Abstract
Genome-wide association studies (GWAS) have identified 45 susceptibility loci associated with lung cancer. Only less than SNPs, small insertions and deletions (INDELs) are the second most abundant genetic polymorphisms in the human genome. INDELs are highly associated with multiple human diseases, including lung cancer. However, limited studies with large-scale samples have been available to systematically evaluate the effects of INDELs on lung cancer risk. Here, we performed a large-scale meta-analysis to evaluate INDELs and their risk for lung cancer in 23,202 cases and 19,048 controls. Functional annotations were performed to further explore the potential function of lung cancer risk INDELs. Conditional analysis was used to clarify the relationship between INDELs and SNPs. Four new risk loci were identified in genome-wide INDEL analysis (1p13.2: rs5777156, Insertion, OR = 0.92, p = 9.10 × 10–8; 4q28.2: rs58404727, Deletion, OR = 1.19, p = 5.25 × 10–7; 12p13.31: rs71450133, Deletion, OR = 1.09, p = 8.83 × 10–7; and 14q22.3: rs34057993, Deletion, OR = 0.90, p = 7.64 × 10–8 ). The eQTL analysis and functional annotation suggested that INDELs might affect lung cancer susceptibility by regulating the expression of target genes. After conducting conditional analysis on potential causal SNPs, the INDELs in the new loci were still nominally significant. Our findings indicate that INDELs could be potentially functional genetic variants for lung cancer risk. Further functional experiments are needed to better understand INDEL mechanisms in carcinogenesis.
Keywords: INDELs, genome-wide association studies, lung cancer
Introduction
Lung cancer is one of the most frequently diagnosed cancers and the leading cause of cancer mortality worldwide.1 It is estimated that nearly 2.1 million new lung cancer cases occurred in 2018, accounting for approximately 11.6% of total cancer diagnoses.2 Although tobacco smoking is a major lung cancer risk factor, genetic factors also play an important role in lung carcinogenesis. According to previous studies, common SNPs can explain approximately 12–21% heritability in lung cancer in Asian and European populations.3, 4 Genome‐wide association studies (GWAS) have previously identified 45 susceptibility loci associated with lung cancer,5 and single nucleotide polymorphisms (SNPs) in the CHRNA3, CHRNA5, TERT and human leukocyte antigen (HLA) regions showed consistent and robust associations in different studies.
To date, the vast majority of studies have focused on the relationship between SNPs and lung cancer. Small insertions and deletions (INDELs), which are another type of variations, also play an important role in lung carcinogenesis. INDELs are defined as short insertions and deletions (ranging from 1 to 10,000 bp) in the human genome.6, 7 As important genetic variations, INDELs are the second most abundant genetic polymorphisms in the human genome, only less than SNPs.8 The final phase of the 1000 Genomes Project (http://www.internationalgenome.org/) has characterized more than 3.4 million INDELs in 88 million variant sites in the human genome, and compared to phase I, the number of INDELs increased by 70%.8 This provides a comprehensive panel to explore the effects of INDELs. INDELs in the genome are highly associated with multiple human diseases; nearly 24% of Mendelian diseases are caused by INDELs based on the Human Gene Mutation Database (HGMD).9 Over the past decade, the development of high‐throughput sequencing has made it possible to detect INDELs in individual genomes. Next‐generation sequencing (NGS) analyses have identified INDELs across multiple cancer types10, 11; however, these INDELs were at the somatic level with low frequency. At the germline level, INDELs have been described as associated with cancers in case–control studies by genotyping or genomic imputation. For example, a single INDEL in the 6q25.3 locus, which is related to the SLC22A1 and SLC22A2 genes, increased the risk of prostate cancer in a multiethnic GWAS.12 Another study in a Chinese population found that a 5‐bp INDEL in the GAS5 gene increased hepatocellular carcinoma risk.13 For lung cancer risk, Sun et al. reported a six‐nucleotide deletion variant in the CASP8 promoter was related to reduced risk of multiple cancers, including lung cancer.14 In addition, Liu et al. found two insertion variants in BRM promoter region were also associated with the increased risk of lung cancer.15 However, limited studies with large‐scale samples have been available to systematically evaluate the effects of INDELs on lung cancer risk. In our study, we aimed to investigate the relationship between INDELs and lung cancer risk at a genome‐wide level. To accomplish this, we conducted a large‐scale case–control study with 23,202 lung cancer cases and 19,048 controls to dissect the associations between INDELs and lung cancer risk among European and Asian populations.
Materials and Methods
Study population
In our study, we integrated three published lung cancer GWAS, including the TRICL‐ILCCO OncoArray European data (The OncoArray Consortium lung cancer GWAS: 43,398 participants in total, European population; http://epi.grants.cancer.gov/oncoarray/),16 the DCEG Lung Cancer Study (the National Cancer Institute lung cancer GWAS: 5,716 cases and 5,821 controls, European population),17 and our published NJMU GWAS data (Nanjing Medical University lung cancer GWAS from Nanjing and Beijing: 2,331 cases and 3,077 controls, Chinese population).18 Briefly, for the TRICL‐ILCCO OncoArray data, we used the same quality control strategies in the previous paper.16 The DCEG Lung Cancer Study was applied from the Genotypes and Phenotypes (dbGAP; https://www.ncbi.nlm.nih.gov/gap) database.17 Considering the duplication of samples within the TRICL‐ILCCO OncoArray data, 3,251 samples were removed when IBD (identity‐by‐descent) >0.45. Consequently, 2,427 cases and 1,944 controls in the DCEG Lung Cancer Study were kept for further analysis. For the NJMU GWAS data, standard sample quality control strategies were also performed according to the original paper.18 Finally, a total of 23,202 cases and 19,048 controls were included for further analysis (Table S1). Each study was approved by the local institutional review board.
Genotype quality control and imputation
The details of the imputation procedures used in the TRICL‐ILCCO OncoArray project have been described previously.16, 19 Briefly, SHAPETIT V2 and IMPUTE2 were used for phasing and imputation, respectively. The 1000 Genomes Project Phase III database (released at October 2014) was used as a reference dataset. After imputation, there were 1,857,403 INDELs in the TRICL‐ILCCO OncoArray data. Then, we performed standard quality control on the imputed INDELs data by excluding the data with the following characteristics: (i) imputation quality INFO <0.9; (ii) genotyping call rate < 95%; (iii) minor allele frequency (MAF) in controls <0.01; or (iv) Hardy–Weinberg equilibrium (HWE) <1 × 10−12 in cases or <1 × 10−7 in controls. We also excluded 17,812 INDELs located in genome segmental duplication regions,20 which may lead to inaccuracy during imputation. Thus, the total number of TRICL‐ILCCO OncoArray INDELs was 694,395. For the DCEG GWAS and NJMU GWAS data, the imputation procedures have been previously described.21, 22 We conducted the same quality control criteria on the DCEG GWAS and NJMU GWAS imputation data. Finally, we obtained 484,196 overlapped INDELs for the subsequent analyses (Fig. S1).
eQTL and differential expression analysis
We used the Genotype‐Tissue Expression (GTEx; http://www.gtexportal.org/home/) Project expression quantitative trait locus (eQTL) database (V7 release) for identified INDELs. We searched for each INDEL‐gene pair eQTL analysis results in lung tissue. Due to lack of information on INDEL rs71450133 in GTEx database, we use SNP rs28435996 which showed high linkage disequilibrium (LD; r2 = 0.94) with rs71450133 as a tagging SNP. Differential expression analyses were performed using data from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) project.23, 24 A total of 106 paired lung tumor tissues and adjacent tissues from the TCGA database were used to performed differential expression analyses using Wilcoxon paired test.
In silico functional annotation and rank scoring system development
We combined multiple sources of public functional annotation databases to explore the potential function of the INDELs, similar strategy was also applied in the recent largest breast cancer GWAS study with the INQUISIT algorithm.25 Genomic regulatory region and functional score were used to evaluate INDELs and SNPs showed high LD with them. Regulatory elements, including promoter, enhancer and transcription factor binding sites (TFBS) data were based on the Encyclopedia of DNA Elements (ENCODE; https://www.encodeproject.org/) Project A549 human lung cancer cell line data.26 Four annotation database, including 3DSNP (http://cbportal.org/3dsnp/),27 Combined Annotation‐Dependent Depletion (CADD; http://cadd.gs.washington.edu/home),28 Phenotype‐Informed Noncoding Element Scoring (PINES; http://genetics.bwh.harvard.edu/pines)29 and RegulomeDB (http://www.regulomedb.org/)30 were also used to identify the potential pathogenicity and function of the INDELs. We developed a rank scoring system to integrate all these data together and INDELs identified in our study, as well as SNPs which showed a high LD (r2 > 0.6) relationship with INDELs were all annotated by this rank scoring system.
We generated binary variables, feature rank, to represent the importance of each variant in each database, 1 defined as more important and 0 defined as less important. For chromatin biofeatures data, as mentioned above, promoter, enhancer and TFBS, if INDELs or SNPs located in the regulatory region, the feature ranks were defined as 1, else as 0. For four annotation databases (3DSNP, CADD, PINES and RegulomeDB) with scores, if a variant’s score in the top 10% of corresponding INDEL LD block, the feature rank was defined as 1 (more important), otherwise it was defined as 0 (less important). Finally, all feature ranks of seven annotations were accumulated as a final score for each INDEL and SNP, ranging from 0 to 7. The variant with the highest score was considered as a potentially causal variant.
Statistical analysis
For the three GWAS studies, the association testing for each INDEL was performed using the SNPTEST (version 2.5.4) software, which is based on a probabilistic dosage model adjusting for age, gender and the first three principal components in the TRICL‐ILCCO OncoArray; age, gender and the first principal component in the DCEG GWAS; and age, gender, pack‐years and the first principal component in the NJMU GWAS. Meta‐analysis (fixed‐effect model) was conducted to combine individual association estimates from the three GWAS datasets. Testing for differences in the genetic effects across the three studies was assessed by using the I2 and p values calculated from Cochran’s Q statistic. Meta‐analysis was conducted using the GWAMA software. Subgroup analysis was performed for baseline characteristics, including age, gender, histology, smoking status and ethnicity. For the conditional analysis, a multivariate logistic regression model adjusting for age, gender, the first three principal components and known lung cancer risk variants was used with the TRICL‐ILCCO OncoArray.
General analyses were performed using the R software (version 3.3.1). p ≤ 0.05 was used as the threshold of statistical significance and all statistical tests were two‐sided. A suggestive threshold of 1.0 × 10−6 was used to present significant INDELs,31, 32 and Bonferroni correction was also applied to account for multiple comparisons (threshold: 0.05/484,196 = 1.03 × 10−7).
Results
Study overview
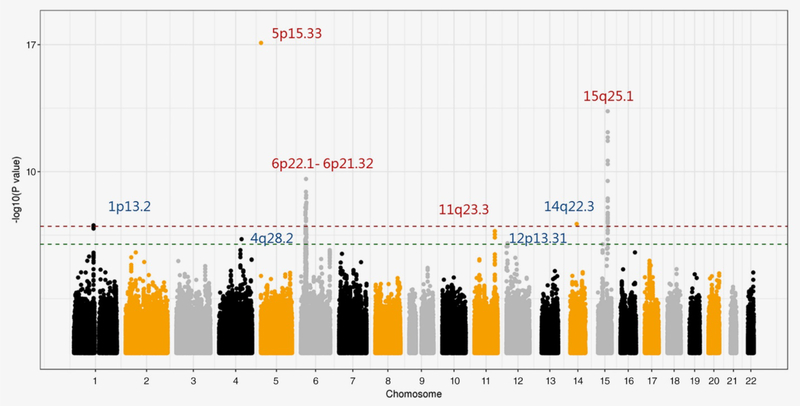
In our study, we imputed a total of 484,196 INDELs based on 23,202 lung cancer cases and 19,048 controls. Nineteen INDELs along with 11 loci were identified as being significantly associated with lung cancer risk at a suggestive threshold of 1.0 × 10−6 (Fig. 1; Tables 1 and 2). Among them, four loci (1p13.2, 4q28.2, 12p13.31 and 14q22.3) were novel risk loci for lung cancer, while seven loci have been previously reported as lung cancer risk loci as indicated by SNPs (5p15.33, 6p21.32, 6p21.33, 6p22.1, 6p22.2, 11q23.3 and 15q25.1). The results of INDELs in three studies were listed in Table S2. Four new risk loci were identified in our genome‐wide INDEL analysis (Table 1, Figure 2), including rs5777156 in 1p13.2 (Insertion, OR = 0.92, 95% CI = 0.89–0.95, p = 9.10 × 10−8); rs58404727 in 4q28.2 (Deletion, OR = 1.19, 95% CI = 1.11–1.28, p = 5.25 × 10−7); rs71450133 in 12p13.31 (Deletion, OR = 1.09, 95% CI = 1.05–1.13, p = 8.83 × 10−7); and rs34057993 in 14q22.3 (Deletion, OR = 0.90, 95% CI = 0.87–0.94, p = 7.64 × 10−8). INDELs rs5777156 and rs34057993 were still significant after Bonferroni correction (p < 1.03 × 10−7). There was no evidence of heterogeneity among the studies for the new risk loci. Subgroup analyses on the four new INDELs from the OncoArray data are summarized in Table S3. No evidence of heterogeneity was observed for the new risk loci among age, gender, smoking status, histology type and ethnicity, which implied the effects of the new risk loci were robust.
Figure 1.
Manhattan plots of INDEL associations with lung cancer risk. The x‐axis represents the chromosomal location and the y‐axis represents the −log10 (p‐value). Red, previously known as loci and blue, new loci identified in this analysis. The red line denotes the Bonferroni correction significance (p = 1.03 × 10−7) and the green line denotes the suggestive significance (p < 1.0 × 10−6). [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
The association between the INDELs in the new regions and lung cancer risk
| Overall results3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | INDEL | Gene | INS/DEL | INFO. | Major | Minor | EUR1 | EAS2 | OR (95%CI) | p | Het p |
| 1p13.2 | rs5777156 | MAGI3 | Insertion | 0.999 | – | A | 0.24 | 0.61 | 0.92 (0.89, 0.95) | 9.10 × 10−8 | 0.837 |
| 4q28.2 | rs58404727 | RP11-184M15.2 | Deletion | 0.999 | T | – | 0.02 | 0.33 | 1.19 (1.11, 1.28) | 5.25 × 10−7 | 0.191 |
| 12p13.31 | rs71450133 | PLEKHG6 | Deletion | 0.986 | AA | – | 0.18 | 0.38 | 1.09 (1.05, 1.13) | 8.83 × 10−7 | 0.990 |
| 14q22.3 | rs34057993 | OTX2-AS1 | Deletion | 0.975 | G | – | 0.17 | 0.27 | 0.90 (0.87, 0.94) | 7.64 × 10−8 | 0.587 |
The effect allele frequencies of the insertion or deletion in 1000 Genomes EUR samples.
The effect allele frequencies of the insertion or deletion in 1000 Genomes EAS samples.
The OR (95% CI) and p-value for the meta-analysis were fixed-effects model.
Abbreviations: INFO., imputation quality info.; Het p: p-value for heterogeneity test.
Table 2.
The association between the INDELs in the known regions and lung cancer risk
| Overall results3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | INDEL | Gene | INS/DEL | Major | Minor | EUR1 | EAS2 | OR (95% CI) | p | Het p |
| 5p15.33 | rs34218850 | TERT | Deletion | C | – | 0.34 | 0.19 | 1.14 (1.11, 1.18) | 7.98 × 10−18 | 0.097 |
| 6p21.32 | rs200675567 | HLA-DQA1 | Deletion | C | – | 0.11 | 0.16 | 0.90 (0.86, 0.96) | 4.03 × 10−2 | 0.412 |
| 6p21.32 | rs9279532 | NOTCH4 | Deletion | G | – | 0.12 | 0.05 | 1.11 (1.07, 0.94) | 2.77 × 10−7 | 0.588 |
| 6p21.33 | rs550239034 | POU5F1 | Deletion | TT | – | 0.25 | 0.46 | 0.92 (0.89, 0.95 | 2.12 × 10−7 | 0.055 |
| 6p21.33 | rs549219764 | HCP5 | Deletion | G | – | 0.20 | 0.02 | 1.11 (1.07, 1.15) | 2.01 × 10−9 | 0.091 |
| 6p22.1 | rs9280949 | RPP21 | Insertion | – | T | 0.09 | 0.06 | 1.16 (1.11, 1.22) | 2.53 × 10−10 | 0.468 |
| 6p22.1 | rs139089584 | UNC00533 | Insertion | – | TTTG | 0.29 | 0.54 | 0.92 (0.89, 0.95) | 2.40 × 10−7 | 0.145 |
| 6p22.1 | rs34832458 | HLA-G | Insertion | – | T | 0.39 | 0.24 | 0.92 (0.89, 0.94) | 1.29 × 10−9 | 0.077 |
| 6p22.1 | rs374787445 | HLA-F-AS1 | Deletion | C | – | 0.18 | 0.23 | 1.11 (1.07, 1.15) | 6.56 × 10−9 | 0.323 |
| 6p22.2 | rs145093187 | BTN2A1 | Insertion | – | T | 0.12 | 0.05 | 0.87 (0.83, 0.91) | 9.44 × 10−9 | 0.478 |
| 11q23.3 | rs139157129 | MPZL2 | Deletion | A | – | 0.48 | 0.43 | 0.93 (0.90, 0.95) | 1.90 × 10−7 | 0.864 |
| 15q25.1 | rs577626090 | CHRNA5 | Deletion | AAAAG | – | 0.37 | 0.03 | 1.29 (1.25, 1.33) | 9.91 × 10−64 | 0.945 |
| 15q25.1 | rs138784116 | CHRNB4 | Deletion | AGG | – | 0.37 | 0.14 | 0.89 (0.86, 0.92) | 4.65 × 10−14 | 0.655 |
| 15q25.1 | rs143284856 | MORF4L1 | Insertion | – | TT | 0.47 | 0.12 | 1.11 (1.08, 1.14) | 1.29 × 10−12 | 0.732 |
| 15q25.1 | rs61655864 | CHRNA5 | Deletion | A | – | 0.29 | 0.77 | 0.81 (0.79, 0.84) | 6.24 × 10−37 | 0.068 |
The effect allele frequencies of the insertion or deletion in 1000 Genomes EUR samples.
The effect allele frequencies of the insertion or deletion in 1000 Genomes EAS samples.
The OR (95% CI) and p-value for the meta-analysis were fixed-effects model.
Abbreviation: Het p, heterogeneity p-value.
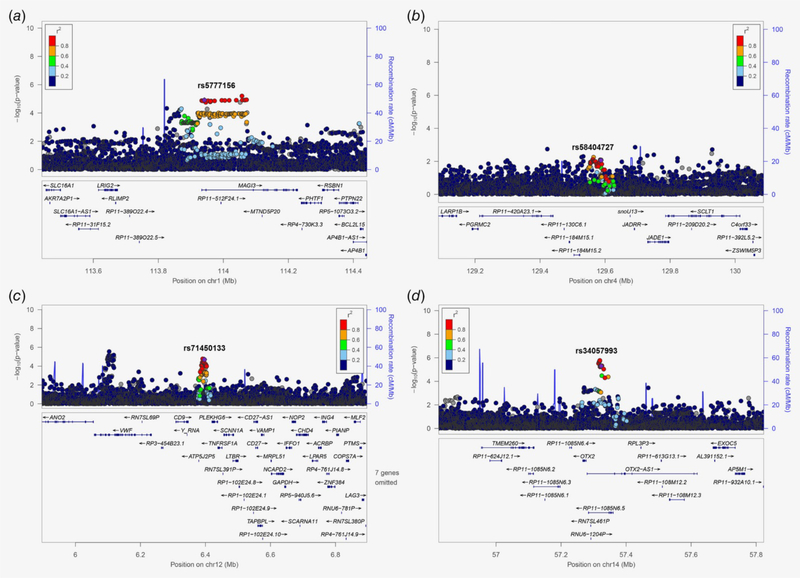
Figure 2.
Regional plots of the 4 new regions, including (a) Chr1p13.2: rs5777156, (b) Chr4q28.2: rs58404727, (c) Chr12p13.31: rs71450133 and (d) Chr14q22.3: rs34057993. The x‐axis shows the chromosomal positions and the left y‐axis shows the –log10 p values from an association test. The INDELs are shown as purple diamonds. The colors of the dots indicate the LD relationship between the most significantly associated INDELs and the remaining SNPs in the 1 Mb region. The right y‐axis shows the recombination rate between the SNPs. The genes within the region‐of‐interest are annotated with arrows indicating the direction of transcription. [Color figure can be viewed at wileyonlinelibrary.com]
INDELs in known lung cancer risk loci
The results for 15 INDELs in known lung cancer risk loci are presented in Table 2. At 15q25.1, a well‐known lung cancer susceptibility locus related to nicotine addiction, INDELs harbored the lowest p‐value (rs577626090, Deletion, OR = 1.29, 95% CI = 1.25–1.33, p = 9.91 × 10−64). INDELs also reached the significance threshold in 5p15.33 and HLA region. We validated the recently reported Oncoarray risk locus, which correlated with 11q23.3 in our analysis (rs139157129, Deletion, OR = 0.93, 95% CI =0.90–0.95, p = 1.90 × 10−7). INDELs in the known loci showed strong effects, and 10 of the 15 INDELs were still significant after Bonferroni correction (p threshold = 1.03 × 10−7).
Functional annotations of new regions
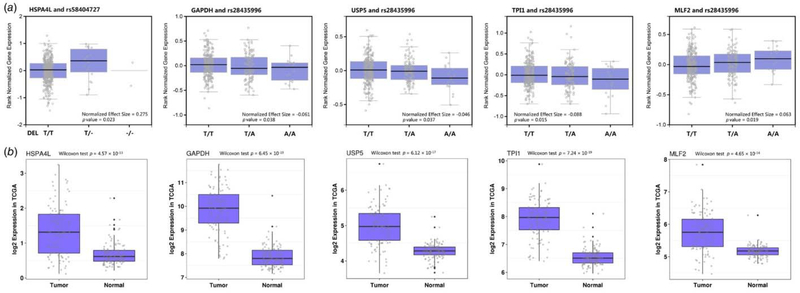
Because the underlying mechanisms of known regions have been well illustrated, we performed functional annotations on the four new loci in our study. To explore the potential functions of the INDELs, we performed eQTL and differential expression analyses based on GTEx lung tissue data and TCGA lung cancer data for these four new regions. In GTEx lung eQTL database, we identified a total of 10 genes that showed significant cis‐eQTL results (p‐value <0.05), and five of them were related to cancer in previous studies. INDEL rs58404727 was a lung cis‐eQTL variant for HSPA4L, which encodes heat shock protein family A (Hsp70) member 4 like. HSPA4L expression was significantly upregulated in lung tumor tissues compared to adjacent lung tissues (p = 4.57 × 10−13; Figure 3). For INDEL rs71450133, its tag SNP rs28435996 was associated with decreased GAPDH, TPI1, USP5 expression and increased MLF2 expression. In the differential expression analysis, GAPDH, TPI1, USP5 and MLF2 were all significantly upregulated in lung tumor tissues compared to adjacent lung tissues (Fig. 3). The full results from the cis‐eQTL and differential expression analyses are presented in Table S4.
Figure 3.
eQTL and differential expression of the INDELs among GTEx lung tissue and TCGA lung cancer data. [Color figure can be viewed at wileyonlinelibrary.com]
To identify the causal variants for the four INDELs regions, we constructed a rank scoring system based on the public functional databases. As shown in Table 3, we found that rs5777156, rs71450133 and rs34057993 were related to multiple regulatory elements (promoter histone marks, enhancer histone marks and TFBS) in multiple tissues or cell lines, while rs58404727 is located in a desert region. Furthermore, rs5777156 was located in the promoter histone marks and enhancer histone marks in the A549 EtOH 0.02pct lung carcinoma cell line in the ENCODE database; rs71450133 also showed enhancer histone marks in the A549 EtOH 0.02pct lung carcinoma cell line and in NHLF lung fibroblast primary cells in the ENCODE database. In the RegulomeDB annotation, the RegulomeDB score for rs5777156 was 3a, suggesting that rs5777156 might affect TF binding at the DNase peak. Meanwhile, rs71450133 may interact with the VWF and CD9 genes through the 3D SNP annotation. The other two INDELs did not show any functional evidence in multiple databases. For these four new signals, we also identified seven candidate causal SNPs based on the rank scoring system (Table S5). At 1p13.2, a noncoding variant, rs12567622 in MAGI3, were predicted as the causal variant. At 4q28.2, the most plausible target SNP was rs72618844, which also showed an enhancer histone mark in the A549 EtOH 0.02pct lung carcinoma cell line. At 12p13.31, the predicted causal SNPs include rs7304688, which is located in the regulatory element site in A549 EtOH 0.02pct lung carcinoma cell line. At 14q22.3, the rs10483677 SNP was a predicted causal variant. Further studies will be required to determine whether these SNPs are truly causal variants for each locus.
Table 3.
Comprehensive functional annotations for the INDELs in the new regions
| Chr. | SNP | Region | INS/DEL | Gene | Enhancer1 | Promoter1 | TFBS1 | 3D score1 | 3D Interaction gene1 | CADD2 | RegulomeDB3 | PINES4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p13.2 | rs5777156 | Intronic | Insertion | MAGI3 | 6 | 1 | 2 | 2.300 | – | 4.264 | 3a | 0.243 |
| 4q28.2 | rs58404727 | Intergenic | Deletion | RP11-184M15.2 | 0 | 0 | 0 | 1.820 | – | 3.743 | 7 | 0.499 |
| 12p13.31 | rs71450133 | Intergenic | Deletion | PLEKHG6 | 11 | 0 | 0 | 3.810 | VWF, CD9 | 6.315 | 6 | 0.089 |
| 14q22.3 | rs34057993 | Intronic | Deletion | OTX2-AS1 | 18 | 1 | 0 | 6.960 | – | 1.310 | 7 | 0.061 |
Enhancer, promoter and TFBS were obtained from 3DSNP based on the ENCODE database. 3DSNP was the overall function score and the interacting gene reflected the three-dimensional interaction genes.
CADD was used to evaluate the relative deleteriousness.
RegulomeDB was used to identify DNA features and regulatory elements in noncoding regions in the human genome.
PINES provided a powerful in silico method to prioritize and finely map the functional noncoding variants. SNPs with lower p values indicated more abundant functions.
The relationship between INDELs and SNPs
To understand the effects of the INDELs or SNPs on lung cancer risk, we examined the relationship between these two types of variations from the same loci. In the known loci, we found that most of the INDELs were in considerable LD with previously reported risk SNPs (r2: 0.5–1.0; Table S6). However, five INDELs in the HLA region did not show high LD with known risk SNPs (r2 < 0.1). We performed a conditional analysis to determine whether those five INDELs exerted independent effects from known SNPs for each locus. INDEL rs145093187 showed an independent signal after adjusting the reported SNPs through Bonferroni correction (OR = 0.86, 95% CI = 0.81–0.91, conditional p = 5.10 × 10−8), while other the INDELs did not reach the suggestive threshold (Table S7). For the new loci, the regional plots provide the LD relationship between the INDELs and SNPs at a 1 Mb window (Fig. 2). We found that although INDELs showed a strong effect on lung cancer risk, there were still SNPs with high LD (r2 > 0.8) showing a stronger effect. We also conducted conditional analysis on the INDELs and top SNPs in each locus. By adding the SNP with the lowest p‐value into the model for each locus, neither the INDEL nor the SNP showed a significant signal (Table S8). Meanwhile, we also performed conditional analyses on the four candidate causal SNPs and four new INDELs in each locus. When we added the candidate causal SNPs to the model, the INDELs showed stronger effects at the statistical level. In half of the four conditional analyses, the INDELs remained nominally significant (p < 0.05; Table S8).
Discussion
In our study, we conducted a genome‐wide meta‐analysis with 23,202 cases and 19,048 controls to systematically explore the associations between INDELs and lung cancer risk. We identified 19 signals for lung cancer risk, and four of them were first reported in lung cancer.
INDEL rs5777156 is an insertion lying in the MAGI3 intron at 1p13.2. MAGI3 acts as a scaffolding protein at cell–cell junctions, regulating various cellular and signaling processes, such as the Ras signaling pathway and PTEN pathway. Previous studies showed that MAGI3 could downregulate Wnt/β‐catenin signaling, suppressing malignant glioma cell phenotypes,33 and competes with NHERF‐2 to negatively regulate LPA2 receptor signaling in colon cancer cells.34 Additionally, INDEL rs5777156 and the predicted causal variant were all present in regulatory elements, including promoter and enhancer histone marks in a lung carcinoma cell line based on the ENCODE database, suggesting that rs5777156 may affect lung cancer risk through transcript regulation.
Our study also identified a new risk locus at 4q28.2 marked by INDEL rs58404727 mapping to 65 kb upstream of RP11‐184M15.2, which is a lncRNA with little functional evidence. However, the predicted causal variant SNP rs72618844 showed promoter and enhancer histone marks in A549 lung carcinoma cell line. INDEL rs58404727 may be a tagging signal at this locus, while rs72618844 affects lung cancer risk.
INDEL rs71450133 is a deletion that maps to 23 kb upstream of PLEKHG6 at 12p13.31. Genetic variants at 12p13.31 have been shown by previous studies to be associated with colorectal cancer risk in East Asians.35 Although the function of PLEKHG6 in tumors is unclear, some studies showed that PLEKHG6 might regulate the invasion activity of breast cancer cells.36, 37 In the eQTL analyses, rs71450133 was associated with the expression of several genes, and four of them were tumor related. GAPDH encodes a member of the glyceraldehyde‐3‐phosphate dehydrogenase protein family and can interact with proteins participating in DNA repair.38 USP5, namely ubiquitin specific peptidase 5, plays an important role in ubiquitination. USP5 expression has been proven to be associated with several cancer types, such as hepatocellular carcinoma, glioblastoma and pancreatic cancer.39–41 Previous studies have shown that USP5 had many cellular targets and stabilizes multiple proteins, such as p53.42 TPI1, triosephosphate isomerase 1, encodes a crucial enzyme in the carbohydrate metabolism, and previous studies have shown its expression level might be associated with several cancer types.43, 44 Another gene, MLF2 or Myeloid Leukemia Factor 2, is related to myeloid leukemia and leukemia, and MLF2 knockdown may reduce tumor initiation and metastasis in breast cancer.45 Functional annotation based on ENCODE suggested that rs71450133 and its high LD SNPs are located in regulatory elements in A549 EtOH 0.02pct lung carcinoma cell line.
Another new susceptibility locus, 14q22.3, was marked by INDEL rs34057993, which is a deletion located in the intron of noncoding RNA OTX2‐AS1, an OTX2 antisense RNA at 14q22.3. OTX2, which encodes a member of the bicoid subfamily of homeodomain‐containing transcription factors, has been implicated as a potential driver of medulloblastoma tumorigenesis.46, 47 Although rs34057993 and its LD SNPs did not show any promoter or enhancer histone marks, genes associated with INDEL rs34057993 were cancer‐related, it is possible that rs34057993 may act by regulating the expression of genes to influence lung cancer risk.
In our study, we found four novel risk loci for lung cancer, as well as illustrated the relationships between INDELs and SNPs. In the reported regions, most of the significant INDELs were correlated with previously reported SNPs, especially in 5p15.33 and 15q25.1. In the HLA region, we found a novel signal that was independent of the previously reported SNPs. Considering the complex LD and haplotype structure in the HLA region,48 the novel INDEL may be a true association. In the new regions, we also observed INDELs that did not harbor the lowest p values and showed high LD with nearby SNPs. The effects of the INDELs were decreased after adjusting for the top SNP in each region. This suggests that the presented SNPs promote more stable effects in both known and new regions. However, it is generally assumed that SNPs with the most significant signal usually tag causal variants with a small effect. After conducting conditional analysis on seven potential causal SNPs, we found that the INDELs in the new loci were still nominally significant. Thus, it is possible that the INDELs may also be both causal and tagging variants. The combination of these variants with small effects together could lead to lung cancer. The functional annotation results confirmed our insights. In the new region, two INDELs, rs5777156 and rs34057993, showed enhancer histone marks in regulatory regions, which may influence enhancer activity in lung cancer. However, the most significant SNPs in those two regions did not show strong functional evidence. This means INDELs could also be a causal variant, which could regulate gene expression and affect the risk of lung cancer. The comprehensive annotation of each locus also identified potential causal variants in high LD with the INDELs. Interestingly, we noticed that all 19 significant INDELs mapped to the noncoding region (intronic or intergenic region). INDELs in the coding region can result in frameshift and nonframeshift mutations, which are relatively severe mutations and more likely to be observed in Mendelian diseases or tumors.9, 11 Overall, the limitation of the present study is that we only evaluated the functional evidence from available databases for the identified INDELs, further functional experiments are needed to better understand INDEL mechanisms in lung cancer carcinogenesis.
In conclusion, we performed a large‐scale case–control study to evaluate INDELs and their risk for lung cancer, and four new risk loci at 1p13.2, 4q28.2, 12p13.31 and 14q22.3 were identified. Our findings indicate that INDELs could be potentially functional genetic variants for lung cancer risk.
Supplementary Material
Acknowledgements
We thank the study participants and research staff for their contributions and commitment to our study. This work was supported by the National Natural Science of China (81521004, 81820108028), the Priority Academic Program for the Development of Jiangsu Higher Education Institutions [Public Health and Preventive Medicine], Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067) and the National Institutes of Health and the National Cancer Institute (CA209414 and CA092824 to D.C.C.).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–86. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3.Sampson JN, Wheeler WA, Yeager M, et al. Analysis of heritability and shared heritability based on genome‐wide association studies for thirteen cancer types. J Natl Cancer Inst 2015; 107: djv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai J, Shen W, Wen W, et al. Estimation of heritability for nine common cancers using data from genome‐wide association studies in Chinese population. Int J Cancer 2017; 140: 329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosse Y, Amos CIA. Decade of GWAS results in lung cancer. Cancer Epidemiol Biomark Prev 2018; 27: 363–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills RE, Luttig CT, Larkins CE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 2006; 16: 1182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullaney JM, Mills RE, Pittard WS, et al. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet 2010; 19: R131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auton A, Brooks LD, Durbin RM, et al. Abecasis GR. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stenson PD, Mort M, Ball EV, et al. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye K, Wang J, Jayasinghe R, et al. Systematic discovery of complex insertions and deletions in human cancers. Nat Med 2016; 22: 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann TJ, Van Den Eeden SK, Sakoda LC, et al. A large multiethnic genome‐wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov 2015; 5: 878–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao R, Hu S, Wang S, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis 2015; 36: 1136–43. [DOI] [PubMed] [Google Scholar]
- 14.Sun T, Gao Y, Tan W, et al. Lin D. a six‐nucleotide insertion‐deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet 2007; 39: 605–13. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Gramling S, Munoz D, et al. Two novel BRM insertion promoter sequence variants are associated with loss of BRM expression and lung cancer risk. Oncogene 2011; 30: 3295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKay JD, Hung RJ, Han Y, et al. Large‐scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet 2017; 49: 1126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landi MT, Chatterjee N, Yu K, et al. A genome‐wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009; 85: 679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Wu C, Shi Y, et al. A genome‐wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011; 43: 792–6. [DOI] [PubMed] [Google Scholar]
- 19.Amos CI, Dennis J, Wang Z, et al. The OncoArray consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol Biomark Prev 2017; 26: 126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Cirulli ET, Han Y, et al. Systematic assessment of imputation performance using the 1000 genomes reference panels. Brief Bioinform 2015; 16: 549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y, Wang C, Zhu M, et al. Targeted sequencing of chromosome 15q25 identified novel variants associated with risk of lung cancer and smoking behavior in Chinese. Carcinogenesis 2017; 38: 552–8. [DOI] [PubMed] [Google Scholar]
- 22.Dong J, Cheng Y, Zhu M, et al. Fine mapping of chromosome 5p15.33 identifies novel lung cancer susceptibility loci in Han Chinese. Int J Cancer 2017; 141: 447–56. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michailidou K, Lindström S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017; 551: 92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9:e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Quan C, Chen H, et al. 3DSNP: a database for linking human noncoding SNPs to their three‐dimensional interacting genes. Nucleic Acids Res 2017; 45: D643–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodea CA, Mitchell AA, Bloemendal A, et al. Phenotype‐specific information improves prediction of functional impact for noncoding variants. bioRxiv 2016; 19: 173. [Google Scholar]
- 30.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early‐onset inflammatory bowel disease. Nat Genet 2009; 41: 1335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013; 340: 1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Q, Yang Y, Feng D, et al. MAGI3 negatively regulates Wnt/beta‐catenin signaling and suppresses malignant phenotypes of glioma cells. Oncotarget 2015; 6: 35851–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee SJ, Ritter SL, Zhang H, et al. MAGI‐3 competes with NHERF‐2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology 2011; 140: 924–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Jia WH, Matsuda K, et al. Large‐scale genetic study in east Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 2014; 46: 533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu D, Asiedu M, Wei Q. Myosin‐interacting guanine exchange factor (MyoGEF) regulates the invasion activity of MDA‐MB‐231 breast cancer cells through activation of RhoA and RhoC. Oncogene 2009; 28: 2219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu D,Haruta A, Wei Q. GIPC1 interacts with MyoGEF and promotes MDA‐MB‐231 breast cancer cell invasion. J Biol Chem 2010; 285: 28643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosova AA, Khodyreva SN, Lavrik OI. Role of Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) in DNA repair. Biochemistry 2017; 82: 643–54. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Wang WM, Lu YF, et al. Usp5 functions as an oncogene for stimulating tumorigenesis in hepatocellular carcinoma. Oncotarget 2017; 8: 50655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izaguirre DI, Zhu W, Hai T, et al. PTBP1‐dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog 2012; 51: 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XY, Wu HY, Mao XF, et al. USP5 promotes tumorigenesis and progression of pancreatic cancer by stabilizing FoxM1 protein. Biochem Biophys Res Commun 2017; 492: 48–54. [DOI] [PubMed] [Google Scholar]
- 42.Dayal S, Sparks A, Jacob J, et al. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem 2009; 284: 5030–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang H, Ma N, Shang Y, et al. Triosephosphate isomerase 1 suppresses growth, migration and invasion of hepatocellular carcinoma cells. Biochem Biophys Res Commun 2017; 482: 1048–53. [DOI] [PubMed] [Google Scholar]
- 44.Linge A, Kennedy S, O’Flynn D, et al. Differential expression of fourteen proteins between uveal melanoma from patients who subsequently developed distant metastases versus those who did not. Invest Ophthalmol Vis Sci 2012; 53: 4634–43. [DOI] [PubMed] [Google Scholar]
- 45.Dave B, Granados‐Principal S, Zhu R, et al. Targeting RPL39 and MLF2 reduces tumor initiation and metastasis in breast cancer by inhibiting nitric oxide synthase signaling. Proc Natl Acad Sci USA 2014; 111: 8838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adamson DC, Shi Q, Wortham M, et al. OTX2 is critical for the maintenance and progression of Shh‐independent medulloblastomas. Cancer Res 2010; 70: 181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bunt J, Hasselt NE, Zwijnenburg DA, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer 2012; 131: E21–32. [DOI] [PubMed] [Google Scholar]
- 48.de Bakker PI, Raychaudhuri S Interrogating the major histocompatibility complex with high‐throughput genomics. Hum Mol Genet 2012; 21: R29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.