Abstract
Influenza pandemics are complex events that have occurred frequently throughout human history, three during the past century alone. Now the world is facing the first 21st century pandemic, and the comparison among them is essential to identify common epidemiologic patterns, clinical characteristics, and outcomes. The evolution of medicine, including diagnostic and treatment options, the critical care advances, and global responses are new interventions that could modify the general outcome of the pandemic. Learning from past and current events could lead to a plan for prompt and efficient response in future pandemics and may be help us to predict the unpredictable.
Keywords: H1N1 Influenza, 2009 pandemic, Influenza epidemics, Influenza ARDS, Influenza critically ill
Introduction
Influenza pandemics have occurred frequently throughout history, and are associated with high rates of morbidity, social disruption, and economic losses [1, 2]; given the historical precedents, the current epidemiologic and ecologic circumstances, and the knowledge about influenza viruses, it was reasonable to assume that additional pandemics would occur [3•]. In 2009, the prediction became real and now the world is facing the first 21st century influenza pandemic [4].
The previous pandemics experiences represent an extraordinary opportunity for a better understanding of the epidemiology and clinical biology of the disease. The aim of this article is to describe the major data generated during the last century pandemics in contrast to the current one.
Epidemiology
The “Spanish flu” pandemic of 1918–1919 was exceptionally severe, with a mortality rate among the infected of more than 2.5%. It became one of the deadliest public health crises in human history, with about 50–100 million deaths worldwide. The outbreaks of the disease swept not only North America and Europe but spread as far as the Alaskan wilderness and the most remote islands of the Pacific. Large proportions of the population became ill; 28% of the US population was probably infected [5].
The pandemic developed during World War I, so the Army and Navy camps suffered severely from the outset. Rarely before in the history of war has infection exhibited a more explosive character or have so large a proportion of troops been infected in camps. The epidemic resulted in an unusually high fatality among the troops. From September 12 to October 18, 1918, 274,745 cases of influenza, 46,286 pneumonias, and 14,616 deaths were reported among the troops in the United States [3].
The influenza pandemic of 1918–1919 was caused by a recombinant influenza A H1N1 virus that was highly virulent. The sequences from the coding regions of hemagglutinin, neuraminidase, nucleoprotein, and matrix proteins have revealed that the virus belongs to the subgroup of strains that infect humans and swine [5, 6•].
The disease manifestations in 1918 and 1919 varied from the common cold to pneumonia. The leading symptoms were sudden-onset, severe headache, chills, back pain, fever, prostration, and neurologic and gastrointestinal disturbances. Most people improved with only supportive care. The virus had a considerably varied period of incubation—in some patients, infections as short as a day or two—and the disease was transmissible prior to the patient developing symptoms [3, 7••].
In some cases an aggressive, fatal bronchopneumonia associated with progressive cyanosis and collapse occurred and had a high mortality in healthy, young adults. The impact was so profound that the average US life expectancy dropped by more than 10 years [5].
The pandemic spread in three rapidly occurring waves over a 9-month period before settling into an annual pattern of seasonal recurrence. The first wave of the 1918 pandemic, as measured by pneumonia and influenza mortality, occurred in only a few, mostly Northern European, countries in the summer of 1918 (July and August), and was associated with modest mortality. The two following waves, seen in most of the world during summer to autumn of 1918 and winter of 1918–1919, were more deadly than the first. Although most of the world was afflicted during the three successive waves of influenza, a few insular regions, mainly in the southwestern Pacific, did not experience the pandemic [8].
The 1957 and 1968 pandemics, which resulted from reassortment between circulating descendants of the 1918 human virus and circulating avian influenza strains, were not as deadly as the 1918 pandemic [9].
During the H2N2 Asian influenza pandemic of 1957, the first virus was isolated in China in February. Subsequently, the virus spread to Australia and Southeast Asia by June, and reached the rest of Europe and South America by July–September. By December 1957, every continent had been infected [10].
The 1968 (Hong Kong) H3N2 influenza pandemic occurred in two waves. The virus spread from China in July, but the explosive outbreak was delayed by 6 months. The second wave was more severe and the virus was more transmissible than it had been in the first wave [9].
During 1957 and 1968, pandemics a substantial increase in hospitalizations and deaths from respiratory illnesses and pneumonias were reported. A case series analysis revealed that the most frequently isolated bacteria from patients with post-influenzal pneumonias were Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus, and “pure” influenza pneumonia was considered the cause of death in 20% of the fatal cases [10, 11].
On April 23, 2009, the Mexican government issued an epidemic alert notifying the world of a new influenza pandemic. The new virus spread quickly around the world and in June 2009, the World Health Organization (WHO) declared the start of the first 21st century pandemic [4]. As of January 17, 2010, worldwide more than 209 countries and overseas territories or communities have reported laboratory-confirmed cases of pandemic influenza H1N1 2009, including at least 14,142 deaths [12].
The 2009 H1N1 influenza pandemic virus is a multiple reassortant hybrid virus containing swine, human, and avian influenza viral genes, generated by two well-established swine influenza virus lineages: a North American descendant of the 1918 human virus that has long circulated in pigs, and a swine virus lineage derived from complete genome adaptation of an avian virus [11, 13••].
Most human infections with pandemic H1N1 2009 viruses result in a self-limiting, uncomplicated, or even asymptomatic disease with a clinical course similar to that observed for seasonal influenza, but also atypical clinical signs such as vomiting and diarrhea. Severe complications and deaths have occurred in individuals with chronic health conditions, obesity, respiratory diseases, and pregnancy and even among healthy young people [14, 15•]. In contrast to the pandemics of the last century, although bacterial colonization has been described in hospitalized patients, bacterial pneumonia complications are rare [13••, 14••, 15•, 16].
The WHO has reported that many countries in the northern hemisphere have passed the peak of their second wave, whereas in others intense influenza activity is still reported [12].
Mexico’s general epidemic curve has two defined waves; during the second wave, which occurred 6 months after the first, many more cases were reported and confirmed, and the mortality rates were higher than during the first wave. However, the influenza activity has not been homogenous within the country. The peak of the epidemic curves among different states differed by several weeks (Fig. 1), an important factor to consider when implementing national public-health policies. For example, at the beginning of the pandemic in Mexico, a national contingency plan was instituted even in regions were the disease activity was low or nonexistent. This approach is costly and could be inefficient if the rate of infection and morbidity is different in different regions.
Fig. 1.
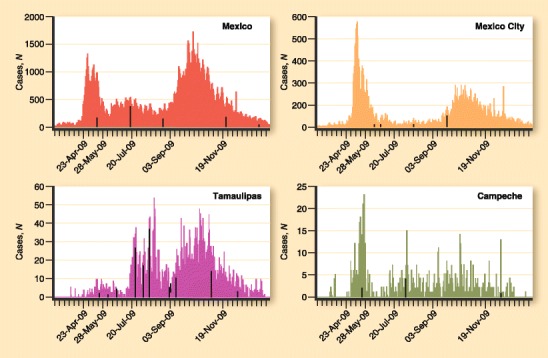
Epidemic curves during the pandemic in Mexico: comparison of general curve versus three different states
The WHO continues to assess as moderate the impact of the influenza pandemic. Accurate assessments of mortality and mortality rates will likely be possible only 1–2 years after the pandemic has peaked, and will rely on methods similar to those used to calculate excess mortality during seasonal influenza epidemics [17].
Diagnosis
The evolution of diagnostic methods during the current pandemic has allowed a more accurate epidemiologic surveillance; however, the usefulness of existing tests in clinical decisions—such as whether to start antiviral drug treatment—is limited because of their general availability, cost, complexity, or low sensitivities. Laboratory identification of human influenza virus infections is commonly performed using direct antigen detection, virus isolation in cell culture, or detection of influenza-specific RNA.
The viral culture provides information regarding influenza subtypes, strains, and potential drug resistance, but because of the cost and technical complexity, is recommended only for research and surveillance purposes [18••].
The sensitivity of influenza antigen detection in respiratory specimens by immunofluorescent staining in comparison to cell culture ranges between 70% to 100%, with a general specificity 80% to 100%, positive predictive value 85% to 94%, and negative predictive value 96% to 100%. It is a complex method that requires special equipment and training, and false-negative results were reported during the 2009 pandemic [19].
In recent years, commercial influenza rapid diagnostic tests became available. These are mostly antigen detection tests, which can produce results within 30 min, but their performance characteristics vary according to assay type, sample, and time of collection [20]. During the 2009 influenza pandemic the sensitivities of diverse rapid influenza antigen tests for pandemic influenza A/H1N1 virus were low to moderate (<55%) and their specificity ranged from 86% to 100%. Thus, the low sensitivity of rapid antigen tests for H1N1 infection tests makes them relatively impractical in patients with influenza-like illness because further testing will frequently be required to make a diagnosis [21, 22].
The real-time reverse-transcriptase polymerase chain reaction (RT-PCR) is the method of choice, relying on the amplification of specific sequences of the genome of A H1N1 viruses. RT-PCR is expensive and requires specific expertise and batch testing. The rising numbers of A H1N1 infection could generate genetic variation, modifying the accuracy of the test; other variables that could change the sensitivity are the time after symptom onset and viral load (<3 days) [23, 24].
Routine testing for pandemic influenza is costly, demanding, and beyond the reach of most of the countries that are affected. Clinical diagnosis has been the only universally available method used to make rapid diagnoses and to initiate treatment decisions during all the influenza pandemics. During periods of influenza prevalence, clinical diagnosis (based on typical presentations) can be highly predictive of influenza [25].
Finally, the histopathologic diagnosis is not specific. Pulmonary pathologic changes found during the 1918 and 2009 pandemics are similar. During the 1918–1919 pandemic, acute aggressive bronchopneumonia featuring epithelial necrosis, microvasculitis/vascular necrosis, hemorrhage, and edema were reported [26]. Recently, Mauad et al. [27•] described the first autopsy report detailing the pulmonary pathology of the 2009 H1N1 pandemic infection: 95.2% (20/21) of patients had severe diffuse alveolar damage with varying degrees of alveolar hemorrhage, necrotizing bronchiolitis, and tracheobronchitis.
Management
The availability of therapeutic options such as antiviral agents and antibiotics, mechanical ventilation, and intensive care supportive measures could play a central role in the epidemiology and outcomes of the current pandemic.
Antiviral Agents
Neuraminidase, one of the glycoproteins on the surface of the influenza virus, is an enzyme that cleaves sialic acid residues from receptors for the virus, enabling the virus to spread throughout the body. Inhibition of this enzyme stops this spread and effectively curtails the infection. Two inhibitors specific for influenza virus neuraminidase are currently being used to control influenza infections (oseltamivir and zanamivir), and two others were under development [28].
The 2009 H1N1 virus is susceptible to neuraminidase inhibitors, but resistant to the adamantines. Observational studies support the benefit of neuraminidase inhibitors in reducing complications, including deaths, among hospitalized patients with 2009 pandemic influenza A (H1N1) [14••, 15•, 16, 29••].
Although most 2009 H1N1 hospitalized patients have been treated with oral oseltamivir, including critically ill patients, there are no comparative studies evaluating pharmacokinetic and clinical response. The doses and duration of treatments have been varied, and some experts have recommended duplicate doses [30••].
On October 23, 2009, Food and Drug Administration Commissioner Margaret Hamburg issued an Emergency Use Authorization for peramivir, a neuraminidase inhibitor that may be effective in selected adult and pediatric hospitalized patients [31]. Parenteral neuraminidase inhibitors (peramivir, zanamivir) might be beneficial for some patients, but clinical trials are needed to further evaluate the effectiveness of these drugs.
The main side effects associated with antiviral drugs are gastrointestinal and include nausea, vomiting, and diarrhea. Bronchospasm can occur with zanamivir, which is a powder for inhalation. It should not be delivered via nebulization because of the presence of lactose, which may compromise ventilator function [32]. We have treated 3,000 ambulatory patients with oseltamivir or zanamivir: The primary side effects reported in our population are nausea and anorexia. Patients also report difficulty using zanamivir. None of our patients have required in-hospital treatment after the initiation of antivirals. More than 100 critically ill patients have been treated with oseltamivir in a double dose for 10 days, and the secondary adverse effects have been similar to those patients with the standard doses.
A major public health concern is the emergence of resistant virus, which could be either associated with sequence changes in the hemagglutinin or with sequence changes affecting the catalytic site on the neuraminidase. Oseltamivir resistance to influenza virus has been a noted in some regions. H1N1 viruses with oseltamivir resistance due to N1 neuraminidase mutation H274Y increased from 3% during 2007–2008 influenza season to 97% before the beginning of the 2009 pandemic. The emergence of oseltamivir-resistant 2009 H1N1 virus has been rarely identified, up to September 24, 2009, 28 resistant viruses have been detected worldwide, characterized and reported to WHO. Twelve of these drug-resistant viruses were from people using oseltamivir for post-exposure prophylaxis. Six were from patients with severe immunosuppression using oseltamivir for treatment. Four were from other patients receiving oseltamivir treatment. Two were from patients who were not taking oseltamivir for either treatment or prophylaxis. In December 2009, WHO reported two clusters of patients infected with oseltamivir-resistant H1N1 viruses. Both clusters, detected in Wales, the United Kingdom, and North Carolina (USA), occurred in a single ward in a hospital, and both involved patients whose immune systems were severely compromised or suppressed. All of the resistant viruses carried the same H275Y mutation, indicating resistance to oseltamivir but susceptibility to the second antiviral drug, zanamivir. Undoubtedly, H1N1 antiviral drug resistance will be a growing problem [33–35].
ICU Management and Outcomes
About 25% of the hospitalized patients have required ICU management with a wide range of mortality rates (7%–58%) [30••, 36, 37••, 38, 39].
Mechanical ventilation and critical care management might be the most important differences among the pandemics. In 1916 Severy and in 1926 Schwake built the first negative-pressure ventilators, but these machines required the patient to stand. Although they incorporated mechanical elements, their practicality for severely ill patients was limited. The first negative pressure ventilator to be used successfully in clinical practice on a widespread basis was the Drinker-Shaw “iron lung” developed in 1928. Positive-pressure ventilation was reintroduced in 1950 after the experience generated with positive-pressure compression devices in 1927. During World War II, the first volume ventilator was introduced. Since then, mechanical ventilation has undergone continuous evolution, with the introduction of multiple new modes and applications [40].
During the pandemics the most frequent serious complications of influenza are pulmonary and include primary viral pneumonia, secondary bacterial pneumonia, pneumonia attributable to unusual pathogens, and exacerbations of chronic underlying diseases [41].
The main causes of death have been hypoxemia, secondary infections, sepsis, and severe acute respiratory distress syndrome (ARDS) [42, 43]. Several studies have described critically ill patients with Pandemic H1N1 2009 Influenza A Virus infection [30••, 36, 37••, 38, 39], which have included cases of rapidly progressive lower respiratory tract disease resulting in respiratory failure, development of ARDS, and prolonged intensive care unit (ICU) admission. Perez-Padilla et al. [14••] reported the first 18 hospitalized patients in Mexico City, with H1N1 2009 Influenza A pandemic infection, 10 patients developed respiratory distress requiring intubation and mechanical ventilation with a mortality rate of 58%.
After evaluating the most relevant series of critically ill patients 70% required mechanical ventilation, 38% vasopressors, 7% prone position, and 5% renal replacement therapy [41]. The mortality rates have been variable. Dominguez-Cherit et al. [30••] described 58 critically ill patients with 2009 influenza A H1N1 virus infection, during the first wave in Mexico. Of these, 24 (41.4%) died. Series reported from the United States, Canada, and Spain, reported a mortality rates of 7%, 17% and 18.7%, respectively [36, 37••, 38]. The risk factors identified in the series were similar; therefore the outcomes differences might be associated with the time between the symptoms onset and the initial treatment.
The use of some protective ventilator strategies has lowered the general ventilation mortality rate. The strategies in patients with ARDS include target tidal volume of 6 mL/kg (predicted body weight) and the initial upper limit goal for plateau pressures ≤30 cm H2O. Invasive ventilation, with a lung-protective strategy, has been recommended as the initial approach for the management of ARDS patients with Pandemic H1N1 Influenza [29••, 44, 45].
Hydrocortisone should be considered in patients with refractory septic shock [46, 47] and should be given in a dose of 200 mg/d in four divided doses or as a bolus of 100 mg followed by a continuous infusion (240 mg/d) [48••]. Marik et al. [48••] investigated the efficacy of low doses of corticosteroids in septic shock patients with or without early ARDS, showing that a 7-day treatment with low doses of corticosteroids was associated with better outcomes in septic shock-associated early ARDS non-responders, but not in responders or in septic shock patients without ARDS. Nonrandomized, controlled studies have been done addressing the utility of steroids in patients with refractory septic shock and ARDS related to Pandemic influenza A H1N1 virus infection.
A large proportion of critically ill patients with pandemic Influenza A H1N1 virus infection who survived required mechanical ventilation and rescue therapies such as high-frequency oscillatory ventilation, prone positioning, neuromuscular blockade, inhaled nitric oxide, and extracorporeal membrane oxygenation [30••, 36, 37••, 38, 39].
Conclusions
Influenza is one of the oldest known epidemic diseases, yet hardly any other infectious disease is more difficult to study. Pandemics have been similar in several aspects, yet each has unique characteristics probably based on the complexity of the viral-host interactions. To predict the next pandemic, we should think about the feasibility of predicting the unpredictable. Many fundamental questions remain unsolved. During the 21st century, pandemic medical progress in antiviral medication and management of critically ill patients has been evident. In addition to availability of antiviral drugs, critical care, and multidisciplinary management teams, more timely and widespread public health interventions might be the key to better outcomes. Learning from the past is necessary to identify the most effective management strategies and to prepare for the future, unavoidable, next pandemic.
Acknowledgments
The authors wish to thank Hugo Lopez-Gatell, MD, PhD, and the subsecretary of prevention and promotion from the Health Minister, for the prevalence information.
Disclosure
No potential conflict of interest relevant to this article was reported.
Contributor Information
Guillermo Domínguez-Cherit, Phone: +52-55-54870900, FAX: +-52-55-56552510, Email: guidom@prodigy.net.mx.
Silvio A. Ñamendys-Silva, Phone: +52-55-54870900, FAX: +52-55-56552510, Email: snamendys@incan.edu.mx
Alethse de la Torre, Phone: +52-55-55738803, FAX: +52-55-55133501, Email: alethse@hotmail.com.
Alejandro E. Macias, Phone: +52-55-55738803, FAX: +52-55-55133501, Email: aaeehhmm@yahoo.com
Jose Angel Cordova-Villalobos, Phone: +52-55-55738803, FAX: +52-55-55133501, Email: jacordova@salud.gob.mx.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
- 1.Perlroth DJ, Glass RJ, Davey VJ, et al. Health outcomes and costs of community mitigation strategies for an influenza pandemic in the United States. Clinical Infectious Diseases. 2010;50:165–174. doi: 10.1086/649867. [DOI] [PubMed] [Google Scholar]
- 2.Fraser C, Donnelly CA, Cauchemez S, et al. (H1N1): Early findings pandemic potential of a strain of influenza A. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soper GA. The influenza-pneumonia pandemic in the American army camps during September. Science. 1918;48:451–456. doi: 10.1126/science.48.1245.451. [DOI] [PubMed] [Google Scholar]
- 4.Chan M. Statement by World Health Organization Director-General, Dr Margaret Chan. 11 June 2009. Available at http://www.who.int/mediacentre/influenzaAH1N1_presstranscript_20090611.pdf. Accessed 12 June 2009.
- 5.Taubenberger JK, Reid AH, Krafft A, et al. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 6.• Tumpey TM, Basler CF, Aguilar PV, et al.: Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 2005, 310, 7:77–80. Experimental evidence about the virulence, lethality and pathogenicity of the 1918 virus. [DOI] [PubMed]
- 7.Soper GA. The lessons of the pandemic. Science. 1919;49:501–506. doi: 10.1126/science.49.1274.501. [DOI] [PubMed] [Google Scholar]
- 8.Brown P, Gajdusek DCE, Morris JA. Virus of the 1918 influenza pandemic era: new evidence about its antigenic character. Science. 1969;166:117–119. doi: 10.1126/science.166.3901.117. [DOI] [PubMed] [Google Scholar]
- 9.Jackson C, Vynnycky E, Mangtani P. Estimates of the transmissibility of the 1968 (Hong Kong) influenza pandemic: evidence of increased transmissibility between successive waves. Am J Epidemiol. 2010;171:465–478. doi: 10.1093/aje/kwp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oxford JS, Sefton A, Jackson R, et al. World War I may have allowed the emergence of “Spanish” influenza. Lancet Infect Dis. 2002;2:111–114. doi: 10.1016/S1473-3099(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Harvey HA, Memoli MJ. The 1918 influenza pandemic: lessons for 2009 and the future. Crit Care Med. 2010;38(Suppl):S1–S11. doi: 10.1097/CCM.0b013e3181ceb25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO Pandemic (H1N1) 2009: update 84. Available at http://www.who.int/csr/don/2010_01_22/en/index.html. Accessed February 2010.
- 13.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;361:1–10. doi: 10.1056/NEJMp0903765. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Padilla R, de la Rosa-Zamboni D, de Leon Ponce S, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;13:680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 15.Jamieson DJ, Honein MA, Rasmussen SA, et al. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet. 2009;374:451–458. doi: 10.1016/S0140-6736(09)61304-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen KF, Gaydos C, Rothman RE. Update on emerging infections: news from the Centers for Disease Control and Prevention. Hospitalized patients with novel influenza A (H1N1) virus infection-California, April–May, 2009. Ann Emerg Med. 2009;54:732–736. doi: 10.1016/j.annemergmed.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO: Comparing deaths from pandemic and seasonal influenza. Available at http://www.who.int/csr/disease/swineflu/notes/briefing_20091222/en/index.html. Accessed February 2010.
- 18.•• WHO: Information for laboratory diagnosis of pandemic (H1N1) 2009 virus in humans- revised. Available at http://www.who.int/csr/resources/publications/swineflu/WHO_Diagnostic_RecommendationsH1N1_20090521.pdf. Accessed February 2010. The WHO´s guidelines about different diagnostic methods for influenza pandemic 2009, indications, and interpretation.
- 19.Kapelusznika L, Patela R, Jaoa J. Severe pandemic (H1N1) 2009 influenza with false negative direct fluorescent antibody assay: case series. J Clin Virol. 2009;46:279–281. doi: 10.1016/j.jcv.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Gavin PJ, Thomson RB. Review of rapid diagnostic tests for influenza. Clin Appl Immunol Rev. 2003;4:151–172. doi: 10.1016/S1529-1049(03)00064-3. [DOI] [Google Scholar]
- 21.Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (swine) influenza A/H1N1. Clinical Infectious Diseases. 2009;49:1090–1093. doi: 10.1086/644743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginocchio CC, Zhang F, Manji R, et al. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 2009;45:191–195. doi: 10.1016/j.jcv.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabbaraju K, Wong S, Wong AA, et al. Design and validation of real-time reverse transcription-PCR assays for detection of pandemic (H1N1) 2009 virus. J Clin Microbiol. 2009;47:3454–3460. doi: 10.1128/JCM.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng PKC, Wong KKY, Mak GC, et al. Performance of laboratory diagnostics for the detection of influenza A (H1N1) virus as correlated with the time after symptom onset and viral load. J Clin Virol. 2010;47:182–185. doi: 10.1016/j.jcv.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 25.Monto AS, Gravenstein S, Elliott M, et al. Clinical signs and symptoms predicting influenza infection. Arch Intern Med. 2000;160:3243–3247. doi: 10.1001/archinte.160.21.3243. [DOI] [PubMed] [Google Scholar]
- 26.Starr Isaac. Influenza in 1918: Recollections of the Epidemic in Philadelphia. Ann Intern Med. 2006;145:138–140. doi: 10.7326/0003-4819-145-2-200607180-00132. [DOI] [PubMed] [Google Scholar]
- 27.Mauad T, Hajjar LA, Callegari GD, et al. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med. 2010;181:72–79. doi: 10.1164/rccm.200909-1420OC. [DOI] [PubMed] [Google Scholar]
- 28.Jefferson T, Demicheli V, Rivetti D, et al. Antivirals for influenza in healthy adults: systematic review. Lancet. 2006;367:303–313. doi: 10.1016/S0140-6736(06)67970-1. [DOI] [PubMed] [Google Scholar]
- 29.•• WHO: Clinical management of human infection with pandemic (H1N1) 2009: revised guidance. Available at http://www.who.int/csr/resources/publications/swineflu/clinical_management/en/. Guidelines and recommended management of the patients infected with the H1N! virus. Accessed February 2010.
- 30.Domínguez-Cherit G, Lapinsky SE, Macias AE, et al. Critically Ill patients with 2009 influenza A(H1N1) in Mexico. JAMA. 2009;302:1880–1887. doi: 10.1001/jama.2009.1536. [DOI] [PubMed] [Google Scholar]
- 31.Birnkrant D, Cox E. The emergency use authorization of peramivir for treatment of 2009 H1N1 influenza. N Engl J Med. 2009;361:2204–2207. doi: 10.1056/NEJMp0910479. [DOI] [PubMed] [Google Scholar]
- 32.Long JK, Mossad SB, Goldman MP. Antiviral agents for treating influenza. Cleve Clin J Med. 2000;67:92–95. doi: 10.3949/ccjm.67.2.92. [DOI] [PubMed] [Google Scholar]
- 33.Kawai N, Ikematsu H, Hirotsu N, et al. Clinical effectiveness of oseltamivir and zanamivir for treatment of influenza A virus subtype H1N1 with the H274Y mutation: a Japanese, multicenter study of the 2007–2008 and 2008–2009 influenza seasons. Clin Infect Dis. 2009;49:1828–1835. doi: 10.1086/648424. [DOI] [PubMed] [Google Scholar]
- 34.WHO: Oseltamivir resistance in immunocompromised hospital patients. Available at http://www.who.int/csr/disease/swineflu/notes/briefing_20091202/en/. Accessed February 2010.
- 35.Baz M, Abed Y, Papanburg J, et al. Emergence of oseltamivir-resistant pandemic H1N1 virus during prophylaxis. N Engl J Med. 2009;361:2296–2297. doi: 10.1056/NEJMc0910060. [DOI] [PubMed] [Google Scholar]
- 36.Jain S, Kamimoto L, Bramley AM, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April–June 2009. N Engl J Med. 2009;361:1935–1944. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- 37.Kumar A, Zarychanski R, Pinto R, et al. Critically ill patients with 2009 influenza A (H1N1) infection in Canada. JAMA. 2009;302:1872–1879. doi: 10.1001/jama.2009.1496. [DOI] [PubMed] [Google Scholar]
- 38.Rello J, Rodríguez A, Ibañez P, et al. Intensive care adult patients with severe respiratory failure caused by Influenza A (H1N1)v in Spain. Crit Care. 2009;13(5):R148. doi: 10.1186/cc8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Australia and New Zealand Extracorporeal Membrane Oxygenation (ANZ ECMO) Influenza Investigators. Davies A, Jones D, Bailey M, et al. Extracorporeal membrane oxygenation for 2009 influenza A(H1N1) acute respiratory distress syndrome. JAMA. 2009;302:1888–1895. doi: 10.1001/jama.2009.1535. [DOI] [PubMed] [Google Scholar]
- 40.Colice GL. Historical perspective on the development of mechanical ventilation. In: Tobin MJ, editor. Principles and Practice of Mechanical Ventilation. 2. New York: McGraw Hill; 2006. pp. 1–36. [Google Scholar]
- 41.Rothberg MB, Haessler SD: Complications of seasonal and pandemic influenza. Crit Care Med 2009 (Epub ahead of print). [DOI] [PubMed]
- 42.New influenza A(H1N1) virus infections: global surveillance summary, May 2009. Wkly Epidemiol Rec 2009, 84:173–179. [PubMed]
- 43.Bernard GR, Artigas A, Brigham KL, et al. The American-European consensus conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149:818–824. doi: 10.1164/ajrccm.149.3.7509706. [DOI] [PubMed] [Google Scholar]
- 44.Hui DS, Lee N, Chan PK: Clinical management of pandemic (H1N1) infection. Chest 2009 (Epub ahead of print). [DOI] [PMC free article] [PubMed]
- 45.Ramsey CD, Funk D, Miller RR 3rd, Kumar A: Ventilator management for hypoxemic respiratory failure attributable to H1N1 novel swine origin influenza virus. Crit Care Med 2009 (Epub ahead of print). [DOI] [PubMed]
- 46.Annane D, Bellissant E, Bollaert PE, et al. Corticosteroids in the treatment of severe sepsis and septic shock in adults: a systematic review. JAMA. 2009;301:2362–2375. doi: 10.1001/jama.2009.815. [DOI] [PubMed] [Google Scholar]
- 47.Annane D, Sébille V, Bellissant E, Ger-Inf-05 Study Group Effect of low doses of corticosteroids in septic shock patients with or without early acute respiratory distress syndrome. Crit Care Med. 2006;34:22–30. doi: 10.1097/01.CCM.0000194723.78632.62. [DOI] [PubMed] [Google Scholar]
- 48.Marik PE, Pastores SM, Annane D, et al. Recommendations for the diagnosis and management of corticosteroid insufficiency in critically ill adult patients: consensus statements from an international task force by the American College of Critical Care Medicine. Crit Care Med. 2008;36:1937–1949. doi: 10.1097/CCM.0b013e31817603ba. [DOI] [PubMed] [Google Scholar]


