Abstract
Single nucleotide polymorphisms (SNPs) of interleukin (IL)-6 are associated with the development of chronic renal disease (CRD). Their impact for sepsis in the field of CRD was investigated. One control cohort of 115 patients with CRD without infection and another case cohort of 198 patients with CRD and sepsis were enrolled. Genotyping at the −174 (rs1800795) and −572 positions of IL-6 (rs1800796) was done by restriction fragment length polymorphism. Circulating IL-6 was measured by an enzyme immunoassay. The GG genotype of rs1800796 was more frequent among cases (78.3 %) than controls (62.6 %). No difference in the genotype frequencies of rs1800795 between cases and controls were found. Odds ratio for sepsis was 2.07 (95%CI 1.24–3.44, p = 0.005) with the GG genotype of rs1800796, which was confirmed by logistic regression analysis taking into consideration the presence of chronic comorbidities. All-cause mortality until day 28 was similar between patients with the GG genotype and the GC/CC genotypes of rs1800796, but death caused from cardiovascular events not-related with infection was more frequent with the GG genotype (14.6 % vs 2.4 %, p = 0.031). Circulating IL-6 was greater among patients of the GC/CC genotypes of rs1800796 and multiple organ dysfunction (p = 0.013). The GG genotype of rs1800796 predisposes to sepsis in CRD and to 28-day mortality by sepsis-unrelated cardiovascular phenomena.
Keywords: Severe Sepsis, Multiple Organ Dysfunction Syndrome, Control Cohort, Chronic Renal Disease, Acute Pyelonephritis
Introduction
Interleukin-6 (IL-6) is a pleiotropic cytokine which plays a crucial role in both innate and adaptive immune responses [1]. It promotes pro-inflammatory phenomena such as stimulation and proliferation of lymphocytes, differentiation of B cells and it is one of the most important mediators of acute phase response. IL-6 is produced by several types of immune cells including activated macrophages, monocytes, and lymphocytes.
There have been several publications about the association of increased circulating IL-6 with diseases such as Alzheimer disease, atherosclerosis, cardiovascular disease and autoimmune disorders. Increased levels of IL-6 are found in patients with chronic renal disease (CRD) particularly those on hemodialysis; increased IL-6 is associated with greater risk of depression, malnutrition, inflammation, atherosclerosis, cardiovascular events and mortality [2–5]. It has been suggested that circulating levels of IL-6 may vary depending on the carriage of single nucleotide polymorphisms (SNPs) at the promoter region of the IL-6 gene [6]. Several studies are published providing conflicting evidence for the role of carriage of minor frequency alleles of these SNPs for genetic predisposition to CRD. The most broadly studied is the G/C SNP at the −174 promoter position of IL-6. Two studies have shown that patients with the GC genotype are susceptible to end stage renal disease (ESRD) [7, 8]. Several studies on the association of this SNP with susceptibility to sepsis have provided contradictory results. More precisely, in one study the CC genotype was associated with increased susceptibility for sepsis acquisition whereas in two other studies the GG genotype was associated with better outcome [9–11].
Another G/C SNP at the −572 position of the promoter of IL-6 has been described (rs1800796); the GG genotype is associated with lower levels of plasma IL-6 and lower risk for CRD in a Japanese population [12]. Since patients with CRD are prone for bacterial superinfections whereas this SNP increases likelihood for CRD per se, we asked if carriage of minor frequency alleles of this SNP may affect susceptibility of patients with CRD to bacterial sepsis.
Patients and methods
Study design
This prospective multicentre study was conducted in 25 departments across Greece and Cyprus from January 2007 to February 2014. The study protocol was approved by the Ethics Committee of the participating hospitals. Written informed consent was provided by the patients or their first-degree relatives for patients unable to consent. Each patient was enrolled once in the study.
Inclusion criteria were: (a) age ≥18 years, (b) Caucasian origin, (c) sepsis due to lower respiratory tract infection, acute pyelonephritis, intra-abdominal infection, primary bacteraemia, or catheter-related bacteremia defined by international criteria [13], (d) CRD defined as creatinine clearance ≤50 ml/min as calculated by the Cockroft-Gault formula [14), and (e) blood sampling within the first 24 h from onset of sepsis. All these patients had sepsis during their hospitalization. Among enrolled patients those on hemodialysis with end-stage renal disease had CLcr <5–10 ml/min.
Exclusion criteria were: (a) infection by the human immunodeficiency virus infection (HIV), (b) neutropenia defined as <1,000 neutrophils/mm³, and (c) chronic intake of corticosteroids defined as any more than 0.4 mg/kg daily oral intake of equivalent prednisone for more than 15 days.
Patients were classified as suffering from sepsis, severe sepsis or septic shock according to standard definitions [15]. Sepsis was defined as infection accompanied by at least two of the following: (a) body temperature >38 °C or <36 °C, (b) heart rate >90/min, (c) respiratory rate >20/min or PaCO2 < 32 mmg, and (d) white blood cell count (WBC) >12,000 mm³ or <4,000 mm³ or >10 % bands. Severe sepsis was defined as sepsis aggravated by at least one organ failure, i.e., acute respiratory distress syndrome, acute coagulopathy and sudden mental deterioration as previously defined [15]. Septic shock was defined as severe sepsis aggravated by systolic blood pressure below 90 mmHg despite adequate fluid resuscitation necessitating the administration of vasopressors. Multiple organ dysfunction syndrome (MODS) was defined as the failure of at least two organs requiring intervention to maintain homeostasis [15]. Renal dysfunction was not considered a separate organ failure for the definition of severe sepsis and MODS in the current study.
For every patient a complete diagnostic work-up was performed comprising history, thorough physical examination, white blood cell count, blood biochemistry, arterial blood gases, blood cultures from peripheral veins and central lines, urine cultures, chest X-ray and chest and abdominal computed tomography if appropriate. If necessary, quantitative cultures of tracheobronchial secretions were performed and evaluated. At the time of admission APACHE II and SOFA scores were calculated for every patient. Survival and cause of death was recorded for 28 days. The cause of death was defined as either death due to infection or as death due to a non-infectious related cardiovascular event like myocardial infarction, sudden arrhythmia, pulmonary embolism and stroke. Clinical and demographic data were recorded in a case report form (CRF).
One control cohort was enrolled. Inclusion criteria for the control cohort were: (a) age ≥18 years, (b) Caucasian origin, (c) lack of history of any bacterial infection in the last two years, and (d) CRD defined as creatinine clearance ≤50 ml/min as calculated by the Cockroft-Gault formula [14]. Patients of the control cohort participated after written informed consent. Among enrolled controls those on hemodialysis with ESRD had CLcr <5–10 ml/min. The same exclusion criteria applied for the control cohort as for the patient cohort.
Laboratory investigation
Ten ml of venous blood were collected after venipuncture of one forearm vein under aseptic conditions during the first 24 h from sepsis onset. Three millilitres were collected into EDTA-coated tubes (Becton Dickinson, Cockeysville, MD) and stored at −80 °C until used for DNA isolation. Another 7 ml were collected into sterile and pyrogen-free tubes (Becton Dickinson) and centrifuged. Serum was stored at −80 °C until processed.
Genomic DNA was extracted by the Purigen Blood Core Kit C (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Typing for the SNP of IL-6 was performed by PCR on a Sensoquest thermal cycler LabCycler using 50 ng of genomic DNA at a final volume of 27 μL with 50 mmol·L−1 of MgCl2, 20 mmol·L−1 of dNTPs and 1 mmol·L−1 of Taq polymerase (all reagents provided by New England BioLabs, Ipswich, MA, USA). For the −174 (G/C) SNP (rs1800795), one 303 bp fragment was amplified using the forward primer 5′-TTG TCA AGA CAT GCC AAA GTG CT-3′ and the reverse primer 5′-GCC TCA GAC ATC TCC AGT CC-3′ [16, 17]. PCR reaction consisted of one initialization step of 95 °C for 4 min followed by 45 cycles; each cycle consisted of one denaturation step of 95 °C for 30 s, one hybridization step of 62 °C for 1 min and one elongation step of 68 °C for 1 min. Then another cycle of 68 °C for 5 min was carried out before termination. Ten μL of the PCR product was digested after incubation for 30 min at 37 °C with 0.5 U of the restriction enzyme NlaIII (New England Bio Labs). Digested PCR products were electrophoresed on 2 % agarose gel and visualized using ultraviolet radiation after ethidium bromide staining, with the application of positive controls. The visualized electrophoretic pattern was one fragment of 233 bp in patients of the GG genotype, two fragments of 233 bp and 122 bp in patients of the GC genotype, and one fragment of 122 bp in patients of the CC genotype.
For the −572 (G/C) SNP (rs1800796), one 163 bp fragment was amplified using the forward primer 5′-GGA GAC GCC TTG AAG TAA CTG C −3′ and the reverse primer 5′-GAG TTT CCT CTG ACT CCA TCG CAG −3′ [18–20]. PCR reaction consisted of one initialization step of 95 °C for 5 min followed by 35 cycles; each cycle consisted of one denaturation step of 95 °C for 1 min, one hybridization step of 55 °C for 1 min and one elongation step of 72 °C for 1 min. Then another cycle of 72 °C for 10 min was carried out before termination. Ten μL of the PCR product was digested after incubation for 30 min at 37 °C with 0.5 U of the restriction enzyme BsrBI (New England Bio Labs). Digested PCR products were electrophoresed on 2 % agarose gel and visualized using ultraviolet radiation after ethidium bromide staining, with the application of positive controls. The visualized electrophoretic pattern had two fragments of 102 bp and 61 bp in patients of the GG genotype; three fragments of 163 bp, 102 bp and 61 bp in patients of the GC genotype; and one fragment of 163 bp in patients of the CC genotype.
Circulating IL-6 was measured in serum using an enzyme immunosorbent assay (eBiosciences). The lowest limit of detection was 20 pg/ml.
Study endpoints
The primary study endpoint was the association of carriage of the studied SNP genotypes with the susceptibility to sepsis. The secondary endpoints were the association of the studied SNP genotypes with (a) the cause of death and (b) circulating IL-6.
Statistical analysis
Comparisons of genotype frequencies and of allele frequencies between cases and controls were done by the X 2 -test. Comparisons of demographic and clinical characteristics between genotype subgroups of patients were done by the X 2 -test for qualitative variables and by the Mann–Whitney U test for quantitative characteristics. Odds ratios (OR) and 95 % confidence intervals (CIs) were calculated by Mantel-Haenszel’s statistics. Logistic regression analysis was done to define factors independently associated with the development sepsis; OR and 95 % CI were calculated. Any value of p below 0.05 was considered significant.
Results
A total of 115 patients with CRD without infection served as controls; 198 patient cases with CRD and sepsis were enrolled. The study flow chart is shown in Fig. 1 and the demographic and clinical characteristics of controls and cases are provided in Table 1. Since the distribution of genotypes did not differ between patients with ESRD and without ESRD, they were both analyzed together. Distribution of genotypes in the control group was in the Hardy-Weinberg equilibrium for both studied SNPs; it was not in equilibrium for rs1800796 in the sepsis cohort (Table 2). The frequency of the GG genotype of rs1800796 was greater in cases than in controls. Carriage of this GG genotype was associated with OR = 2.07 for sepsis (95%CI 1.24–3.44, p = 0.005). The frequency of the G allele was greater among cases than controls.
Fig. 1.
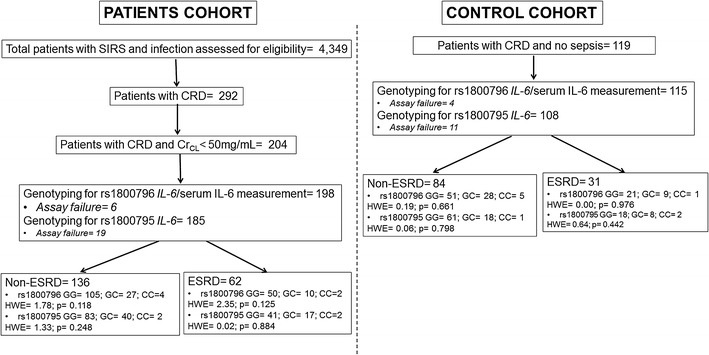
Study flow chart. Cr CL creatinine clearance, ESRD end-stage renal disease, HWE Hardy-Weinberg equilibrium, IL interleukin, SIRS systemic inflammatory response syndrome
Table 1.
Demographic and clinical characteristics of controls and cases enrolled in the study
| Characteristic | Controls (n = 115) | Patients (n = 198) | p-value |
|---|---|---|---|
| Age (years, mean ± SD) | 69.1 ± 11.8 | 67.7 ± 19.5 | 0.445 |
| Male gender (%) | 60 (52.2) | 117 (59.1) | 0.238 |
| End-stage renal disease (%) | 31 (26.9) | 62 (31.3) | 0.355 |
| Type 2 diabetes mellitus (%) | 52 (45.2) | 89 (44.9) | 1.000 |
| Chronic heart failure | 32 (27.8) | 79 (39.9) | 0.037 |
| Chronic obstructive pulmonary disorder (%) | 3 (2.6) | 34 (17.2) | <0.0001 |
| Solid tumor malignancy (%) | 8 (7.0) | 29 (14.6) | 0.046 |
| IL-6 (pg/ml, median-range) | <20 (10–1400) | 50 (20–124000) | <0.0001 |
Table 2.
Genotype and allele frequencies of rs1800795 and rs1800796 of IL-6 among control patients with chronic renal disease and patients with sepsis and chronic renal disease
| Genotype and allele frequencies | Controls | Patients | p-valueb | |
|---|---|---|---|---|
| rs1800795a | GG genotype (n, %) | 79 (73.1) | 123 (66.5) | |
| GC genotype (n, %) | 26 (24.1) | 57 (30.8) | 0.465 | |
| CC genotype (n, %) | 3 (2.8) | 5 (2.7) | ||
| HWE = 0.23 | HWE = 0.28 | |||
| p of HWE = 0.631 | p of HWE = 0.597 | |||
| G allele (n, %) | 184 (85.2) | 303 (81.9) | 0.304 | |
| C allele (n, %) | 32 (14.8) | 67 (18.1) | ||
| rs1800796a | GG genotype (n, %) | 72 (62.6) | 155 (78.3) | |
| GC genotype (n, %) | 37 (32.2) | 37 (18.7) | 0.011 | |
| CC genotype (n, %) | 6 (5.2) | 6 (3.0) | ||
| HWE = 0.19 | HWE = 3.78 | |||
| p of HWE = 0.664 | p of HWE = 0.050 | |||
| G allele (n, %) | 181 (78.7) | 347 (87.6) | <0.0001 | |
| C allele (n, %) | 49 (21.3) | 49 (12.4) | ||
HWE Hardy-Weinberg equilibrium
aSuccessful genotyping of rs1800795 was done for 108 controls and 185 patients and of rs1800796 for 115 controls and 198 patients
b Determined by the chi-square test
However, as shown in Table 1, the frequencies of chronic heart failure, of chronic obstructive pulmonary disorder and of solid tumor malignancy were greater in cases than in controls. To confirm that presence of the GG genotype of rs1800796 was an independent factor associated with sepsis, logistic regression analysis was performed. The above three comorbidities entered the equation with the GG genotype. Analysis proved that the GG genotype of rs1800796 was independently associated with the development of sepsis (Table 3).
Table 3.
Logistic regression analysis of factors related with the development of sepsis
| Factor | Odds ratio | 95 % confidence intervals | p-value |
|---|---|---|---|
| GG genotype of rs1800796 of IL-6 | 2.01 | 1.18–3.42 | 0.010 |
| Chronic heart failure | 1.49 | 0.89–2.51 | 0.129 |
| Chronic obstructive pulmonary disorder | 6.99 | 2.07–23.62 | 0.002 |
| Solid tumor malignancy | 2.20 | 0.94–5.51 | 0.069 |
We next asked the question if this association of the GG genotype of rs1800796 with increased susceptibility for sepsis among patients with CRD also modulated disease phenotype. To this end, we compared the clinical characteristics between cases with the GG genotype and those with the GC and CC genotypes (Table 4). The major finding was that although all-cause mortality on the first 28 days was similar between the two subgroups of genotypes, death of patients with the GG genotype of rs1800796 was caused from cardiovascular events not-related with infection at a greater frequency than patients with the GC and CC genotypes of rs1800796. More precisely, among the 80 patients of the sepsis cohort who died by day 28, death was due to infection and sepsis in 22 out of the 62 patients who were carriers of the GG genotype (35.5 %) and in 17 out of 18 patients who were carriers of the GC and CC genotypes (94.4 %, p = 0.016). The OR for death due to infection-unrelated cardiovascular events for carriers of the GG genotype of rs1800796 was 9.35 (95 %CI 1.16–75.05, p = 0.035).
Table 4.
Demographic and clinical characteristics of enrolled patients in relation to genotyping of rs1800796 of IL-6
| Characteristics | GG (n = 155) | GC and CC (n = 43) | p |
|---|---|---|---|
| Male gender (n, %) | 96 (61.9) | 21 (48.9) | 0.039 |
| Age (years, mean ± SD) | 74.5 ± 13.5 | 76.3 ± 10.7 | 0.497 |
| White blood cells (/mm³, mean ± SD) | 17,989.3 ± 14,121.3 | 15,443.5 ± 7,356.9 | 0.274 |
| APACHE II (mean ± SD) | 21.7 ± 6.7 | 20.5 ± 6.7 | 0.434 |
| SOFA score (mean ± SD) | 7.73 ± 3.84 | 7.48 ± 4.44 | 0.791 |
| Severe sepsis/shock (n, %) | 111 (71.6) | 25 (58.1) | 0.138 |
| Multiple organ dysfunction on day 1 (n, %) | 70 (45.2) | 13 (30.2) | 0.084 |
| Underlying infection (n, %) | |||
| Acute pyelonephritis | 64 (41.3) | 20 (46.5) | |
| Community-acquired pneumonia | 29 (18.7) | 12 (27.9) | |
| Intraabdominal abscess | 14 (9.0) | 2 (4.7) | |
| Primary bacteremia | 25 (16.1) | 3 (7.0) | 0.702 |
| Ventilator-associated pneumonia | 9 (5.8) | 3 (7.0) | |
| Hospital-acquired pneumonia | 8 (5.2) | 2 (4.6) | |
| Catheter-related infection | 6 (3.9) | 1 (2.3) | |
| Isolated microorganisms from blood (n, %) | |||
| Escherichia coli | 21 (13.5) | 5 (11.6) | |
| Acinetobacter baumannii | 8 (5.2) | 0 (0) | |
| Staphylococcus aureus | 7 (4.5) | 3 (6.9) | 0.803 |
| Klebsiella pneumoniae | 6 (3.9) | 1 (2.3) | |
| Pseudomonas aeruginosa | 5 (3.2) | 0 (0) | |
| Other | 8 (5.2) | 1 (2.3) | |
| Isolated microorganisms from urine (n, %) | |||
| Escherichia coli | 25 (16.1) | 11 (25.6) | |
| Klebsiella pneumoniae | 10 (6.5) | 2 (4.7) | |
| Pseudomonas aeruginosa | 5 (3.2) | 1 (2.3) | 0.633 |
| Proteus mirabilis | 5 (3.2) | 2 (4.7) | |
| Other | 3 (1.9) | 0 (0.0) | |
| Underlying conditions (n, %) | |||
| End-stage chronic renal disease | 50 (32.5) | 12 (27.9) | 0.711 |
| Type 2 diabetes mellitus | 67 (43.2) | 22 (51.2) | 0.389 |
| Chronic heart failure | 63 (40.6) | 16 (37.2) | 0.728 |
| Chronic obstructive pulmonary disorder | 28 (18.1) | 6 (14.0) | 0.650 |
| Solid tumor malignancy | 20 (12.9) | 9 (20.9) | 0.223 |
| All-cause 28-day mortality (n, %) | 62 (40.0) | 18 (41.9) | 0.862 |
| Death due to infection | 22 (14.6) | 17 (39.5) | |
| Death due to non-infectious related cardiovascular events | 40 (25.8) | 1 (2.4) | 0.031 |
Median IL-6 was 40 pg/ml (range <20–12,400 pg/ml) for patients with GG genotype of rs1800796 and 32.5 pg/ml (range <20–9,600 pg/ml) for patients with GC and CC genotypes of rs1800796 (p = 0.365). However, analysis of Table 4, which showed a borderline significantly greater frequency of MODS among patients with the GG genotype of rs1800796 than patients with the GC and CC genotypes of rs1800796, led to compare circulating IL-6 separately for patients who were presented on the first day with MODS and for those who did not have MODS on initial admission. Circulating IL-6 was greater among patients of the GC and CC genotypes and MODS than among patients with the GG genotype and MODS. A similar difference was not found among patients without MODS (Fig. 2a and b). This finding corroborates that at the early presentation of sepsis with MODS, the GC and CC genotypes were associated with a more pro-inflammatory response. This is in accordance with the causality of death to the initial sepsis episode at a greater frequency among patients with the GC and CC genotypes of rs1800796 than among patients with the GG genotype of rs1800796. Since the overall mortality did not differ between patients with the GG genotype and patients of the GC and CC genotypes, it was not surprising to find that IL-6 levels were greater in non-survivors and in survivors irrespective of the genotype (Fig. 2c).
Fig. 2.
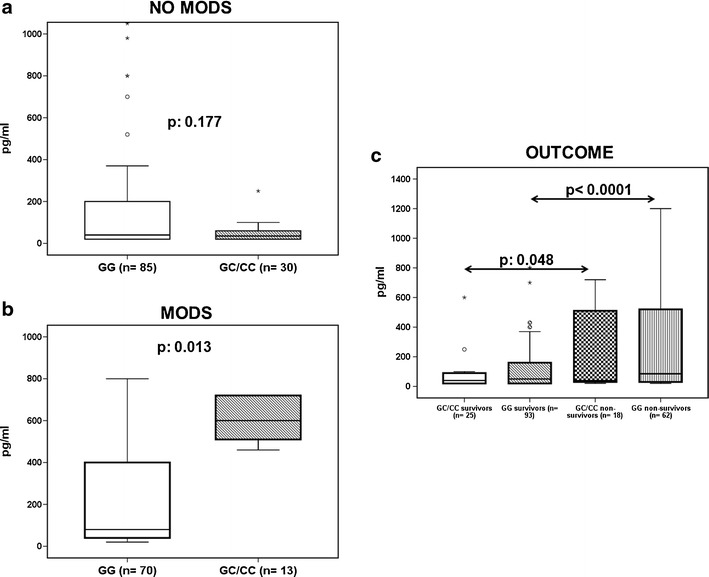
Circulating interleukin-6. a Patients without multiple organ dysfunction (MODS) on admission. b Patients with MODS on admission. P values indicate comparisons between carriers of only G alleles (i.e., GG genotypes) and carriers of at least one C allele of rs1800796 of IL-6. Circles denote extremes and asterisks denote outliers. c IL-6 admission levels in relation to genotype of rs1800796 of IL-6 and final outcome; p-values of indicated comparisons are provided
Discussion
The biological significance of the SNP at the −572 promoter position of IL-6 has not been broadly studied. Current findings suggest for the first time that carriage of the GG genotype of the wild-type alleles of the SNP has an independent association with predisposition for sepsis development among patients with CRD. When sepsis develops, carriage of this genotype is less pro-inflammatory, as evidenced by lower circulating IL-6 in most severe cases with MODS. However, carriage of this GG genotype is associated with death due to cardiovascular events not related to sepsis. This finding is in accordance with one previous publication showing that the GG genotype of rs1800796 of IL-6 increases the likelihood of death due to cardiovascular events [12].
Available studies on the role of SNPs of IL-6 for predisposition for infection mainly focus on sepsis and chronic hepatitis C (HCV). Those in HCV infected patients extensively study the role of the −572 SNP. In a study enrolling 424 Italian patients, no role of the −572 SNP on the physical course of HCV was found. Instead the frequencies of SNPs at positions −597 and −174 of the promoter of IL-6 were decreased compared to healthy controls and associated with worse histology scores [19]. In another study, 121 patients with mild HCV infection and persistently normal transaminases were longitudinally evaluated for 120 months by liver biopsies; carriage of −572 SNP C alleles was associated with earlier progression into liver fibrosis [20]. In another study, it was found that haplotypes of IL-6 containing the wild-type G allele of the −572 SNP were associated with a greater chance for achieving sustained viral responses after treatment [21].
Only one study has been published on the role of the −572 SNPs in severe infections. This involves a cohort of 421 consecutive admissions of children in intensive care units; although results are borderline suggestive for a role of this SNP for sepsis development no association with disease severity is found [22]. All other studies in sepsis patients focus on the SNP at the −174 promoter position of IL-6; their results generally agree that homozygosity for the wild-type G allele is protective against organ failure. More precisely, among 1,227 patients with CAP, those bearing the GG genotype were protected from acute respiratory distress syndrome (ARDS) [11]. In another study on 100 patients with CAP due to Streptococcus pneumoniae, it was found that the GG genotype was associated with fewer cases of extrapulmonary bacterial dissemination [23]. In a study where haplotyping of IL-6 was done in 228 critically ill patients; haplotype clades containing the −174 minor frequency C allele were strongly associated with increased mortality and MODS [24].
Two limitations of the present study should be recognized: (1) there was a limited number of patients as a result of a study design on targeting a specific subpopulation of sepsis developing in the field of CRD; however, in our setting statistically significant differences of the genotype distribution of rs1800796 between controls and cases were found, and (2) the use of restriction fragment length polymorphism (RFLP) as methodology instead of sequencing of the PCR products. The applied RFLP approaches for genotyping of rs1800795 and rs1800796 have been broadly successfully used in many other studies as well [16–20, 22].
Most case–control association studies in infections enroll patients with the studied infections and matched controls. However, matching does not take into consideration the predisposition a SNP brings for an underlying disorder that can predispose to severe infection. To this end, lack of matching for an underlying disorder can be a confounding factor leading to erroneous results. The present study does well beyond these limitations and explores the add-on effect of the −572 G/C SNP of IL-6 that confers to the development of CRD per se [12] on the development of sepsis. This is of paramount importance because CRD is a well-recognized entity generating sepsis predisposition [25]. Results show for the first time that although the GG genotype of rs1800796 predisposes for sepsis development, it paves the road for unfavorable outcome due to cardiovascular events not-related with sepsis per se.
Acknowledgments
The study was supported by the Hellenic Institute for the Study of Sepsis. The funder had no involvement in study design, data collection, data analysis, drafting the manuscript and decision to publish.
Compliance with ethical standards
Conflict of interests
None of the authors has conflict to disclose related to the submitted manuscript.
References
- 1.Stenvinkel P, Ketteler M, Johnson JRJ, Lindholm B, Pecoits-Filho R, Riella M, Heimbürger O, Cederholm T, Girndt M. IL-10, IL-6, and TNF-a. Central factors in the altered cytokine network of uremia. The good, the bad, and the ugly. Kidney Int. 2005;67:1216–1233. doi: 10.1111/j.1523-1755.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- 2.Hasuike Y, Nonoguchi H, Ito K, Naka M, Kitamura R, Nanami M, Tokuyama M, Kida A, Otaki Y, Kuragano T, Nakanishi T. Interleukin-6 is a predictor of mortality in stable hemodialysis patients. Am J Nephrol. 2009;30:389–398. doi: 10.1159/000235687. [DOI] [PubMed] [Google Scholar]
- 3.Wetmore JB, Lovett DH, Hung AM, Cook-Wiens G, Mahnken JD, Sen S, Johansen KL. Associations of interleukin-6, C-reactive protein and serum amyloid A with mortality in haemodialysis patients. Nephrology. 2008;13:593–600. doi: 10.1111/j.1440-1797.2008.01021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda H, Qureshi AR, Heimburger O, Barany P, Wang K, Pecoits-Filho R, Stenvinkel P, Lindholm B. Serum albumin, C-reactive protein, interleukin 6, and fetuin A as predictors of malnutrition, cardiovascular disease, and mortality in patients with ESRD. Am J Kidney Dis. 2006;47:139–148. doi: 10.1053/j.ajkd.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Sonikian M, Metaxaki P, Papavasileiou D, Boufidou F, Nikolaou C, Vlassopoulos D, Vlahakos DV. Effects of interleukin-6 on depression risk in dialysis patients. Am J Nephrol. 2010;31:303–308. doi: 10.1159/000285110. [DOI] [PubMed] [Google Scholar]
- 6.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittal RD, Manchanda PK. Association of interleukin (IL)-4 intron-3 and IL-6-174 G/C gene polymorphism with susceptibility to end-stage renal disease. Immunogenetics. 2007;59:159–165. doi: 10.1007/s00251-006-0182-6. [DOI] [PubMed] [Google Scholar]
- 8.Buraczynska M, Jozwiak L, Ksiazek P, Borowicz E, Mierzicki P. Interleukin-6 gene polymorphism and faster progression to end-stage renal failure in chronic glomerulonephritis. Trans Res. 2007;150:101–105. doi: 10.1016/j.trsl.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Balding J, Healy CM, Livingstone WJ, White B, Mynett-Johnson L, Cafferkey M, Smith OP. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes Immun. 2003;4:533–540. doi: 10.1038/sj.gene.6364020. [DOI] [PubMed] [Google Scholar]
- 10.Schluter B, Raufhake C, Erren M, Erren M, Schotte H, Kipp F, Rust S, Van Aken H, Assmann G, Berendes E. Effect of the interleukin-6 promoter polymorphism (−174 G/C) on the incidence and outcome of sepsis. Crit Care Med. 2002;30:32–37. doi: 10.1097/00003246-200201000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Loeches I, Sole-Violan J, Rodriguez de Castro F, García-Laorden MI, Borderías L, Blanquer J, Rajas O, Briones ML, Aspa J, Herrera-Ramos E, Marcos-Ramos JA, Sologuren I, González-Quevedo N, Ferrer-Agüero JM, Noda J, Rodríguez-Gallego C. Variants at the promoter of interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intensive Care Med. 2012;38:256–262. doi: 10.1007/s00134-011-2406-y. [DOI] [PubMed] [Google Scholar]
- 12.Okada R, Wakai K, Naito M. Pro-/anti-inflammatory cytokine gene polymorphisms and chronic kidney disease: a cross-sectional study. BMC Nephrol. 2012;13:2. doi: 10.1186/1471-2369-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calandra T, Cohen J. The international sepsis forum consensus definitions of infections in the intensive care unit. Crit Care Med. 2005;33:1639–1648. doi: 10.1097/01.CCM.0000168253.91200.83. [DOI] [PubMed] [Google Scholar]
- 14.Scrutinio D, Passantino A, Santoro D, Cacciapaglia E, Farinola G. Prognostic value of formulas estimating excretory renal function in the elderly with systolic heart failure. Age Ageing. 2009;38:296–301. doi: 10.1093/ageing/afp006. [DOI] [PubMed] [Google Scholar]
- 15.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 16.Yalçin S, Kayalti Z, Söylemezoğlu T. Role of interleukin-6 -176G/C promoter polymorphism in trace metal levels of autopsy kidney and liver tissues. Int J Hyg Envir Health. 2011;214:219–224. doi: 10.1016/j.ijheh.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Yalcin S, Demirbas S, Onguru O. Interleukin-6 (−174G/C) promoter polymorphism in patients infected by hepatitis B virus. J Exp Inter Med. 2012;2:173–177. [Google Scholar]
- 18.Zavaleta-Muñiz SA, Martín-Márquez BT, Gonzalez-Lopez L et al. (2013) The -174G/C and -572G/C interleukin 6 promoter gene polymorphisms in Mexican patients with rheumatoid arthritis: a case–control study. Clin Develop Immunol 2013:959084 [DOI] [PMC free article] [PubMed]
- 19.Cussigh A, Falleti E, Fabris C, Gonzalez-Montoya NG, Díaz-Toscano ML, Ponce-Guarneros JM, Ruiz-Padilla AJ, Vázquez-Del Mercado M, Maldonado-González M, Fafutis-Morris M, Flores-Martínez SE, Martínez-García EA, Gamez-Nava JI. Interleukin promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics. 2011;63:33–41. doi: 10.1007/s00251-010-0491-7. [DOI] [PubMed] [Google Scholar]
- 20.Falleti E, Fabris C, Vandelli C, Colletta C, Cussigh A, Smirne C, Fontanini E, Cmet S, Minisini R, Bitetto D, Toniutto P, Pirisi M. Genetic polymorphisms of interleukin-6 modulate fibrosis progression in mild chronic hepatitis C. Hum Immunol. 2010;71:999–1004. doi: 10.1016/j.humimm.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Yee LJ, Im K, Borg B, Yang H, Liang TJ. Interleukin-6 haplotypes and the response to therapy of chronic hepatitis C virus infection. Genes Immun. 2009;10:365–372. doi: 10.1038/gene.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michalek J, Svetlikova P, Fedora M, Klimovic M, Klapacova L, Bartosova D, Hrstkova H, Hubacek JA. Interleukin-6 gene variants and the risk of sepsis development in children. Hum Immunol. 2007;68:756–760. doi: 10.1016/j.humimm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Schaaf B, Rupp J, Muller-Steinhardt M, Kruse J. The interleukin-6 -174 promoter polymorphism is associated with extrapulmonary bacterial dissemination in Streptococcus pneumoniae infection. Cytokine. 2005;31:324–328. doi: 10.1016/j.cyto.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland AM, Walley KR, Manocha S, Rusell JA. The association of interleukin 6 haplotype clades with mortality in critically ill adults. Arch Intern Med. 2005;165:75–82. doi: 10.1001/archinte.165.1.75. [DOI] [PubMed] [Google Scholar]
- 25.Sarnak MJ, Jaber BL. Mortality caused by sepsis in patients with end-stage renal disease compared with the general population. Kidney Int. 2000;58:1758–1764. doi: 10.1111/j.1523-1755.2000.00337.x. [DOI] [PubMed] [Google Scholar]


