worswick DA, Izzo AA, et al. Vaccine prophylaxis of abattoir-associated Q fever: eight years’ experience in Australian abattoirs. Epidemiol Infect 1990;104:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ruiz S, Wolfe DN. Vaccination against Q fever for biodefense and public health indications. Front Microbiol 2014;5:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Reeves PM, Paul SR, Sluder AE, Brauns TA, Poznansky MC. Q-vaxcelerate: A distributed development approach for a new Coxiella burnetii vaccine. Hum Vaccin Immunother 2017;13:2977–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bond KA, Franklin LJ, Sutton B, Firestone SM. Q-Vax Q fever vaccine failures, Victoria, Australia 1994–2013. Vaccine 2017;35:7084–7. [DOI] [PubMed] [Google Scholar]
- [34].Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA 2003;100:5455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frazier ME, Heinzen RA, Stiegler GL, Mallavia LP. Physical mapping of the Coxiella burnetii genome. Acta Virol 1991;35:511−8. [PubMed] [Google Scholar]
- [36].Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the Genus Coxiella. Infect Immun 2009;77:642–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Beare PA, Samuel JE, Howe D, Virtaneva K, Porcella SF, Heinzen RA. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J. Bacteriol 2006;188:2309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Larson CL, Martinez E, Beare PA, Jeffrey B, Heinzen RA, Bonazzi M. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol 2016;11:919–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 2009;106:4430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 2011;77:3720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Omsland A, Hackstadt T, Heinzen RA. Bringing culture to the uncultured: Coxiella burnetii and lessons for obligate intracellular bacterial pathogens. PLoS Pathog 2013;9:e1003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Beare PA. Genetic Manipulation of Coxiella burnetii. Adv Exp Med Biol 2012;984:249–71. [DOI] [PubMed] [Google Scholar]
- [43].Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 2012;78:4580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 2009;191:1369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jager C, Lautenschlager S, Willems H, Baljer G. Coxiella burnetii plasmid types QpDG and QpH1 are closely related and likely identical. Vet Microbiol 2002;89:161–6. [DOI] [PubMed] [Google Scholar]
- [46].Lautenschlager S, Willems H, Jager C, Baljer G. Sequencing and characterization of the cryptic plasmid QpRS from Coxiella burnetii. Plasmid 2000;44:85–8. [DOI] [PubMed] [Google Scholar]
- [47].Willems H, Ritter M, Jager C, Thiele D. Plasmid-homologous sequences in the chromosome of plasmidless Coxiella burnetii Scurry Q217. J Bacteriol 1997; 179:3293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Valkova D, Kazar J. A new plasmid (QpDV) common to Coxiella burnetii isolates associated with acute and chronic Q fever. FEMS Microbiol Lett 1995;125:275–80. [DOI] [PubMed] [Google Scholar]
- [49].Ning Z, Yu SR, Quan YG, Xue Z. Molecular characterization of cloned variants of Coxiella burnetii isolated in China. Acta Virol 1992;36:173–83. [PubMed] [Google Scholar]
- [50].Minnick MF, Heinzen RA, Douthart R, Mallavia LP, Frazier ME. Analysis of QpRS-specific sequences from Coxiella burnetii. Ann N Y Acad Sci 1990;590:514–22. [DOI] [PubMed] [Google Scholar]
- [51].Samuel JE, Frazier ME, Mallavia LP. Stability of plasmid sequences in an acute Q-fever strain of Coxiella burnetii. J Gen Microbiol 1988;134:1795–805. [DOI] [PubMed] [Google Scholar]
- [52].Samuel JE, Frazier ME, Kahn ML, Thomashow LS, Mallavia LP. Isolation and characterization of a plasmid from phase I Coxiella burnetii. Infect. Immun 1983;41:488–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Samuel JE, Frazier ME, Mallavia LP. Correlation of plasmid type and disease caused by Coxiella burnetii. Infect Immun 1985;49:775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Glazunova O, Roux V, Freylikman O, Sekeyova Z, Fournous G, Tyczka J, et al. Coxiella burnetii genotyping. Emerg Infect Dis 2005;11:1211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Vishwanath S, Hackstadt T. Lipopolysaccharide phase variation determines the complement-mediated serum susceptibility of Coxiella burnetii. Infect Immun 1988;56:40–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hackstadt T Antigenic variation in the phase I lipopolysaccharide of Coxiella burnetii isolates. Infect. Immun 1986;52:337–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hackstadt T, Peacock MG, Hitchcock PJ, Cole RL. Lipopolysaccharide variation in Coxiella burnetti: intrastrain heterogeneity in structure and antigenicity. Infect Immun 1985;48:359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Peng Y, Zhang Y, Mitchell WJ, Zhang G. Development of a Lipopolysaccharide-Targeted Peptide Mimic Vaccine against Q Fever. J Immunol 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shannon JG, Howe D, Heinzen RA. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc. Natl. Acad. Sci. U. S. A 2005;102:8722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Beare PA, Jeffrey BM, Long CM, Martens CM, Heinzen RA. Genetic mechanisms of Coxiella burnetii lipopolysaccharide phase variation. PLoS Pathog 2018;14:e1006922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bewley KR. Animal models of Q fever (Coxiella burnetii). Comp Med 2013;63:469–76. [PMC free article] [PubMed] [Google Scholar]
- [62].Andoh M, Zhang G, Russell-Lodrigue KE, Shive HR, Weeks BR, Samuel JE. T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are crucial for disease development in Coxiella burnetii infection in mice. Infect Immun 2007;75:3245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].van Schaik EJ, Case ED, Martinez E, Bonazzi M, Samuel JE. The SCID Mouse Model for Identifying Virulence Determinants in Coxiella burnetii. Front Cell Infect Microbiol 2017;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Russell-Lodrigue KE, Zhang GQ, McMurray DN, Samuel JE. Clinical and pathologic changes in a guinea pig aerosol challenge model of acute Q fever. Infect Immun 2006;74:6085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].La Scola B, Lepidi H, Maurin M, Raoult D. A guinea pig model for Q fever endocarditis. J Infect Dis 1998;178:278–81. [DOI] [PubMed] [Google Scholar]
- [66].Ruble DL, Elliott JJ, Waag DM, Jaax GP. A refined guinea pig model for evaluating delayed-type hypersensitivity reactions caused by Q fever vaccines. Lab Anim Sci 1994;44:608–12. [PubMed] [Google Scholar]
- [67].Kazar J, Lesy M, Propper P, Valkova D, Brezina R. Comparison of virulence for guinea pigs and mice of different Coxiella burnetii phase I strains. Acta Virol 1993;37:437–48. [PubMed] [Google Scholar]
- [68].Long CM, Beare PA, Cockrell DC, Larson CL, Heinzen RA. Comparative virulence of diverse Coxiella burnetii strains. Virulence 2019;10:133–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Norville IH, Hartley MG, Martinez E, Cantet F, Bonazzi M, Atkins TP. Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology 2014;160:1175–81. [DOI] [PubMed] [Google Scholar]
- [70].Bastos RG, Howard ZP, Hiroyasu A, Goodman AG. Host and Bacterial Factors Control Susceptibility of Drosophila melanogasterto Coxiella burnetii Infection. Infect Immun 2017;85:e00218–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 2007;9:829–40. [DOI] [PubMed] [Google Scholar]
- [72].Dragan AL, Kurten RC, Voth DE. Characterization of Early Stages of Human Alveolar Infection by the Q Fever Agent Coxiella burnetii. Infect Immun 2019;87:e00028–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 2013;11:561–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Graham JG, Winchell CG, Kurten RC, Voth DE. Development of an Ex Vivo Tissue Platform To Study the Human Lung Response to Coxiella burnetii. Infect Immun 2016;84:1438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].McDonough JA, Newton HJ, Roy CR. Coxiella burnetii Secretion Systems. Adv Exp Med Biol 2012;984:171–97. [DOI] [PubMed] [Google Scholar]
- [76].Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol 2003;49:965–76. [DOI] [PubMed] [Google Scholar]
- [77].Qiu J, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 2017;15:591–605. [DOI] [PubMed] [Google Scholar]
- [78].Martinez E, Allombert J, Cantet F, Lakhani A, Yandrapalli N, Neyret A, et al. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc Natl Acad Sci U S A 2016;113:E3260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kohler LJ, Reed Sh C, Sarraf SA, Arteaga DD, Newton HJ, Roy CR. Effector Protein Cig2 Decreases Host Tolerance of Infection by Directing Constitutive Fusion of Autophagosomes with the Coxiella-Containing Vacuole. MBio 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Graham JG, Winchell CG, Sharma UM, Voth DE. Identification of ElpA, a Coxiella burnetii pathotype-specific Dot/Icm type IV secretion system substrate. Infect Immun 2015;83:1190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Maturana P, Graham JG, Sharma UM, Voth DE. Refining the Plasmid-Encoded Type IV Secretion System Substrate Repertoire of Coxiella burnetii. J Bacteriol 2013;195:3269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, et al. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 2011;193:1493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol 2011;2:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, et al. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 2014;10:e1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, et al. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 2009;191:4232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 2008;320:1651–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Crabill E, Schofield WB, Newton HJ, Goodman AL, Roy CR. Dot/Icm-Translocated Proteins Important for Biogenesis of the Coxiella burnetii-Containing Vacuole Identified by Screening of an Effector Mutant Sublibrary. Infect Immun 2018;86:e00758–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol 2004;186:7344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Coleman SA, Fischer ER, Cockrell DC, Voth DE, Howe D, Mead DJ, et al. Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect Immun 2007;75:290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mulye M, Zapata B, Gilk SD. Altering lipid droplet homeostasis affects Coxiella burnetii intracellular growth. PLoS One 2018;13:e0192215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Larson CL, Beare PA, Howe D, Heinzen RA. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 2013;110: E4770–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, et al. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 2015;83:661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Latomanski EA, Newton P, Khoo CA, Newton HJ. The Effector Cig57 Hijacks FCHO-Mediated Vesicular Trafficking to Facilitate Intracellular Replication of Coxiella burnetii. PLoS Pathog 2016;12:e1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Weber MM, Faris R, van Schaik EJ, McLachlan JT, Wright WU, Tellez A, et al. The Type IV Secretion System Effector Protein CirA Stimulates the GTPase Activity of RhoA and Is Required for Virulence in a Mouse Model of Coxiella burnetii Infection. Infect Immun 2016;84:2524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Winchell CG, Dragan AL, Brann KR, Onyilagha FI, Kurten RC, Voth DE. Coxiella burnetii Subverts p62/Sequestosome 1 and Activates Nrf2 Signaling in Human Macrophages. Infect Immun 2018;86:e00608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mansilla Pareja ME, Gauron MC, Robledo E, Aguilera MO, Colombo MI. The cAMP effectors, Rap2b and EPAC, are involved in the regulation of the development of the Coxiella burnetii containing vacuole by altering the fusogenic capacity of the vacuole. PLoS One 2019;14:e0212202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 2005;7:981–93. [DOI] [PubMed] [Google Scholar]
- [98].Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010;221:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect. Immun 2002;70:5816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Larson CL, Sandoz KM, Cockrell DC, Heinzen RA. Noncanonical Inhibition of mTORC1 by Coxiella burnetii Promotes Replication within a Phagolysosome-Like Vacuole. MBio 2019;10:e02816–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Winchell CG, Graham JG, Kurten RC, Voth DE. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun 2014;82:2229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Winchell CG, Steele S, Kawula T, Voth DE. Dining in: intracellular bacterial pathogen interplay with autophagy. Curr Opin Microbiol 2016;29:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Latomanski EA, Newton HJ. Interaction between autophagic vesicles and the Coxiella-containing vacuole requires CLTC (clathrin heavy chain). Autophagy 2018;14:1710–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Rubins JB. Alveolar macrophages: wielding the double-edged sword of inflammation. Am J Respir Crit Care Med 2003;167:103–4. [DOI] [PubMed] [Google Scholar]
- [105].Guth AM, Janssen WJ, Bosio CM, Crouch EC, Henson PM, Dow SW. Lung environment determines unique phenotype of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol 2009;296:L936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Byrne AJ, Mathie SA, Gregory LG, Lloyd CM. Pulmonary macrophages: key players in the innate defence of the airways. Thorax 2015;70:1189–96. [DOI] [PubMed] [Google Scholar]
- [107].Benoit M, Barbarat B, Bernard A, Olive D, Mege JL. Coxiella burnetii, the agent of Q fever, stimulates an atypical M2 activation program in human macrophages. Eur J Immunol 2008;38:1065–70. [DOI] [PubMed] [Google Scholar]
- [108].Capo C, Lindberg FP, Meconi S, Zaffran Y, Tardei G, Brown EJ, et al. Subversion of monocyte functions by Coxiella burnetii: impairment of the cross-talk between avb3 integrin and CR3. J. Immunol 1999;163:6078–85. [PubMed] [Google Scholar]
- [109].Capo C, Moynault A, Collette Y, Olive D, Brown EJ, Raoult D, et al. Coxiella burnetii avoids macrophage phagocytosis by interfering with spatial distribution of complement receptor 3. J Immunol 2003;170:4217–25. [DOI] [PubMed] [Google Scholar]
- [110].Tujulin E, Macellaro A, Lilliehook B, Norlander L. Effect of endocytosis inhibitors on Coxiella burnetii interaction with host cells. Acta Virol 1998;42:125–31. [PubMed] [Google Scholar]
- [111].Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 2014;10:e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Cherla R, Zhang Y, Ledbetter L, Zhang G. Coxiella burnetii Inhibits Neutrophil Apoptosis by Exploiting Survival Pathways and Antiapoptotic Protein Mcl-1. Infect Immun 2018;86:e00504–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Macdonald LJ, Graham JG, Kurten RC, Voth DE. Coxiella burnetii exploits host cAMP-dependent protein kinase signalling to promote macrophage survival. Cell Microbiol 2014;16:146–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Klingenbeck L, Eckart RA, Berens C, Luhrmann A. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 2013;15:675–87. [DOI] [PubMed] [Google Scholar]
- [115].Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, et al. Dot/Icm Type IVB Secretion System Requirements for Coxiella burnetii Growth in Human Macrophages. MBio 2011;2:e00175–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Vazquez CL, Colombo MI. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ 2010;17:421–38. [DOI] [PubMed] [Google Scholar]
- [117].Luhrmann A, Nogueira CV, Carey KL, Roy CR. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 2010;107:18997–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Voth DE, Heinzen RA. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 2009;77:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Vazquez CL, Colombo MI. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ 2009. [DOI] [PubMed] [Google Scholar]
- [120].Luhrmann A, Roy CR. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun 2007;75:5282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Voth DE, Howe D, Heinzen RA. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun 2007;75:4263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, et al. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun 2014;82:2763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Chen XL, Kunsch C. Induction of cytoprotective genes through Nrf2/antioxidant response element pathway: a new therapeutic approach for the treatment of inflammatory diseases. Curr Pharm Des 2004;10:879–91. [DOI] [PubMed] [Google Scholar]
- [124].Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 2013;51:618–31. [DOI] [PubMed] [Google Scholar]
- [125].Ishimura R, Tanaka K, Komatsu M. Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett 2014;588:822–8. [DOI] [PubMed] [Google Scholar]
- [126].Reddy NM, Kleeberger SR, Kensler TW, Yamamoto M, Hassoun PM, Reddy SP. Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice. J Immunol 2009;182:7264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Babior BM. NADPH oxidase: an update. Blood 1999;93:1464–76. [PubMed] [Google Scholar]
- [128].Panday A, Sahoo MK, Osorio D, Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol 2015;12:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Brennan RE, Russell K, Zhang G, Samuel JE. Both inducible nitric oxide synthase and NADPH oxidase contribute to the control of virulent phase I Coxiella burnetii infections. Infect. Immun 2004;72:6666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Siemsen DW, Kirpotina LN, Jutila MA, Quinn MT. Inhibition of the human neutrophil NADPH oxidase by Coxiella burnetii. Microbes Infect 2009;11:671–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Hill J, Samuel JE. Coxiella burnetii acid phosphatase inhibits the release of reactive oxygen intermediates in polymorphonuclear leukocytes. Infect Immun 2011;79:414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Akporiaye ET, Baca OG. Superoxide anion production and superoxide dismutase and catalase activities in Coxiella burnetii. J Bacteriol 1983;154:520–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Brennan RE, Kiss K, Baalman R, Samuel JE. Cloning, expression, and characterization of a Coxiella burnetii Cu/Zn Superoxide dismutase. BMC Microbiol 2015;15:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sandoz KM, Popham DL, Beare PA, Sturdevant DE, Hansen B, Nair V, et al. Transcriptional Profiling of Coxiella burnetii Reveals Extensive Cell Wall Remodeling in the Small Cell Variant Developmental Form. PLoS One 2016;11:e0149957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Cunha LD, Ribeiro JM, Fernandes TD, Massis LM, Khoo CA, Moffatt JH, et al. Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat Commun 2015;6:10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A 2007;104:11430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Willey RF, Matthews MB, Peutherer JF, Marmion BP. Chronic cryptic Q-fever infection of the heart. Lancet 1979;2:270–2. [DOI] [PubMed] [Google Scholar]
- [138].Hackstadt T. The role of lipopolysaccharides in the virulence of Coxiella burnetii. Ann N Y Acad Sci 1990;590:27–32. [DOI] [PubMed] [Google Scholar]
- [139].Meghari S, Bechah Y, Capo C, Lepidi H, Raoult D, Murray PJ, et al. Persistent Coxiella burnetii infection in mice overexpressing IL-10: an efficient model for chronic Q fever pathogenesis. PLoS Pathog 2008;4:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 2013;15:1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Fernandes TD, Cunha LD, Ribeiro JM, Massis LM, Lima-Junior DS, Newton HJ, et al. Murine Alveolar Macrophages Are Highly Susceptible to Replication of Coxiella burnetii Phase II In Vitro. Infect Immun 2016;84:2439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]