Abstract
Background
Ethanol production through fermentation of gas mixtures containing CO, CO2 and H2 has just started operating at commercial scale. However, quantitative schemes for understanding and predicting productivities, yields, mass transfer rates, gas flow profiles and detailed energy requirements have been lacking in literature; such are invaluable tools for process improvements and better systems design. The present study describes the construction of a hybrid model for simulating ethanol production inside a 700 m3 bubble column bioreactor fed with gas of two possible compositions, i.e., pure CO and a 3:1 mixture of H2 and CO2.
Results
Estimations made using the thermodynamics-based black-box model of microbial reactions on substrate threshold concentrations, biomass yields, as well as CO and H2 maximum specific uptake rates agreed reasonably well with data and observations reported in literature. According to the bioreactor simulation, there is a strong dependency of process performance on mass transfer rates. When mass transfer coefficients were estimated using a model developed from oxygen transfer to water, ethanol productivity reached 5.1 g L−1 h−1; when the H2/CO2 mixture is fed to the bioreactor, productivity of CO fermentation was 19% lower. Gas utilization reached 23 and 17% for H2/CO2 and CO fermentations, respectively. If mass transfer coefficients were 100% higher than those estimated, ethanol productivity and gas utilization may reach 9.4 g L−1 h−1 and 38% when feeding the H2/CO2 mixture at the same process conditions. The largest energetic requirements for a complete manufacturing plant were identified for gas compression and ethanol distillation, being higher for CO fermentation due to the production of CO2.
Conclusions
The thermodynamics-based black-box model of microbial reactions may be used to quantitatively assess and consolidate the diversity of reported data on CO, CO2 and H2 threshold concentrations, biomass yields, maximum substrate uptake rates, and half-saturation constants for CO and H2 for syngas fermentations by acetogenic bacteria. The maximization of ethanol productivity in the bioreactor may come with a cost: low gas utilization. Exploiting the model flexibility, multi-objective optimizations of bioreactor performance might reveal how process conditions and configurations could be adjusted to guide further process development.
Keywords: Ethanol, Bioreactor simulation, Biothermodynamics, Syngas fermentation
Background
Gas mixtures containing CO2, H2 and CO are commonly known as syngas, which is typically produced by two processes, i.e., thermochemical conversion (gasification) of carbonaceous materials like coal and oil, and reforming of natural gas [1]. Lignocellulosic biomass, food, municipal and packaging wastes are alternative raw materials that can also be used for gasification [2, 3] and could lead to production processes with improved sustainability attributes as compared to fossil-based feedstocks [4, 5]. For this reason, syngas from non-fossil sources is considered a key feedstock for the circular economy.
Driven by technical and sustainability limitations of Fischer–Tropsch synthesis [6], experiments performed since the late 1980s explored the potential of certain types of autotrophic acetogenic bacteria to catabolize the three main components of syngas into ethanol [7]. Although with generally low productivities, these microorganisms are also able to produce a variety of other substances of commercial importance, e.g, 2,3-butanediol, butanol and butyric and lactic acids [8, 9]; however ethanol is the first commercialized bioproduct [7].
Acetogens convert carbon into acetyl-CoA through the Wood–Ljungdahl metabolic pathway (WLP) [10]. In the reductive direction, the WLP is considered the most efficient non-photosynthetic and the only linear CO2 fixation pathway to acetyl-CoA [1, 11]. Two molecules of CO2 and/or CO are fixated following two separate branches, the methyl (eastern) and the carbonyl (western) branches. Thorough descriptions on the configuration of the WLP and its link with the particular energy conservation strategies of acetogens can be found elsewhere [1, 9, 12, 13]. The WLP is able to use CO as a source of energy and carbon [14–17], whereas H2 has to be combined with a carbon source that can be CO2 [9, 18]. It has been proposed that CO fermentation would yield higher amounts of Gibbs free energy and adenosine triphosphate (ATP) than H2 [19, 20], while H2 fermentation offers advantages on improved mass transfer due to higher solubilities and diffusion rates than those for CO [20]. Yet, the influence of the gas composition on the technical, economic and environmental performances of the fermentation process still remains quantitatively uncertain, basically due to the inaccuracy of currently available models of the metabolism of acetogenic bacteria.
Several types of mathematical models have been proposed for understanding and predicting the behavior of microorganisms in gas fermentations [21–27]; other simpler models have been used for estimating process performance [4, 5, 28–31]. The most popular of the modeling strategies employed recently by researchers is the genome-scale modeling (GSM), which has been used for assessing several features of the intracellular processes in C. ljungdahlii and C. autoethanogenum during syngas fermentations, e.g, the influence of the link between energy conservation and carbon metabolism on the selectivity between ethanol and acetic acid [25, 32–34], the co-factor specificity of certain enzymes linked to energy conservation [32, 33, 35], the formation of biofilms [26], the possibility of boosting ATP production by supplying arginine [36], and the feasibility of gene knock-out to reach overproduction of native and non-native products of acetogens [37]. Alternatively, with issues generally regarding on the accuracy of the quantitative predictions, GSM has also been used to assess the behavior of simulated microorganism inside large-scale bioreactors [21, 26, 37, 38]; the main cause for these latter issues may be credited to the interlinking between the intracellular processes and the environmental conditions given by the bioreactor, besides GSM’s large dependency on the objective function and the constraints applied to solve the intracellular rates of reaction [25]. The low detail of intracellular kinetics is viewed as another limitation of GSM [39] that becomes relevant given the fact that microorganisms do not reach steady-state inside large-scale bioreactors [40].
Moreover, in most publications reporting models of gas fermentations, scarce effort was invested on comparing the simulation results with experimental data reported by other research groups. Such task is challenging considering the large variety of microbial strains, gas compositions, process conditions used in the reported experiments and the high strain-specificity of more complex models of microbial metabolism. Thus, a general model that focuses on the basic thermodynamic interactions driving the catabolism of CO, CO2 and H2 by a hypothetical strain of acetogenic bacteria (such as in [28]) might be able to consolidate the diversity of reported results.
On the same line, the present study focuses on reinforcing the quantitative aspects of a previously published model [4] by validating the stoichiometric and kinetic parameters of microbial reactions with data and observations reported in scientific literature. The model is then applied to the simulation of an industrial ethanol production case and used to assess the influence of dissolved gas concentrations on bioreactor performance, gas flow profiles, supported accumulation of cells and energy requirements of the fermentation plus downstream processing of the alcohol. The model is intended to be sufficiently flexible and accurate that its results could guide further process design and optimization through model-based scaled-down experiments [41]. Finally, the model construction scheme here presented could be adapted to other process configurations, modes of fermentation and after further refining, coupled to GSM’s, intracellular kinetics and Euler–Lagrange modeling strategies [42].
Results and discussion
This section begins with an assessment of the estimations delivered by the thermodynamics-based black-box model of microbial reactions; the analysis focuses on the predictions’ quantitative reliability by comparing the estimations with data and observations reported in literature for microorganisms that perform similar metabolic reactions. Then the analysis is extended to the characterization of two bioreactor operation regimes in terms of gas flow profiles, supported biomass accumulations, restrictions suggested by thermodynamic feasibility of catabolic reactions at different heights of the bioreactor and finally, process performance. The analysis is lastly closed with the influence of the kinetic parameters on process performance.
Analysis of black-box model of microbial reactions
Gibbs free energy change of catabolic reactions
Figure 1 shows the dependence of the Gibbs free energy change in the catabolic reactions () (see Eqs. 1 and 2 in Table 1) on dissolved gas concentrations; results are presented for independent catabolism of CO and H2/CO2 in Fig. 1a and b, respectively. They show that at dissolved concentrations of the electron donors () lower than 1 mM, the amount of energy harvested from CO catabolism is larger than that from H2/CO2 catabolism. Consequently, H2 threshold concentrations ( = 9.1–15 kJ mol−1) for catabolic ethanol production fall between 3 × 10−3 and 3 × 10−1 mM and lower than 4 × 10−4 mM for CO, depending on the CO2 concentration (). Increasing () diminishes the amount of energy harvested from CO catabolism where CO2 is a product; whereas is beneficial for energy production in H2/CO2 catabolism where CO2 is the carbon source (see Eqs. 1 and 2).
Fig. 1.
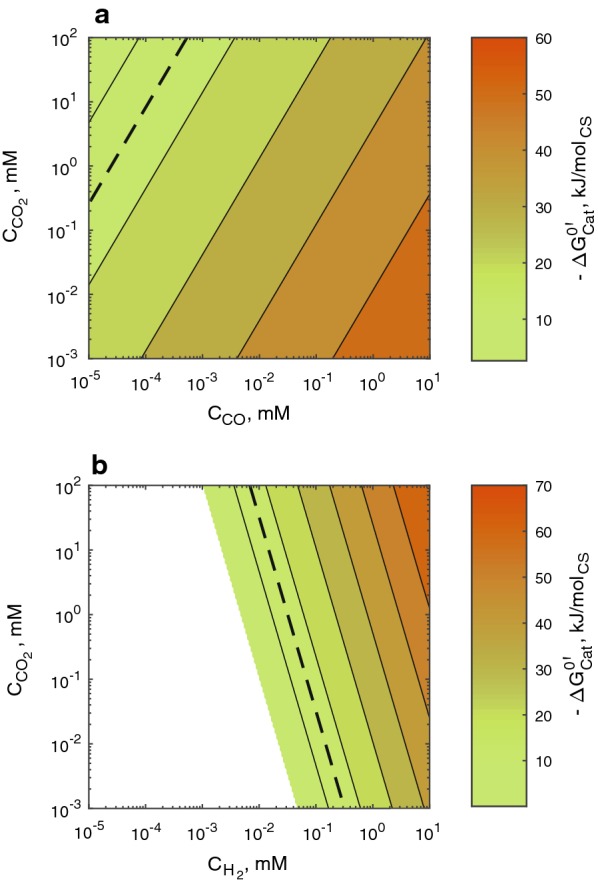
Gibbs free energy generation through independent a) CO and b) H2/CO2 catabolism for ethanol production. The dashed lines indicate where = – 15 kJ mol−1CS. H2/CO2 catabolism is cut-off at = 0, where the catabolic reaction would be at equilibrium and no energy could be released from it; the white region represents the gas concentrations where the inverse reaction (H2/CO2 production from ethanol) would be spontaneous
Table 1.
Catabolic reactions leading to the production of ethanol and acetate and related standard changes in Gibbs free energy and enthalpy
| Reactiona | Eq nr. | ||||
|---|---|---|---|---|---|
| kJ mol−1CS | kJ mol−1bP | kJ mol−1CS | kJ mol−1bP | ||
| − 37.4 | − 224.4 | − 57.4 | − 344.0 | (1) | |
| − 52.3 | − 104.6 | − 178.8 | − 357.6 | (2) | |
| − 33.6 | − 134.3 | − 65.0 | − 260.0 | (3) | |
| − 28.1 | − 56.2 | − 134.7 | − 269.5 | (4) | |
aThe stoichiometry of catabolic reactions and the energy changes are defined to satisfy balances on all elements involved, charge and degree of reduction. Standard Gibbs free energy and enthalpy of formation of the compounds involved in Eqs. (1–4) were retrieved from the supplementary material in [43]
bResults are expressed per mole of product, i.e., the product in Eqs. 1 and 2 is ethanol while acetate is the product in Eqs. 3 and 4
Neither CO nor H2 threshold concentrations have been reported for acetogens during solventogenesis; however, there are reports for acetogens during acetogenesis. In one of these reports, CO uptake (see Eq. 3) by Carboxydothermus hydrogenoformans at 65 °C stopped at a CO partial pressure () of 3.9 × 101 Pa when CO2 was allowed to accumulate in the overhead (reaching 1.3 × 105 Pa); when CO2 was instead withdrawn from the overhead ( of 3.5 × 102 Pa), CO uptake stopped at of 2.0 × 10−1 Pa [44]. At these two points, estimated with Eq. (5) is − 21 and − 15 kJ mol−1CS, respectively (see Additional file 1: Figure S1); intracellular acetate concentration is assumed at 10 mM and the total pressure is 2 × 105 Pa. Similarly, another report mentions that Acetobacterium woodii started growing on H2 and CO2 at 30 °C only after was higher than 2.5 × 102 Pa while was 2.0 × 104 Pa [45]. Assuming A. woodii H2/CO2 catabolism followed Eq. (4), is estimated by Eq. (5) at − 17.9 kJ mol−1CS (see Additional file 1: Figure S2); the intracellular acetate concentration is assumed at 10 mM and the reported total pressure is 1 × 105 Pa. This brief analysis shows that Eq. (5) may be used to predict threshold concentrations for acetate production from gas fermentations with an acceptable level of approximation. Therefore, since energy conservation in acetogenic bacteria is possible during solventogenesis [19], then threshold concentrations might as well be predicted by Eq. (5) for catabolic ethanol production from CO and H2:
| 5 |
The importance of predicting threshold concentrations relies on the fact that the large-scale bioreactor should be designed to avoid reaching such concentrations.
Biomass yields
Since biomass yields () depend on (see Eq. 6) they are a direct function of and follow a similar trend as when plotted against and (see Additional file 1: Figure S3). Biomass yields for CO catabolism are estimated between 0.022 and 0.080 Cmolx mol−1CS which are slightly higher than those estimated for H2/CO2 (0.015–0.067 Cmolx mol−1CS) mainly due to the larger amounts of Gibbs free energy dissipated by cells growth using CO2 as CS (see Eq. 2).
| 6 |
Similar to threshold concentrations, biomass yields have not been reported for gas fermentations during solventogenesis. However, there are specific rates of CO and H2 consumption reported for continuous fermentations at steady-state [32, 33, 46] that can be used to estimate biomass yields. Since in those reports, CO and H2 were simultaneously consumed whereas CO2, acetic acid and ethanol were produced, the biomass yield is estimated by dividing the dilution rate (assuming the rate of cell lysis is negligible) by the reported specific CO uptake rate (). The estimated biomass yields range between 0.044 and 0.090 Cmolx mol−1CS (the specific data used for his calculation is shown in Additional file 1: Table S1) which are slightly higher (yet, within the same order of magnitude) than the estimations given by Eq. (6) for ethanol catabolic production.
Assessment of kinetic parameters
Maximum specific substrate uptake and growth rates
Regarding the predicted kinetic parameters, the thermodynamics-based black-box model returns a maximum substrate uptake rate () of − 4.4 molD Cmol−1x h−1 (see Eq. 7) for both catabolic energy sources, CO and H2. The result is the same for both electron donors since they have the same degree of reduction (2 mole− mol−1D). In addition, as explained in section “Methods”, the maximum consumption and production rates of all compounds involved in the microbial reactions are estimated by linearly relating the predicted stoichiometry with the maximum substrate uptake rates. As consequence, the maximum growth rate () was estimated at 0.29 and 0.19 h−1 for CO and for H2/CO2 fermentations, respectively. for H2/CO2 fermentation is two times lower than CO fermentation because although is the same on both cases, (per mole of electron donor) for H2/CO2 fermentation is also two times lower than the (see Eqs. 1–4 and Additional file 1: Figure S3).
| 7 |
Mohammadi et al. [47] calculated a and a of − 0.87 molD Cmol−1x h−1 (assuming the same molar mass for cell material as here) and 0.195 h−1, respectively. was estimated by fitting batch dissolved CO concentrations (calculated using a method similar to [48]) into the kinetic equation for CO uptake (see section “Thermodynamics-based black-box model of microbial reactions”), while was found by fitting batch growth data from the same experiment. In that study, syngas with a H2/CO ratio of 1 was used and H2 consumption was acknowledged; however was calculated without accounting for the electrons taken up from H2 and the carbon from CO2. If H2 and CO2 uptake would have been considered, the maximum electron uptake rate would be twice as large as reported, i.e., − 1.74 molD Cmol−1x h−1. That value is 2.5 times lower than that predicted by Eq. (7). In addition, their reported is close to the value estimated with the black-box model. Thus, it could be argued that C. ljungdahlii has a 60% reduced electron uptake capacity compared to the maximum estimated for E. coli [49]; yet, more steady-state data with different carbon sources is needed to confirm such conclusion. Accounting for this uncertainty, the bioreactor simulation is performed using the value estimated with Eq. (7) and the influence of a reduced maximum uptake rate on the operation of the gas fermentor is explored as part of the sensitivity analysis (see section “Sensitivity analysis”).
In addition, since growth rates estimated by genome-scale reconstructions of C. ljungdahlii and C. autoethanogenum [25, 37] used the same value reported by [47], their estimations are also comparable with those made by the biothermodynamics-based black-box model (see Additional file 1: Figure S4).
CO and H2 half-saturation constants
A value of 1.7 × 10−2 mM has been calculated for the CO half-saturation constant in () C. ljungdahlii from CO consumption curve fitting [47], whereas has been estimated to range between 4 × 10−2 and 3 × 10−1 mM from assays using enzymatic extracts from acetogens [50–53]. In nature, wetland peats and marine waters oxidize CO with values ranging from 1 × 10−6 to 4 × 10−5 mM [54, 55], while the averaged CO concentration in Earth’s troposphere is equivalent to a concentration of 5 × 10−7 mM in pure water [56]. Similarly, for H2 consumption by soils and methanogenic sludge has been estimated between 7 × 10−8 and 1 × 10−6 mM [57, 58], while the equivalent saturation from H2 concentration in the troposphere is 4 × 10−7 mM [59].
As consumption of 1 mol of CO results in higher Gibbs free energy gains than 1 mol of H2, it could be postulated that cells in nature control metabolic activity at low dissolved gas concentrations by stimulating H2 uptake (with higher affinity, < ). This argument is in accordance with the affinities by which microbes consume H2 and CO in nature. However, most H2 consumption studies have focused on methanogens. H2 threshold concentrations for catabolic methane production (see Eq. 8) can be as low as 5 × 10−6 mM, assuming pre-industrial atmospheric concentrations for CO2 and CH4 (230 ppm and 540 ppb, respectively [60]) and using Eq. (5). Moreover, evidence suggests that acetogens may promote H2 production (from ions) to avoid harmful concentrations of reduced energy carriers when feeding on CO [33, 61]. Therefore, might not necessarily be lower than , in agreement with the ranges of H2 and CO threshold concentrations estimated for ethanol production (see section “Gibbs free energy change of catabolic reactions”) and reported data for acetogens.
| 8 |
It has been argued that half-saturation constants of poorly soluble substances can be overestimated by two orders of magnitude if they are derived from the fitting of consumption curves obtained under mass transfer or other rate limitations [58]. Therefore, the and values of 4 × 10−2 and 5 × 10−3 mM, respectively, were randomly picked aiming to reconcile the information reported in literature (which is prone to overestimation) with the threshold concentrations criteria. Thus, the value for falls midway between the estimated threshold range (see Fig. 1) and the reported value for C. ljungdahlii, while is located in the middle of the threshold range while simultaneously agreeing with the values determined from enzymatic extracts. Nevertheless, the effect of the value of substrate half-saturation constants on the operation of the gas fermentor is discussed in detail as part of the sensitivity analysis (see section “Sensitivity analysis”).
Main limitation of the black-box model of microbial reactions
Since the black-box model of microbial reactions is based on the electron transfer from one electron donor to one electron acceptor, it is not compatible with the simultaneous uptake of more than one electron donor or the generation of more than one product. Thus, the documented influence of process conditions such as pH [46, 62], acetic acid concentration [63], gas compositions [33] and gas dissolved concentrations [32] on the selectivity for either electron donor or for the production of ethanol and acetic acid, could not be reproduced. To perform such analysis, the black-box may be opened and include the mechanisms by which cells adjust the amounts of Gibbs free energy used for ATP production, depending on the specific requirements for growth, maintenance, transport of metabolites across the membrane and motile functions [49, 64, 65].
Analysis of mass transfer-based model of the large-scale bioreactor
Basis of analysis
Although the algorithm that links the black-box model of the microbial reactions with the mass transfer-based model of the large-scale bioreactor uses and as independent variables, dissolved gas concentrations are not independent during bioreactor operation. Moreover, within this section bioreactor performance is discussed from the perspective of a non-dimensional specific uptake rate of the electron donors (, see Eq. 9) in order to partly bypass the uncertainties related to the value of :
| 9 |
The dependence of ethanol productivity and gas utilization on reveals the existence of two operational regimes of the bioreactor at steady-state: (i) one where mass transfer is suboptimal and (ii) one where mass transfer is sufficient. Sections “The ‘suboptimal’ operation regime” and “The ‘optimal’ operation regime” describe the features of each regime.
The ‘suboptimal’ operation regime
The suboptimal regime is characterized by a low performance of the bioreactor and a approaching to 1 in H2/CO2 fermentation; due to the inhibition term used on the CO uptake kinetic equation, approaches to zero as CO concentration is highest (see Fig. 2a) resulting in a dual solution for as function of . According to the mass balances (see Additional file 1: Table S2), bioreactor productivity linearly depends on the mass transfer rate of the electron donors. Mass transfer rate concurrently depends on the mass transfer coefficient () and the driving force (dissolved gas concentration gradient). is determined by bioreactor design, the composition of the liquid phase, gas flow rate and gas sparging method [66], while the driving force is ruled by and the solubility of the gas components. As biomass concentrations () are low within the suboptimal regime (see Fig. 2a, c), is close to saturation. An elevated causes two unfavorable effects over bioreactor operation: (i) in the case of CO fermentation, it might inhibit CO consumption (see section “Gibbs free energy change of catabolic reactions”) mainly at the lower regions of the liquid column where the partial pressure is highest; and (ii) it limits the mass transfer driving force, which consequently hampers gas utilization and bioreactor productivity (see Fig. 2b). Yet, bioreactor performance improves sharply as increases and decreases to approximately 0.7, where mass transfer rate achieves 90% of its estimated maximum. This point marks the start of the optimal regime.
Fig. 2.
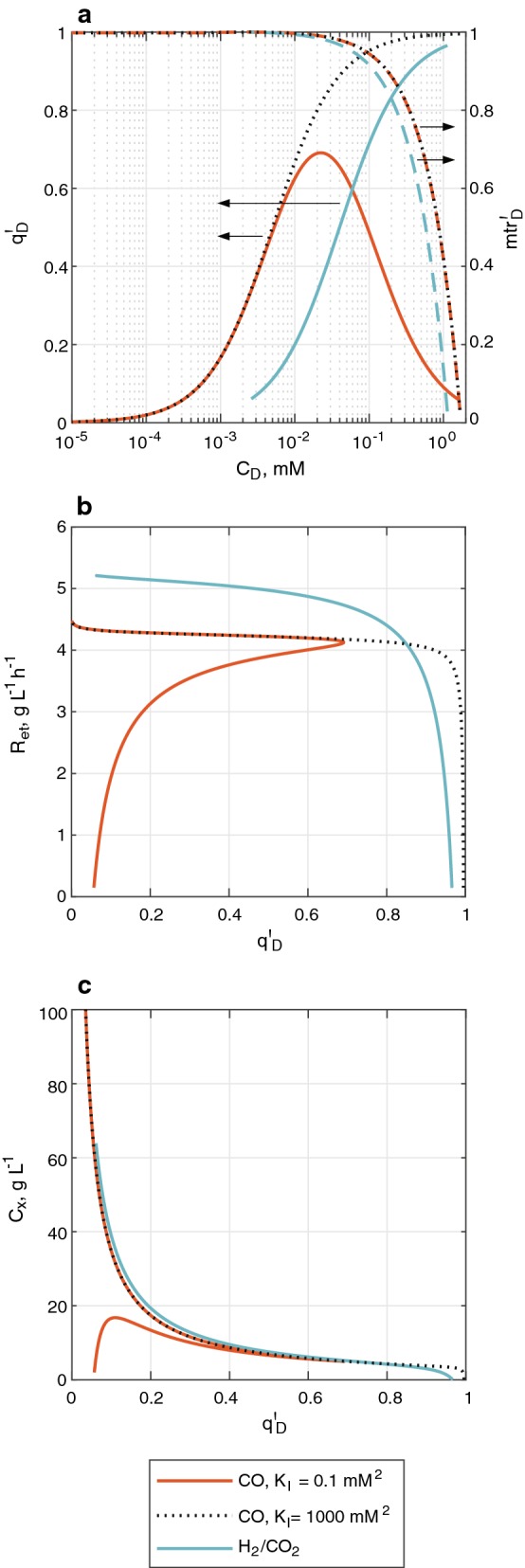
Relations between parameters used to describe bioreactor operational regimes for H2/CO2 and CO fermentations. a Dependency of non-dimensional electron donor uptake rate () and a non-dimensional mass transfer rate () on dissolved electron donor concentration (); b relation between ethanol volumetric productivity () and ; c estimated biomass concentration () as function of . The figure includes curves with black dotted lines that represent the operation of CO fermentation when the effect of substrate inhibition in the kinetic model is minimized by maximizing the value of
The ‘optimal’ operation regime
The optimal regime runs from a of 0.7 until it approaches zero. Within this range, mass transfer rate, ethanol volumetric productivity and gas utilization are above the 90% of their estimated maximum values (see Fig. 2a, b and Additional file 1: Figure S5). Since the value of (see Fig. 2c) is calculated by the optimization algorithm to linearize the term with respect to the mass transfer rate (see Additional file 1: Table S2), large increments on (or equivalently, large reductions on and ) would only return moderate improvements in bioreactor performance (see Fig. 2b). Therefore, working within the optimal regime would be desirable for continuous operation of the large-scale gas fermentor.
Influence of gas composition on bioreactor performance
This section compares bioreactor performance and the features of its operation between CO and H2/CO2 fermentations within the optimal regime. It may be assumed that a process fed by a gas with a composition falling in a determined point between the two composition boundaries (100% CO and 100% 3:1 H2:CO2 mixture), will behave proportionately to the contribution of each boundary.
, and
One parameter that showed to have a significant influence over bioreactor operation for CO and not for H2/CO2 fermentation is the liquid outflow rate (), which as well as is adjusted by the optimization algorithm to fulfill mass balances. allows the removal of cells and the ethanol fraction that was not evaporated and transferred to the offgas. In CO fermentation, is controlled by the biomass production rate for most of the range; this causes excessive ethanol removal along the liquid, preventing its accumulation within the bioreactor. Consequently, the ethanol concentration does not reach 45 g L−1 only until is as low as 0.10 (where is 1 × 10−3 mM and approximates to 20 g L−1) where biomass production rate lowers sufficiently; this could become another challenge for the fermentation process development since sustaining such low values can be difficult [32]. In H2/CO2 fermentation the relatively lower biomass production rates indirectly allow ethanol accumulation throughout the whole optimal regime due to the lower biomass yields (per mole of electron donor).
In order to lower the influence of over bioreactor operation, biomass withdrawal may be decoupled from the removal of fermentation broth. Biomass retention within biofilms is known for increasing bioreactor productivity, yet in prolonged periods could lead to clogging [67]. If the biofilms were shaped into granules instead, clogging may be avoided and larger hydraulic loads can be handled by gas-lift bioreactors due to the high settling velocity of the granules [68]. Up to date there is no report on a gas fermentation set-up that uses biomass retention within granules, however a recent publication showed that C. ljungdahlii produces biofilms under stress by NaCl [69].
In general, the estimated biomass concentrations fermentation may seem unrealistic since the maximum reported for a continuous syngas fermentation using cell recycle is 10 g L−1 [46]. Although an explanation for this limitation has not been given in literature for gas fermentation, one hypothesis may be formulated based on the fact that the abiotic phase in a bioreactor undergoes spatial and temporal variations on the intensities of mixing and mass transfer [42]. If at one given moment inside one portion of the bubble column, the local value of or was on the order of 0.01 mM, a 6% decrease in the local mass transfer coefficient (caused by the turbulent flow of the liquid phase) is enough to cause cells in a concentration of 10 g L−1 to lower that local by tenfold in approximately 0.03 s. Such variations may cause cells to temporarily (yet frequently) circulate through zones where the approaches to their thresholds, causing starvation. The detrimental effects of starvation on product generation and cells viability have been linked to the depletion of certain metabolic pools in fungi [40]. Considering the fact that depletion of the acetyl-CoA pool prevented C. autoethanogenum from achieving higher than 1.4 g L−1 in [32], it could be argued that the achievement of high values of in gas fermentations is not limited by the averaged rates of mass transfer, but instead to the slight spatial and temporal variations on those rates of mass transfer.
Table 2 shows a summary of relevant parameters estimated within the optimal regime of CO and H2/CO2 fermentations; the values in the table describe the bioreactor operation at the liquid column height were the mean log pressure is found. The operation points shown were selected from the to satisfy the following conditions: (i) mass transfer is above the 90% of its maximum; (ii) CO does not inhibit its consumption at the bottom of the vessel (see Fig. 3a); (iii) the rate at which microbial biomass is being produced allows to reach 45 g L−1 in the liquid; (iv) H2 does not reach threshold concentrations at the top of the liquid column (see Fig. 3b), and (v) the concentration of biomass is not higher than 10 g L−1.
Table 2.
Summary of relevant parameters of bioreactor operation and process performance for H2/CO2 and CO fermentations
| Variable | Symbol | Unit | CO fermentation | H2/CO2 fermentation |
|---|---|---|---|---|
| Performance indicators | ||||
| Ethanol volumetric productivity | g L−1 h−1 | 4.25 | 5.1 | |
| Gas utilization | % | 17.1 | 22.9 | |
| Gas outflow composition | ||||
| Hydrogen | mol mol−1 | 0.00 | 0.71 | |
| Carbon dioxide | mol mol−1 | 0.11 | 0.24 | |
| Carbon monoxide | mol mol−1 | 0.84 | 0.00 | |
| Ethanol | mol mol−1 | 0.01 | 0.01 | |
| Water | mol mol−1 | 0.04 | 0.04 | |
| Concentrations in the fermentation brotha,b | ||||
| Hydrogen |
() |
mol m−3 | 0.00 (0.00) |
0.025 {0.033; 0.018} (1.15 {1.63; 0.78}) |
| Carbon dioxide |
() |
mol m−3 | 0.32 {0.00; 4.22} |
12.46 {17.11; 8.94} (13.09 {18.51; 8.86}) |
| Carbon monoxide |
() |
mol m−3 |
2.7 × 10−3 {3.6 × 10−3; 2.0 × 10−3} (1.62 {2.45; 1.01}) |
0.000 (0.000) |
| Ethanol | mol L−1 (g L−1) | 0.96 (44.3) | 0.98 (45.0) | |
| Biomass | Cmol m−3 (g L−1) | 395 (10.0) | 399 (10.1) | |
| Parameters estimated with thermodynamicsa | ||||
| Catabolic energy production | kJ mol−1CS | − 29.2 {− 48.2; − 24.0} | − 19.9 {− 23.0 − 16.45} | |
| Biomass yield | Cmolx mol−1CS | 0.041 | 0.020 | |
| Biomass specific consumption/production rates (logarithmic mean) | ||||
| Hydrogen | mol Cmol−1x h−1 | 0.00 | − 1.67 | |
| Carbon dioxide | mol Cmol−1x h−1 | 1.00 | − 0.56 | |
| Carbon monoxide | mol Cmol−1x h−1 | − 1.52 | 0.00 | |
| Ethanol | mol Cmol−1x h−1 | 0.23 | 0.28 | |
| Water | mol Cmol−1x h−1 | − 0.73 | 0.84 | |
| Cells | h−1 | 0.06 | 0.01 | |
| Non-dimensional electron donor uptake rate | – | 0.35 | 0.38 | |
| Streams entering and leaving the bioreactor | ||||
| Gas flow rate at the top |
() |
mol s−1 (m3 s−1) | 462 (7.8) | 418 (7.1) |
| Gas flow rate at the bottom | () | mol s−1 (m3 s−1) | 479 (4.0) | 528 (4.4) |
| Liquid outflow rate | m3 h−1 | 30.0 | 39.3 | |
| Fresh syngas | mol s−1 | 80.0 | 118 | |
| Parameters regarding gas and liquid flows and mixing (logarithmic mean) | ||||
| Gas flow rate | () | mol s−1 (m3 s−1) | 471 (5.6) | 471 (5.6) |
| Superficial gas velocity (pressure-corrected) | m s−1 | 0.14 | 0.14 | |
| Liquid flow rate | m3 s−1 | 26.9 | 26.9 | |
| Mixing time | s |
60.4c 54.3d |
61.2c 54.3d |
|
| Mass transfer coefficients (logarithmic mean) | ||||
| Hydrogen | s−1 | 0.000 | 0.164 | |
| Carbon dioxide | s−1 | 0.000 | 0.098 | |
| Carbon monoxide | s−1 | 0.104 | 0.000 | |
aThe average value is shown first, followed by the values at the top and the bottom of the liquid column between curly brackets
bThe values between round brackets represent the saturation concentrations of CO, H2 and CO2
cSimulated using the 9 vertically stacked compartments model
dCalculated with Eq. (21)
Fig. 3.
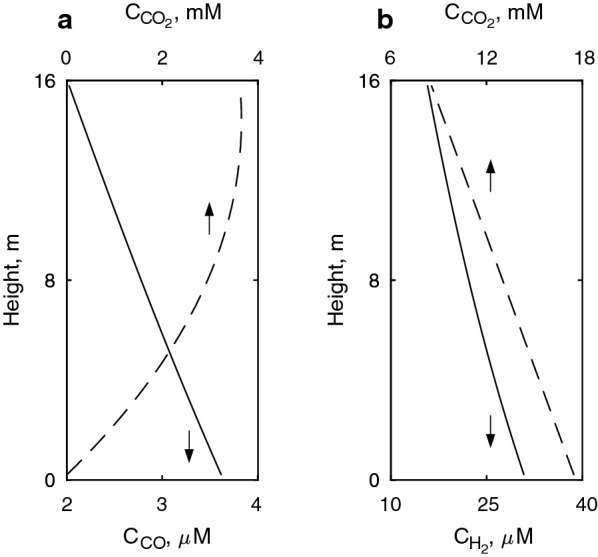
Gas concentration profiles along the liquid column for a CO fermentation and b H2/CO2 fermentation
In such points of operation, H2/CO2 fermentation returns a 19% higher ethanol productivity (5.1 g L−1 h−1) than fermentation of CO (4.3 g L−1 h−1); this difference is attributed to a higher mass transfer rate in H2/CO2 fermentation mainly because the higher H2 diffusivity in water (see section “Thermodynamics-based black-box model of microbial reactions“) makes in 58% higher for H2 compared to CO (see Table 2). Considering that ethanol productivities have reportedly reached 8 g L−1 h−1 through fermentation of CO-rich syngas [70], and that commercial sugar-based fermentations commonly fall between 1.5 and 2.0 g L−1 h−1 [71], it can be argued the estimations made in this study do not fall out of context and even more important, they could be subjected to further improvement.
Moreover, the higher mass transfer rates in H2 have a similar effect on gas utilization (, see section “Thermodynamics-based black-box model of microbial reactions”) in relation to CO fermentation. However, does only reach 23% in absolute terms, which makes the gas recycling step (see section “Process configuration”) a necessity to guarantee full use of the fresh gas fed to the process. Therefore, an upstream operation for gas composition control is essential to avoid the accumulation of gases within the gas recycle.
Lastly, the CO, H2 and CO2 mass transfer coefficients used in the present work range between 0.05–0.21, 0.08–0.33 and 0.05–0.20 s−1, respectively. Unfortunately, mass transfer coefficients for the transfer of CO, H2 and CO2 are available only for laboratory-scale bioreactors [72–74] where the heterogeneous bubbling regime may not be achieved [66] and therefore, the predicted values cannot be compared with reported experiments. However, the estimated ranges agree well with the experimental data (corrected with the gas diffusivities in water, see section “Mass transfer-based model of the industrial bioreactor”) reported for oxygen transfer within large bubble columns by [75].
Energy requirements
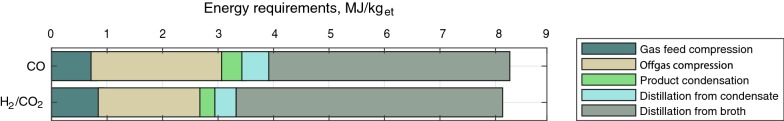
When CO is fed to the reactor, roughly 60% of its carbon goes to CO2. This causes the molar gas flow rate across the reactor to slightly decrease (see Table 2). Contrarily, when the 3:1 H2:CO2 mixture is fed to the fermentor, the two gases are consumed and none is produced; thus the molar gas flow diminishes by 20%. This difference in gas flow profiles impacts significantly on the two largest contributors to total energy requirements, i.e., compression of the gas streams (in agreement with [21]) and product distillation (see Fig. 4).
Fig. 4.
Breakdown of energy requirements of the proposed process configuration
In H2/CO2 fermentation less power is needed to compress the recycling offgas compared to CO fermentation. Furthermore, as the productivity in H2/CO2 fermentation is higher and the offgas’ ethanol evaporation capacity lower (due to lower offgas flow rate), the liquid outflow in the chosen point of operation (see Table 4) is larger than in CO fermentation. As consequence, the distillation of the diluted broth consumes more energy in the H2/CO2 fermentation. All in all, the total absolute energy requirements are higher for the H2/CO2 fermentation; however due to the higher ethanol productivity, the energy needed per unit of ethanol produced is lower than in CO fermentation (see Fig. 5).
Table 4.
Model parameters for bubble column bioreactor design and operation during gas fermentation
| Parameter | Unit | Value |
|---|---|---|
| Operation conditions | ||
| Temperature | K | 310.15 |
| Top pressure | Pa | 1.52 × 105 |
| Gas hold-up | m3G m−3G+L | 0.15 |
| pH | – | 5.0 |
| Maximum ethanol concentrationa | mol m−3 | 1304 |
| Bioreactor dimensions | ||
| Volume | m3 | 700 |
| Height | m | 20 |
| Aspect ratio | – | 3.0 |
| Diameter | m | 6.7 |
| Overhead space | % | 20 |
| Height of gas–liquid mixture | m | 16 |
| Relevant gas properties for the mass transfer model (at 37 °C) | ||
| Diffusivitiesb | ||
|
O2 CO H2 CO2 |
m2 s−1 |
3.21 × 10−9 2.88 × 10−9 4.55 × 10−9 2.70 × 10−9 |
| Henry’s coefficientc | ||
|
CO H2 CO2 |
molS m−3 Pa−1 |
0.79 × 10−5 0.72 × 10−5 24.6 × 10−5 |
aLiquid–vapor equilibria data for the ethanol/water system were estimated using the non-random two-liquid model for calculating activity coefficients (see Additional file 1: Table S3)
bEstimated according to the method presented by Wilke and Chang [84]
cEstimated according to the method presented by Sander [85]
Fig. 5.
Influence of half-saturation constants on ethanol productivity in the large-scale gas fermentor
Sensitivity analysis
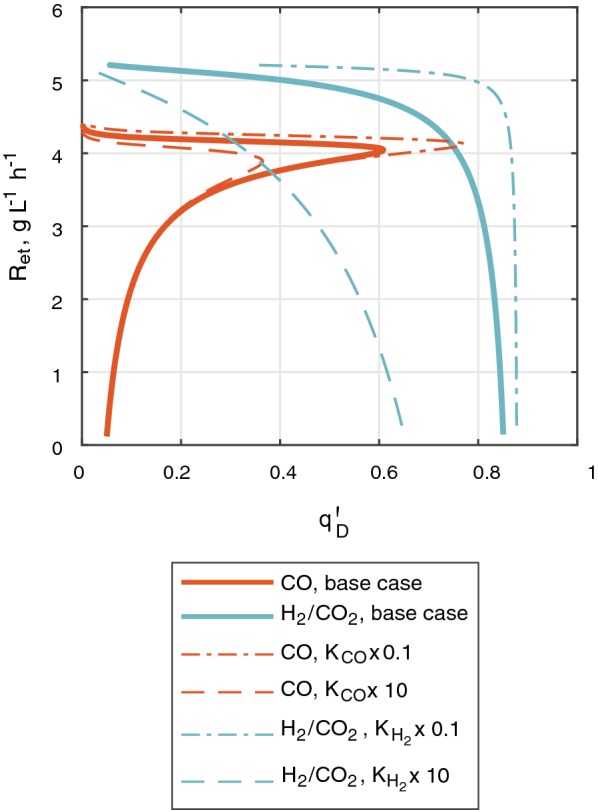
The sensitivity analysis assesses the impact of the value that half-saturation constants and maximum substrate uptake rate would apply on bioreactor performance. If and decreased by tenfold, the appearance of the optimal regime of operation would occur at higher (see Fig. 6), which in an industrial setting could improve bioreactor operation robustness to withstand fluctuations on and . In the opposite case, if and were 10 times higher than what was fixed in section “CO and H2 half-saturation constants”, the preservation of a stable optimal regime could require delicate control of low . This last case could severely affect H2/CO2 fermentation as needs to be kept at relatively higher values (see Table 2) to avoid reaching a threshold concentration at the top of the fermentor. Therefore, as well as need to be in the order of 1 × 10−2 mM or lower.
Fig. 6.
General structure of the calculation process for optimizing productivity. The figure is based on [88]
When the maximum uptake rate is decreased by 60% from the value estimated with Eq. (7), a negligible effect is seen on bioreactor productivity and gas utilization within the optimal regime of bioreactor operation.
Further, as the relation between the rates of mass transfer and consumption of the electron donors is highly linear (see mass balances in Additional file 1: Table S2), a 100% improvement on the mass transfer coefficients (with respect to the values shown in Table 2) would result in an 86% improvement on bioreactor productivity. That means that ethanol productivity for CO and H2/CO2 fermentations could be as high as 7.9 and 9.4 g L−1 h−1, respectively. Similarly, due to the improvement on gas transfer to the liquid, gas utilization would rise to 31 and 38% for CO and H2/CO2 fermentations, respectively. If contrarily, mass transfer coefficients were 50% smaller than predicted for gas transfer to pure water (see section “Mass transfer-based model of the industrial bioreactor”) both ethanol productivity and gas consumption would roughly be cut by half.
The effect of an increased microbial tolerance to ethanol such that may be maintained at 80 g L−1, is only reflected on the energy requirements which in the case of H2/CO2 fermentation would decrease by 30% to 5.8 MJ kg−1et. A similar decrease in energy requirements would be seen for the fermentation of CO a as long as the concentration of biomass climbed to 25 g L−1 at a of 0.13, where is low enough to allow ethanol concentration to rise from 45 g L−1.
Moreover, if the aspect ratio was increased to 10 while maintaining the volume at 700 m3 (vessel height and diameter will be 44.7 and 4.5 m, respectively), the gas utilization will climb to 35 and 44% for CO and H2/CO2 fermentations, respectively, mainly due to the increased gas retention times. As a consequence of the increased gas utilization, the ethanol productivity may slightly rise to 5.3 and 6.7 g L−1 h−1 for the same two cases as previously. Finally, since the hydrostatic pressure at the bottom of the bioreactor will increase with the height of the liquid column, the total power requirements will consequently rise by an average of 11% for the two gas compositions because increase need for gas compression. Furthermore, since the relation between the gas hold-up and the productivity is also highly linear, a 100% increase in the hold-up to 0.30 m3G m−3G+L will be reflected as an equivalent increase on ethanol productivity to 8.6 and 9.4 g L−1 h−1 for CO and H2/CO2 fermentations, respectively. However, due to the reduced gas retention time, the gas utilization will fall to 11 and 14% for the two fermentation cases. Therefore, a balance may have to be established between the height of the bioreactor and the gas hold-up to guarantee that productivity rises without significantly affecting the gas consumption or the energetic demands of the overall process.
Finally, if the gases provided to the fermentation were not pure, as it is likely in an industrial setting, the gas recycling will not be possible. Such process configuration may cause the energy requirements to rise by 15% to 9.6 and 9.5 MJ kg−1et for CO and H2/CO2 fermentations, respectively. The reason behind this result is the fact that the energy savings on the offgas compression are not sufficiently high to counter the extra expenses derived from the compression of higher amounts of gas at the bioreactor inlet. Industrial sources of CO, H2 and CO2, such as syngas and steel manufacturing offgases, which contain impurities that if recycled may accumulate inside the bioreactor, may have to deal with the extra energy expenses of not using gas recycle. On the other hand, the fermentation of mixtures between H2 produced for example, through the electrolysis of water, and CO2 recovered from a generic combustion process may be benefited by the energetic advantage of using gas recycling.
Conclusions
An alternative model for simulating gas fermentation within a large-scale bubble column bioreactor was developed. The model coupled a thermodynamics-based black-box model of main microbial reactions with a mass transfer-based model of the bioreactor. A significant amount of effort was put on validating the black-box model predictions with trends and data found in literature for acetogens or microorganisms using similar catabolic processes:
The estimated threshold concentrations for CO, H2 and CO2 agreed with reported data for acetogens during early and late growth stages at different temperatures.
Predicted biomass specific uptake rates for CO and H2 consumption surpassed reported values by 250% suggesting that there might exist a potential for strain improvement.
Estimation of substrate half-saturation constants form threshold concentrations proved to yield results comparable with data reported for CO and H2 consumption by acetogenic and methanogenic microorganisms.
The large-scale gas fermentor simulation showed that ethanol productivities may reach between 4.3 and 5.1 g L−1 h−1 and CO and H2 utilization per step may not surpass 23%. If instead, mass transfer coefficients were 100% higher than the estimated by the model developed for oxygen transfer to pure water, then productivities may achieve 7.9 and 9.4 g L−1 h−1 while gas utilization may climb to 38%. Such performance indicators are obtained if H2 does not achieve threshold concentrations at the top of the liquid column, CO consumption is not inhibited at the bottom of the bioreactor, ethanol concentration reaches 45 g L−1 and if biomass withdrawal from the bioreactor was decoupled from the fermentation broth removal.
It is recommended that multi-objective optimizations are done to further validate the bioreactor performance results by comparing them with reported data and process configurations that have been proposed and patented. The model could also be used to acquire a broader view on how process performance, especially gas utilization, could be further improved. In addition, the black-box model may be extended to include intracellular processes relevant for energy conservation and guide further understanding on the factors influencing the selectivity between ethanol and acetic acid in acetogens.
Methods
This section describes the structure of the hybrid model, the estimation of relevant parameters and how process performance is assessed.
The hybrid model
A deterministic model for simulating ethanol production in a large-scale gas fermentor is proposed. The model consists of two main parts: (i) a thermodynamics-based black-box model of the microbial reactions and (ii) a model of the fermentor hydrodynamics. Two gas compositions are evaluated separately to represent the two boundaries of possible compositions that acetogenic bacteria are able to catabolize, i.e, pure CO and a 3:1 mixture of H2 and CO2, which, respectively, may be obtained industrially from the offgas of steel manufacturing [70] and by mixing the CO2 recovered from a generic combustion process with H2 produced from, for instance, the electrolysis of water [76].
The simulation of the large gas fermentor is here done by assuming that a generic acetogenic bacterial strain has been adapted, modified or that the conditions in the bioreactor are such that the net rate of acetic acid production is zero, although it is commonly reported that the main product of acetogens is acetic acid while ethanol is generally a co-product. The assumption is sustained on the fact that ethanol production form CO and H2 as electron donors is thermodynamically feasible on its own, disregarding the current understanding of the physiology of acetogens and the limitations of gene editing techniques applied to these bacteria.
Thermodynamics-based black-box model of microbial reactions
The microbial metabolism is considered to be formed by catabolism and anabolism. Ethanol is a product of CO or H2/CO2 catabolism, while cells are the product of anabolism starting from the same energy and carbon sources; CO and H2 are the energy sources or electron donors (), while CO and CO2 are the carbon sources (CS). Table 1 shows the stoichiometry of catabolism (Eqs. 1–4), whereas Eqs. (10) and (11) show the stoichiometries of anabolism. If thermodynamically feasible (), the amount of Gibbs free energy released by catabolism is mainly used to support cell growth. Biomass yields ( in Eq. 6) are then calculated from the ratio between Gibbs free energy dissipation during growth ( in Eq. (12), where is the number of carbon atoms in the CS and is its degree of reduction—electrons available for redox exchange) and free energy change in catabolism ( in Eq. 5) plus the ratio between the degrees of reduction of biomass material and of [77]. In Eq. (6), the term represents the amount of CS needed to produce the necessary amount of free energy to produce 1 Cmol of biomass (); the term represents the amount of CS required for stoichiometrically building 1 Cmolx. Finally, the term (stoichiometric coefficient of in anabolism) in Eq. (6), is used since is not the CS in H2 catabolism. Moreover, the stoichiometric coefficient of any th component in the metabolic reactions (molj Cmol−1x) is determined by adding the contributions of the catabolic and anabolic reactions (see Eq. 13).
| 10 |
| 11 |
| 12 |
| 13 |
Since the Gibbs free energy change ( in Eq. 5) is calculated at physiological conditions, its magnitude and the parameters derived from it (e.g, biomass yield and stoichiometry of metabolic reactions) will depend on temperature and the activity of the m products and substrates at the intracellular space. It has been argued that “the choice of the species used for calculation of of a reaction should be based on the species for which the activity is closest to the reference activity (i.e, 1 mol L−1 for aqueous species, 1 atm for gases)” [43]. Therefore, the value of in Eq. (7) corresponds to the aqueous concentrations for: ethanol, NH4+ and H+ ions, while for the gases: CO, H2 and CO2, their partial pressures are used.
Water intervenes as product and reactant in the catabolic reactions (see Eqs. 1–4). However, the concentration of electron donors, ethanol, CO2 and H+ ions are generally very low and catabolic reactions take place within a “dilute aqueous system” [77]. Therefore, the concentration of water is not considered in the calculation of with Eq. (5) [49, 77].
As CO, H2 and CO2 are uncharged gases, they can freely diffuse across the cell membrane and thus their concentrations are assumed to be the same inside and outside the cells. The same is assumed for ethanol whose concentration () is used as a fixed value at 45 g L−1, which approximates to the highest concentration achieved in a syngas fermentation [78].
The concentration of the dissolved gases is assumed to vary within a large range since, as it will be explained in section “Interlink between both models”, they are the link between the model of microbial reactions and the mass transfer model and largely influence the bioreactor performance. Table 3 summarizes the values of intracellular concentrations of the reactants and products of catabolic reactions (Eqs. 1–4).
Table 3.
Intracellular concentrations of substance involved in catabolic reactions
| Substance | Concentration, mol L−1 |
|---|---|
| Fixed values | |
| H+ ions | 1.0 × 10−7 [64] |
| NH4+ ions | 1.0 × 10−1 [79]a |
| Ethanol | 9.8 × 10−1 |
| Ranges of valuesb | |
| CO | 1 × 10−8–1 × 10−3 |
| H2 | 1 × 10−8–1 × 10−3 |
| CO2 | 1 × 10−6–1 × 10−1 |
aDefined from a 0.1 M ionic strength
bRanges of dissolved gas concentrations were defined based on the corresponding range of partial pressures between 1 × 10−5 to 1 atm
The effect of ionic strength is neglected from the calculation of Gibbs free energy changes since an ionic strength as high as 0.1 M would result in variations of maximum 0.6 kJ mol−1 for the reactions shown in Table 1 [79]. Therefore, the activity coefficients of all substrates involved in the considered microbial reactions are rounded to 1.
The reaction rates of microbial metabolism are calculated by linearly linking their stoichiometry to the hyperbolic substrate uptake kinetics of the electron donors (i.e, CO and H2—see Eqs. 14 and 15). Such kinetic relations were reported to be applicable to CO consumption by C. ljungdahlii [47] and H2 consumption by C. ragsdalei P11 [50]. In Eqs. (14) and (15), and are the half-saturation constants while is the inhibition constant for CO (0.1 mol2 m−6 [47]); section “Model validation” describes the procedure followed to assess and set the values for the half-saturation constants. The maximum substrate uptake rate () is calculated for CO and H2 from the theoretical maximum rate of electron consumption by cells, as shown in Eq. (7) [49]. is a function of the temperature and the degree of reduction of the electron donor. Equation (7) was formulated based on a “maximum rate of electron transport in the catabolic energy production” of 3 mole− Cmol−1x h−1 at 20 °C and which was found to fit uptake data from E. coli growing on different substrates [49].
| 14 |
| 15 |
Model validation
The thermodynamics-based black-box model of microbial reactions is validated by comparing its results against general tendencies observed in reported experiments and data. The estimations of the Gibbs free energy change of catabolic reactions is compared to published experimental data in terms of threshold concentrations of the gases, i.e., the concentrations at which the catabolic reaction returns a minimum amount of energy necessary to power the proton motive force (− 15 kJ mol−1H+ [77], although it could be as low as − 9.1 kJ mol−1H+ in acetogens [80]).
In addition, it has been suggested that the value of the substrate half-saturation constants are likely close to the threshold concentrations of that substance; further in poorly soluble substances, the same constants can also be expected to approximate to their solubility in the aqueous phase [77]. Thus, the values of the and found in literature are first judged against those two criteria and when necessary, modified to a value in accordance with the restrictions.
Lastly, the estimated values for maximum substrate uptake rates and biomass yields are also compared with published data for acetogens or microorganisms that use similar catabolic routes.
Mass transfer-based model of the industrial bioreactor
The height and volume of the industrial bubble column bioreactor are both fixed at 20 m and 700 m3, respectively. The stated height is common in industry [81] while the volume is relatively large yet regarded as cost efficient [82]. Considering that the reactor will have a 20% overhead space and a gas hold-up fixed at 15% [83], the fermentation broth (liquid phase) will occupy 476 m3. More details of the model parameters for the bubble column are shown in the Table 4. In addition, since the aspect ratio and the gas hold-up are parameters which can adopt different values in industry, the effect of different values that those shown in Table 4, is assessed within the sensitivity analysis.
The mass transfer model is defined considering the coexistence of two phases inside the bioreactor: (i) a liquid phase which initially is assumed to have a homogeneous composition, and (ii) a gaseous phase which behaves as a plug flow. Although bacterial cells would constitute a third phase within the bioreactor, they are considered to occupy a negligible volume and to be homogeneously distributed within the liquid phase; thus biomass would not influence mass transfer. As a first approach, the fermentation broth is assumed to be a coalescing liquid despite the fact that ethanol inhibits water coalescence depending on the alcohol concentration [86]. For simplicity, the liquid dynamically behaves as pure water under a heterogeneous bubbling regime inside the bubble column [66]; bubbles are thus assumed as coarse with a 6 mm average diameter [66]; with these assumptions, the volumetric mass transfer coefficients () and the gas hold-up () are estimated from the pressure-corrected superficial gas velocity () using Eqs. (16) and (17) [66, 83]; such equations have been derived by fitting experimental data obtained in bubble columns with a diameter and height between 0.08–11.6 m and 0.3–21 m, respectively [66]. is estimated from the logarithmic mean of the volumetric gas flow rates across the bioreactor calculated at normal conditions of temperature and pressure (see Additional file 1: Table S3 for the specific equations used for the calculation). The gas flow rates and the estimated are ultimately constrained to return a fixed at 0.15. Although the gas hold-up is not a good design closing criteria, it was chosen to limit further maximization of mass transfer coefficients and subsequent minimization of gas use (see section “Technical performance”).
| 16 |
| 17 |
Mass balances are established around the gas fermentor for all species involved in the metabolic reactions leading to ethanol and bacterial biomass production (see Additional file 1: Table S2). Furthermore in the energy balances, the power requirement for compression of the gas feed and the offgas for gas recycle is estimated assuming adiabatic operation of compressors with a mechanical efficiency of 0.7 [28]. The heat required for gas cooling is obtained using average heat capacities within the applied temperature ranges, assuming that condensation will occur only when the final cooling temperature has been reached; a refrigeration coefficient of performance of 3.7 is assumed [87]. Additional file 1: Table S4 shows the specific equations used for estimating energy requirements.
Interlink between both models
The thermodynamics-based model of microbial reactions as well as the mass transfer-based model of the bioreactor converge in the concentration of the dissolved gases and ethanol in the liquid phase. The dissolved concentration of the electron donor ( where is either, CO or H2) is fixed before solving the mass balances to avoid unnecessary issues with convergence into a solution. Thus, a range of steady-state operation points of the gas fermentor are estimated for a range of values of . Although the system has no degrees of freedom (see the list of decision variables below and equations SI1 to SI8 in the Additional file), it does have multiple solutions; therefore, an optimization is used to obtain a solution that maximizes the ethanol volumetric productivity ( in Eq. 18) while minimizing power consumption.
| 18 |
The optimization uses the fraction of ethanol that exits the bioreactor along the liquid phase as objective function; whereas, the biomass concentration, the molar gas inflow and outflow rates, the CO, H2 and CO2 contents in the offgas, the concentration of ethanol and CO2 (only in the case of H2/CO2 fermentation) in the liquid phase and the liquid outflow rate from the bioreactor are the decision variables. The optimization is executed by a sequential quadratic programming (‘sqp’) algorithm implemented in the ‘fmincon’ function in MatLab R2017b. The system of mass balance equations outlaid for the bioreactor model and thermodynamic feasibility of the catabolic reactions are used as constraints within the optimization. The calculation process is schematized in the Fig. 6.
Gas concentration profiles
It is important to assure that at a determined operation point, the microbial uptake of electron donors does not meet gas concentrations that lead to either unfeasible catabolic reactions at the top of the liquid column or inhibition at the bottom (in the case of CO). Thus the dissolved CO, CO2 and H2 concentration profiles along the height of the liquid column are estimated by linearly discretizing the y-axis into 9 ideally mixed compartments stacked vertically. The initial assumption of the liquid phase having a homogenous composition is corrected by this calculation.
The approach is similar to that used in [89], where the height of the liquid column () has three mixing cells stacked vertically (see Eq. 19 [89]); then each cell is subdivided into three compartments stacked horizontally which recreate the effect of dispersion on the radial direction assuming the liquid flow will follow a helicoidal stream line.
| 19 |
Mass balance equations are constructed in each compartment assuming the liquid phase will be exchanged between the adjacent segments at a flow rate determined by Eq. (20) [66]; biomass concentration is assumed homogeneously distributed within the whole liquid volume. The gas phase is assumed to behave as a plug flow. Mass transfer properties in each segment are found using the same methodology as explained in section “Mass transfer-based model of the industrial bioreactor” since it is assumed that each compartment would behave as a shallow bubble column.
| 20 |
The compartmentalization scheme is validated by calculating the mixing time using two approaches, i.e., (i) using Eq. (21) [89] and, (ii) simulating a mixing time-determination experiment, in which a tracer is injected at the top compartment; the mixing time is defined as the time it takes for the tracer concentration at the top compartment to reach 95% of the final concentration.
| 21 |
Process configuration
Fresh gas is first mixed with a stream of recycled offgas and is then fed to the large-scale bioreactor. The fermentable gas is consumed during fermentation and ethanol is produced. At the exit, the offgas is compressed to 3.1 × 105 Pa (a pressure equal to the bioreactor bottom pressure—see Table 4) and cooled to − 6 °C in order to condense nearly 100% of the water–ethanol mixture. This condensate stream along with the fermentation broth is sent to distillation. Figure 7 shows the conceptualized process configuration.
Fig. 7.
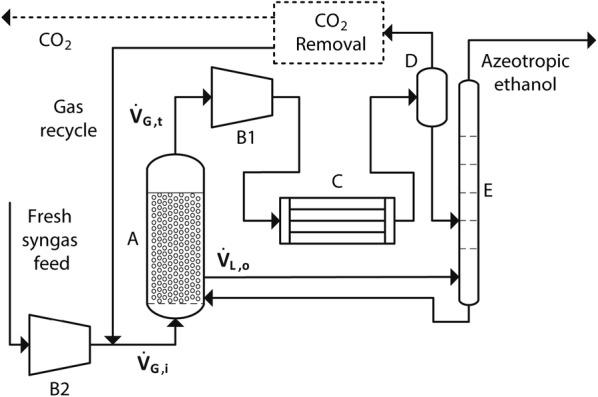
Conceptual process configuration; A: bubble column bioreactor, B1 and B2: gas compression, C: cooling and condensation, D: flash separation, E: azeotropic distillation. For the case in which the H2/CO2 mixture is fed into the bioreactor, the CO2 removal unit is not needed
When pure CO is fed to the fermentation, the dry offgas undergoes CO2 removal prior to recycling; this operation is not needed if H2/CO2 was used as feedstock. Since gas recycling has not been included in process designs reported in literature [90], the possible effects of not including it are discussed within the sensitivity analysis.
Process assessment
Technical performance
In addition to , the process technical performance is evaluated from the perspective of the gas utilization ‘per step’ () inside the bioreactor (Eq. 22) and the energy requirements of the fermentation plus the ethanol separation processes. is calculated as the ratio between the amounts of depleted across the reactor and the fed (fresh plus recycled). Energy requirements account for: (i) power for compression of the gas feed and the offgas, (ii) power for condensation of evaporated ethanol and water and, (iii) heat for the ethanol azeotropic distillation from both: the fermentation broth outflow and the stream recovered from the offgas (data taken from [91]). The power requirements for cooling of bioreactor contents and compressed gases is not accounted for since the utility that would be used is cooling water at ambient temperature and thus its energetic burden is negligible.
| 22 |
The MatLab codes developed within this study for simulating the CO and the H2/CO2 fermentation processes are available as Additional files 2 and 3, respectively.
Sensitivity analysis
Process performance is evaluated at different values of the constants governing the electron donor uptake kinetics (Eqs. 14 and 15), i.e, half-saturation constants () and the maximum specific uptake rates (). This analysis is made due to (i) uncertainty generated by the scarce information available in literature about these parameters, (ii) uncertainties associated with the methodologies used to fix these parameters (see section “Model validation”) and (iii) their paramount importance for the model reliability in quantitative terms.
Additionally, bioreactor performance is also assessed using different values of the mass transfer coefficients, the concentration of ethanol, the height of the liquid column and the gas hold-up. Specifically, 100% higher and 50% lower values of the mass transfer coefficients for CO, H2 and CO2 (as predicted by Eq. 16) are used. The range of possible values was selected considering that: (i) the possible presence of surfactants that may hamper mass transfer by 70% at concentrations as low as 10 ppm [92]; (ii) the uncertain effect of ethanol concentration on mass transfer since it has been reported that ethanol at a wide range of concentrations may rise the gas hold-up by four times [86] while at 50 g L−1, the mass transfer coefficient would increase by 50% [93] and (iii) the proven ability of C. ljungdahlii and C. carboxydivorans [69, 74, 94, 95] to form biofilms which, if shaped into granules may enhance mass transfer by 30% due to intraparticle liquid circulation forced by pressure gradients caused by the circulation of bacteria inside the bioreactor [96].
Similarly, the maintenance of ethanol concentrations at 80 g L−1 are assessed considering that long-term adaptation experiments of C. thermocellum led to a 100% increase in its tolerance to both, ethanol and n-butanol [97], promoted in part by a change in the structural composition of its membrane.
Finally, disregarding possible conflicts with legislation and safety measures, the effect of an increased aspect ratio of the bubble column, form 3 to 10, is also assessed. Moreover, considering that 15 and 30% are regraded as standard values for large bubble column bioreactors to maintain high productivites [83], the effects of using a gas hold-up value of 30% on the process and the bioreactor performance are also assessed in the sensitivity analysis.
Supplementary information
Additional file 1. Supplementary information about the equations used for the construction of the model and supporting information about examples given within the main document.
Additional file 2. MatLab codes for the simulation of CO fermentation.
Additional file 3. MatLab codes for the simulation of H2/CO2 fermentation.
Acknowledgements
The authors thank Joseph Heijnen and Wouter van Winden for their respective critical assessments on the construction of the black-box model of microbial reactions and the validation of the 9-compartment model for estimating concentration profiles.
Latin letters
- ATP
Adenosine triphosphate
Concentration in fermentation broth, mol m−3
Carbon source, i.e., CO or CO2
Electron donor of catabolism, i.e, CO, H2
Diffusivity in pure water, m2 s−1
- F
Molar flow rate, mol s−1
- g
Gravity’s acceleration, 9.8 m s−1
Height of gas–liquid column the bioreactor, m
Gas–liquid mass transfer coefficient, s−1
Half-saturation constant, mol m−3
CO inhibition constant, mol m−3
Mixing number, 16 [91]
Number of mixing cells in the bioreactor
Pressure, Pa
Biomass specific production/consumption rate, mol Cmol−1x h−1
Volumetric productivity, gethanol L−1 h−1
Ideal gas constant, 8.134 m3 Pa mol−1 K−1
Process temperature, K
Mixing time, s
Gas utilization, %
Pressure-corrected superficial gas velocity, m s−1
Volumetric flow rate, m3 s−1
- WLP
Wood–Ljungdahl pathway
Molar fraction in gas phase
Biomass yield per mole of carbon source, Cmolx mol−1CS
Greek letters
Degree of reduction, mole− mol−1
Standard Gibbs free energy change, kJ mol−1
Gibbs free energy dissipation in anabolism, kJ mol−1
Gibbs free energy change at physiological conditions, kJ mol−1
Standard enthalpy change, kJ mol−1
Hold-up
Biomass growth rate, h−1
Diameter of bioreactor vessel, m
Stoichiometric coefficient; positive for products, negative for reactants
Subscripts and superscripts
Anabolism
Calculated at bottom of fermentor
Catabolism
Theoretically consumed
Ethanol
Electron
Gas
At gas inlet
Liquid fermentation broth
Maximum
Metabolism
At bioreactor outlet
Reaction
Gas components, i.e, CO, H2, CO2
Calculated at top of fermentor
Water
Dry microbial biomass
Calculated at standard conditions, 101 kPa and 0 °C
Authors’ contributions
EAB, HN, RMF and JAP contributed to the design of the study, the construction of the model and the editing of the manuscript. EAB wrote the MatLab code, carried out the simulations and wrote the manuscript. HN assessed the definition of assumptions. JAP conceived the final structure of the manuscript. All authors read and approved the final manuscript.
Funding
The authors thank DSM and the BE-Basic Foundation for the financial support provided in the form of a Ph.D. scholarship for the author EAB. This work is part of a Dual Degree Ph.D. project under the agreement between Unicamp and TU Delft.
Availability of data and materials
The MatLab codes created and used for the current study have been provided as additional files and are also available in the Zenodo repository.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-020-01695-y.
References
- 1.Liew F, Martin ME, Tappel RC, Heijstra BD, Mihalcea C, Köpke M. Gas fermentation—A flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wieckert C, Obrist A, von Zedtwitz P, Maag G, Steinfeld A. Syngas production by thermochemical gasification of carbonaceous waste materials in a 150 kWth packed-bed solar reactor. Energy Fuels. 2013;27(8):4770–4776. doi: 10.1021/ef4008399. [DOI] [Google Scholar]
- 3.Couto ND, Silva VB, Monteiro E, Rouboa A. Assessment of municipal solid wastes gasification in a semi-industrial gasifier using syngas quality indices. Energy. 2015;93:864–873. doi: 10.1016/j.energy.2015.09.064. [DOI] [Google Scholar]
- 4.Almeida Benalcázar E, Deynoot BG, Noorman H, Osseweijer P, Posada JA. Production of bulk chemicals from lignocellulosic biomass via thermochemical conversion and syngas fermentation: a comparative techno-economic and environmental assessment of different site-specific supply chain configurations: techno-economic and environmental assessment of bulk chemicals production though biomass gasification and syngas fermentation. Biofuels Bioprod Biorefining. 2017;11(5):861–886. doi: 10.1002/bbb.1790. [DOI] [Google Scholar]
- 5.de Medeiros EM, Posada JA, Noorman H, Osseweijer P, Maciel Filho R. Hydrous bioethanol production from sugarcane bagasse via energy self-sufficient gasification-fermentation hybrid route: simulation and financial analysis. J Clean Prod. 2017;168:1625–1635. doi: 10.1016/j.jclepro.2017.01.165. [DOI] [Google Scholar]
- 6.Griffin DW, Schultz MA. Fuel and chemical products from biomass syngas: a comparison of gas fermentation to thermochemical conversion routes. Environ Prog Sustain Energy. 2012;31(2):219–224. doi: 10.1002/ep.11613. [DOI] [Google Scholar]
- 7.Daniell J, Köpke M, Simpson S. Commercial biomass syngas fermentation. Energies. 2012;5(12):5372–5417. doi: 10.3390/en5125372. [DOI] [Google Scholar]
- 8.Kopke M, Mihalcea C, Liew F, Tizard JH, Ali MS, Conolly JJ, et al. 2,3-butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl Environ Microbiol. 2011;77(15):5467–5475. doi: 10.1128/AEM.00355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, et al. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci. 2010;107(29):13087–13092. doi: 10.1073/pnas.1004716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljungdahl L. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu Rev Microbiol. 1986;40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 11.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta. 2008;1784(12):1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latif H, Zeidan AA, Nielsen AT, Zengler K. Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr Opin Biotechnol. 2014;27:79–87. doi: 10.1016/j.copbio.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol. 2014;12(12):809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 14.Abubackar HN, Veiga MC, Kennes C. Carbon monoxide fermentation to ethanol by Clostridium autoethanogenum in a bioreactor with no accumulation of acetic acid. Bioresour Technol. 2015;186:122–127. doi: 10.1016/j.biortech.2015.02.113. [DOI] [PubMed] [Google Scholar]
- 15.Kerby R, Zeikus JG. Growth of Clostridium thermoaceticum on H2/CO2 or CO as energy source. Curr Microbiol. 1983;8(1):27–30. doi: 10.1007/BF01567310. [DOI] [Google Scholar]
- 16.Xie B-T, Liu Z-Y, Tian L, Li F-L, Chen X-H. Physiological response of Clostridium ljungdahlii DSM 13528 of ethanol production under different fermentation conditions. Bioresour Technol. 2015;177:302–307. doi: 10.1016/j.biortech.2014.11.101. [DOI] [PubMed] [Google Scholar]
- 17.Hurst KM, Lewis RS. Carbon monoxide partial pressure effects on the metabolic process of syngas fermentation. Biochem Eng J. 2010;48(2):159–165. doi: 10.1016/j.bej.2009.09.004. [DOI] [Google Scholar]
- 18.Straub M, Demler M, Weuster-Botz D, Dürre P. Selective enhancement of autotrophic acetate production with genetically modified Acetobacterium woodii. J Biotechnol. 2014;178:67–72. doi: 10.1016/j.jbiotec.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Bertsch J, Müller V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol Biofuels. 2015;8(1):210. doi: 10.1186/s13068-015-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noorman HJ, Heijnen JJ. Biochemical engineering’s grand adventure. Chem Eng Sci. 2017;170:677–693. doi: 10.1016/j.ces.2016.12.065. [DOI] [Google Scholar]
- 21.Chen J, Gomez JA, Höffner K, Barton PI, Henson MA. Metabolic modeling of synthesis gas fermentation in bubble column reactors. Biotechnol Biofuels. 2015;8(1):89. doi: 10.1186/s13068-015-0272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atiyeh HK, Philips JR, Huhnke R. Fermentation control for optimization of syngas utilization. Patent WO/2016/077778; 2016.
- 23.Phillips J, Huhnke R, Atiyeh H. Syngas fermentation: a microbial conversion process of gaseous substrates to various products. Fermentation. 2017;3(2):28. doi: 10.3390/fermentation3020028. [DOI] [Google Scholar]
- 24.Hu P, Bowen SH, Lewis RS. A thermodynamic analysis of electron production during syngas fermentation. Bioresour Technol. 2011;102(17):8071–8076. doi: 10.1016/j.biortech.2011.05.080. [DOI] [PubMed] [Google Scholar]
- 25.Norman ROJ, Millat T, Schatschneider S, Henstra AM, Breitkopf R, Pander B, et al. Genome-scale model of C. autoethanogenum reveals optimal bioprocess conditions for high-value chemical production from carbon monoxide. Eng Biol. 2019;3(2):32–40. doi: 10.1049/enb.2018.5003. [DOI] [Google Scholar]
- 26.Chen J, Gomez JA, Höffner K, Phalak P, Barton PI, Henson MA. Spatiotemporal modeling of microbial metabolism. BMC Syst Biol. 2016;10(1):21. doi: 10.1186/s12918-016-0259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu JK, Lloyd C, Al-Bassam MM, Ebrahim A, Kim J-N, Olson C, et al. Predicting proteome allocation, overflow metabolism, and metal requirements in a model acetogen. PLoS Comput Biol. 2019;15(3):e1006848. doi: 10.1371/journal.pcbi.1006848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redl S, Sukumara S, Ploeger T, Wu L, Ølshøj Jensen T, Nielsen AT, Noorman HJ. Thermodynamics and economic feasibility of acetone production from syngas using the thermophilic production host Moorella thermoacetica. Biotechnol Biofuels. 2017;10(1):150. doi: 10.1186/s13068-017-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camacho Ardila Y, Figueroa JEJ, Lunelli B, Maciel Filho R, Wolf Maciel MR. Simulation of ethanol production via fermentation of the synthesis gas using aspen plus. Chem Eng Trans. 2014;37:637–642. [Google Scholar]
- 30.Pardo-Planas O, Atiyeh HK, Phillips JR, Aichele CP, Mohammad S. Process simulation of ethanol production from biomass gasification and syngas fermentation. Bioresour Technol. 2017;245:925–932. doi: 10.1016/j.biortech.2017.08.193. [DOI] [PubMed] [Google Scholar]
- 31.de Medeiros EM, Posada JA, Noorman H, Maciel Filho R. Dynamic modeling of syngas fermentation in a continuous stirred-tank reactor: multi-response parameter estimation and process optimization. Biotechnol Bioeng. 2019;116(10):2473–2487. doi: 10.1002/bit.27108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valgepea K, de Lemgruber R, Meaghan K, Palfreyman RW, Abdalla T, Heijstra BD, et al. Maintenance of ATP homeostasis triggers metabolic shifts in gas-fermenting acetogens. Cell Syst. 2017;4(5):505–515. doi: 10.1016/j.cels.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Valgepea K, de Souza PintoLemgruber R, Abdalla T, Binos S, Takemori N, Takemori A, et al. Drives metabolic rearrangements in gas-fermenting Clostridium autoethanogenum. Biotechnol Biofuels. 2018;11(1):55. doi: 10.1186/s13068-018-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter H, Molitor B, Wei H, Chen W, Aristilde L, Angenent LT. Ethanol production in syngas-fermenting Clostridium ljungdahlii is controlled by thermodynamics rather than by enzyme expression. Energy Env Sci. 2016;9(7):2392–2399. doi: 10.1039/C6EE01108J. [DOI] [Google Scholar]
- 35.Nagarajan H, Sahin M, Nogales J, Latif H, Lovley DR, Ebrahim A, et al. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb Cell Factories. 2013;12(1):118. doi: 10.1186/1475-2859-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valgepea K, Loi KQ, Behrendorff JB, de Lemgruber SP, Plan M, Hodson MP, et al. Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum. Metab Eng. 2017;41:202–211. doi: 10.1016/j.ymben.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Henson MA. In silico metabolic engineering of Clostridium ljungdahlii for synthesis gas fermentation. Metab Eng. 2016;38:389–400. doi: 10.1016/j.ymben.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Griffin D, Li X, Henson MA. Incorporating hydrodynamics into spatiotemporal metabolic models of bubble column gas fermentation. Biotechnol Bioeng. 2019;116(1):28–40. doi: 10.1002/bit.26848. [DOI] [PubMed] [Google Scholar]
- 39.Gu C, Kim GB, Kim WJ, Kim HU, Lee SY. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20(1):121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haringa C, Tang W, Wang G, Deshmukh AT, van Winden WA, Chu J, van Gulik W, Heijnen JJ, Mudde RF, Noorman HJ. Computational fluid dynamics simulation of an industrial P. chrysogenum fermentation with a coupled 9-pool metabolic model: towards rational scale-down and design optimization. Chem Eng Sci. 2018;175:12–24. doi: 10.1016/j.ces.2017.09.020. [DOI] [Google Scholar]
- 41.Noorman H. An industrial perspective on bioreactor scale-down: what we can learn from combined large-scale bioprocess and model fluid studies. Biotechnol J. 2011;6(8):934–943. doi: 10.1002/biot.201000406. [DOI] [PubMed] [Google Scholar]
- 42.Lapin A, Müller D, Reuss M. Dynamic behavior of microbial populations in stirred bioreactors simulated with Euler−Lagrange methods: traveling along the Lifelines of Single Cells †. Ind Eng Chem Res. 2004;43(16):4647–4656. doi: 10.1021/ie030786k. [DOI] [Google Scholar]
- 43.Kleerebezem R, Van Loosdrecht MCM. A generalized method for thermodynamic state analysis of environmental systems. Crit Rev Environ Sci Technol. 2010;40(1):1–54. doi: 10.1080/10643380802000974. [DOI] [Google Scholar]
- 44.Henstra AM. CO metabolism of Carboxydothermus hydrogenformans and Archaeoglobus fulgidus [PhD Thesis]. Wageningen University; 2006. http://edepot.wur.nl/34992. Accessed on 4 Sep 2018.
- 45.Poehlein A, Schmidt S, Kaster A-K, Goenrich M, Vollmers J, Thürmer A, et al. An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS ONE. 2012;7(3):e33439. doi: 10.1371/journal.pone.0033439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richter H, Martin M, Angenent L. A two-stage continuous fermentation system for conversion of syngas into ethanol. Energies. 2013;6(8):3987–4000. doi: 10.3390/en6083987. [DOI] [Google Scholar]
- 47.Mohammadi M, Mohamed AR, Najafpour GD, Younesi H, Uzir MH. Kinetic studies on fermentative production of biofuel from synthesis gas using Clostridium ljungdahlii. Sci World J. 2014;2014:1–8. doi: 10.1155/2014/910590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Kok S, Meijer J, van Loosdrecht MCM, Kleerebezem R. Impact of dissolved hydrogen partial pressure on mixed culture fermentations. Appl Microbiol Biotechnol. 2013;97(6):2617–2625. doi: 10.1007/s00253-012-4400-x. [DOI] [PubMed] [Google Scholar]
- 49.Heijnen JJ. Bioenergetics of microbial growth. In: Flickinger MC, Drew SW, editors. Encyclopedia of bioprocess technology. Hoboken: Wiley; 2002. pp. 265–291. [Google Scholar]
- 50.Skidmore BE, Baker RA, Banjade DR, Bray JM, Tree DR, Lewis RS. Syngas fermentation to biofuels: effects of hydrogen partial pressure on hydrogenase efficiency. Biomass Bioenergy. 2013;55:156–162. doi: 10.1016/j.biombioe.2013.01.034. [DOI] [Google Scholar]
- 51.Ahmed A, Lewis RS. Fermentation of biomass-generated synthesis gas: effects of nitric oxide. Biotechnol Bioeng. 2007;97(5):1080–1086. doi: 10.1002/bit.21305. [DOI] [PubMed] [Google Scholar]
- 52.Adams MWW, Mortensen LE. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. A comparison with hydrogenase I. J Biol Chem. 1984;259(11):7045–7055. [PubMed] [Google Scholar]
- 53.Dobrindt U, Blaut M. Purification and characterization of a membrane-bound hydrogenase from Sporomusa sphaeroides involved in energy-transducing electron transport. Arch Microbiol. 1996;165(2):141–147. doi: 10.1007/s002030050309. [DOI] [PubMed] [Google Scholar]
- 54.Rich JJ, King GM. Carbon monoxide consumption and production by wetland peats. FEMS Microbiol Ecol. 1999;28(3):215–224. doi: 10.1111/j.1574-6941.1999.tb00577.x. [DOI] [Google Scholar]
- 55.Xie H, Bélanger S, Demers S, Vincent WF, Papakyriakou TN. Photobiogeochemical cycling of carbon monoxide in the southeastern Beaufort Sea in spring and autumn. Limnol Oceanogr. 2009;54(1):234–249. doi: 10.4319/lo.2009.54.1.0234. [DOI] [Google Scholar]
- 56.MacCallum SN. Tropospheric carbon monoxide: satellite observations and their applications [PhD Thesis]. University of Edinburgh; 2010. http://hdl.handle.net/1842/4342. Accessed on 20 Sep 2018.
- 57.Greening C, Constant P, Hards K, Morales SE, Oakeshott JG, Russell RJ, et al. Atmospheric hydrogen scavenging: from enzymes to ecosystems. Appl Environ Microbiol. 2015;81(4):1190–1199. doi: 10.1128/AEM.03364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giraldo-Gomez E, Goodwin S, Switzenbaum MS. Influence of mass transfer limitations on determination of the half saturation constant for hydrogen uptake in a mixed-culture CH4-producing enrichment. Biotechnol Bioeng. 1992;40(7):768–776. doi: 10.1002/bit.260400704. [DOI] [PubMed] [Google Scholar]
- 59.Novelli PC, Lang PM, Masarie KA, Hurst DF, Myers R, Elkins JW. Molecular hydrogen in the troposphere: global distribution and budget. J Geophys Res Atmos. 1999;104(D23):30427–30444. doi: 10.1029/1999JD900788. [DOI] [Google Scholar]
- 60.Intergovernmental Panel on Climate Change. Summary for policymakers. In: Stocker TF, Qin D, Plattner G, Tignor M, Allen S, Boschung J, et al., editors. Climate Change 2013: The Physical Science Basis Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; 2013. http://www.climatechange2013.org/images/report/WG1AR5_SPM_FINAL.pdf. Accessed on 4 Oct 2018.
- 61.Wang S, Huang H, Kahnt J, Mueller AP, Kopke M, Thauer RK. NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J Bacteriol. 2013;195(19):4373–4386. doi: 10.1128/JB.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abubackar HN, Fernández-Naveira Á, Veiga MC, Kennes C. Impact of cyclic pH shifts on carbon monoxide fermentation to ethanol by Clostridium autoethanogenum. Fuel. 2016;178:56–62. doi: 10.1016/j.fuel.2016.03.048. [DOI] [Google Scholar]
- 63.Gaddy J, Arora D, Ko CW, Phillips JR, Basu R, Wikstrom C, et al. Methods for increasing the production of ethanol from microbial fermentation. Patent US 7,285,402 B2; 2007.
- 64.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/MMBR.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roels JA. Energetics and kinetics in biotechnology. Amsterdam: Elsevier Biomedical Press; 1983. p. 330. [Google Scholar]
- 66.Heijnen JJ, Van’t Riet K. Mass transfer, mixing and heat transfer phenomena in low viscosity bubble column reactors. Chem Eng J. 1984;28(2):B21–B42. doi: 10.1016/0300-9467(84)85025-X. [DOI] [Google Scholar]
- 67.Weber FJ, Hartmans S. Prevention of clogging in a biological trickle-bed reactor removing toluene from contaminated air. Biotechnol Bioeng. 1996;50(1):91–97. doi: 10.1002/(SICI)1097-0290(19960405)50:1<91::AID-BIT10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 68.Lettinga G, van Velsen AFM, Hobma SW, de Zeeuw W, Klapwijk A. Use of the upflow sludge blanket (USB) reactor concept for biological wastewater treatment, especially for anaerobic treatment. Biotechnol Bioeng. 1980;22(4):699–734. doi: 10.1002/bit.260220402. [DOI] [Google Scholar]
- 69.Philips J, Rabaey K, Lovley DR, Vargas M. Biofilm formation by Clostridium ljungdahlii is induced by sodium chloride stress: experimental evaluation and transcriptome analysis. PLoS ONE. 2017;12(1):e0170406. doi: 10.1371/journal.pone.0170406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molitor B, Richter H, Martin ME, Jensen RO, Juminaga A, Mihalcea C, et al. Carbon recovery by fermentation of CO-rich off gases—turning steel mills into biorefineries. Bioresour Technol. 2016;215:386–396. doi: 10.1016/j.biortech.2016.03.094. [DOI] [PubMed] [Google Scholar]
- 71.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315(5813):801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 72.Chang IS, Kim BH, Lovitt RW, Bang JS. Effect of CO partial pressure on cell-recycled continuous CO fermentation by Eubacterium limosum KIST612. Process Biochem. 2001;37(4):411–421. doi: 10.1016/S0032-9592(01)00227-8. [DOI] [Google Scholar]
- 73.Park S-J, Moon S-H, Lee H-J, Lim J-J, Kim J-M, Seo J, et al. A comparison of human cord blood- and embryonic stem cell-derived endothelial progenitor cells in the treatment of chronic wounds. Biomaterials. 2013;34(4):995–1003. doi: 10.1016/j.biomaterials.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 74.Shen Y, Brown R, Wen Z. Enhancing mass transfer and ethanol production in syngas fermentation of Clostridium carboxidivorans P7 through a monolithic biofilm reactor. Appl Energy. 2014;136:68–76. doi: 10.1016/j.apenergy.2014.08.117. [DOI] [Google Scholar]
- 75.Kataoka H, Takeuchi H, Nakao K, Yagi H, Tadaki T, Otake T, et al. Mass transfer in a large bubble column. J Chem Eng Jpn. 1979;12(2):105–110. doi: 10.1252/jcej.12.105. [DOI] [Google Scholar]
- 76.dos Santos KG, Eckert CT, De Rossi E, Bariccatti RA, Frigo EP, Lindino CA, et al. Hydrogen production in the electrolysis of water in Brazil, a review. Renew Sustain Energy Rev. 2017;68:563–571. doi: 10.1016/j.rser.2016.09.128. [DOI] [Google Scholar]
- 77.Heijnen J. A thermodynamic approach to predict black box model parameters for microbial growth. In: von Stockar U, editor. Biothermodynamics. Switzerland: EPFL Press; 2013. pp. 443–473. [Google Scholar]
- 78.Phillips JR, Klasson KT, Clausen EC, Gaddy JL. Biological production of ethanol from coal synthesis gas: medium development studies. Appl Biochem Biotechnol. 1993;39–40(1):559–571. doi: 10.1007/BF02919018. [DOI] [Google Scholar]
- 79.Maskow T, von Stockar U. How reliable are thermodynamic feasibility statements of biochemical pathways? Biotechnol Bioeng. 2005;92(2):223–230. doi: 10.1002/bit.20572. [DOI] [PubMed] [Google Scholar]
- 80.Müller V, Hess V. The minimum biological energy quantum. Front Microbiol. 2017;8:2019. doi: 10.3389/fmicb.2017.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaikh A, Al-Dahhan M. Scale-up of bubble column reactors: a review of current state-of-the-art. Ind Eng Chem Res. 2013;52(24):8091–8108. doi: 10.1021/ie302080m. [DOI] [Google Scholar]
- 82.Humbird D, Davis R, McMillan JD. Aeration costs in stirred-tank and bubble column bioreactors. Biochem Eng J. 2017;127:161–166. doi: 10.1016/j.bej.2017.08.006. [DOI] [Google Scholar]
- 83.Riet K, van’t Tramper J. Basic bioreactor design. New York: M. Dekker; 1991. p. 465. [Google Scholar]
- 84.Wilke CR, Chang P. Correlation of diffusion coefficients in dilute solutions. AIChE J. 1955;1(2):264–270. doi: 10.1002/aic.690010222. [DOI] [Google Scholar]
- 85.Sander R. Compilation of Henry’s law constants (version 4.0) for water as solvent. Atmos Chem Phys. 2015;15(8):4399–4981. doi: 10.5194/acp-15-4399-2015. [DOI] [Google Scholar]
- 86.Guo K, Wang T, Yang G, Wang J. Distinctly different bubble behaviors in a bubble column with pure liquids and alcohol solutions: distinctly different bubble behaviors in a bubble column. J Chem Technol Biotechnol. 2017;92(2):432–441. doi: 10.1002/jctb.5022. [DOI] [Google Scholar]
- 87.Dalkilic AS, Wongwises S. A performance comparison of vapour-compression refrigeration system using various alternative refrigerants. Int Commun Heat Mass Transf. 2010;37(9):1340–1349. doi: 10.1016/j.icheatmasstransfer.2010.07.006. [DOI] [Google Scholar]
- 88.Almeida Benalcázar E, Noorman H, Maciel Filho R, Posada-Duque J. Hybrid Model for ethanol production via syngas fermentation: coupling between a thermodynamics-based black-box model of bacterial reactions and mass transfer in a large-scale bubble column bioreactor. Proc 27th Eur Biomass Conf Exhib. 2019;2019(27–30):1408–1417. [Google Scholar]
- 89.Groen D. Macromixing in bioreactors [PhD Thesis]. Delft University of Technology; 1994. https://repository.tudelft.nl/islandora/object/uuid%3A3ac019f1-d19a-4853-9a29-554f1149bd5b?collection=research. Accessed 24 Jul 2019.
- 90.Handler RM, Shonnard DR, Griffing EM, Lai A, Palou-Rivera I. Life cycle assessments of ethanol production via gas fermentation: anticipated greenhouse gas emissions for cellulosic and waste gas feedstocks. Ind Eng Chem Res. 2016;55(12):3253–3261. doi: 10.1021/acs.iecr.5b03215. [DOI] [Google Scholar]
- 91.Vane LM. Separation technologies for the recovery and dehydration of alcohols from fermentation broths. Biofuels Bioprod Biorefining. 2008;2(6):553–588. doi: 10.1002/bbb.108. [DOI] [Google Scholar]
- 92.Vasconcelos JMT, Rodrigues JML, Orvalho SCP, Alves SS, Mendes RL, Reis A. Effect of contaminants on mass transfer coefficients in bubble column and airlift contactors. Chem Eng Sci. 2003;58(8):1431–1440. doi: 10.1016/S0009-2509(02)00675-9. [DOI] [Google Scholar]
- 93.Sun Y, Nozawa T, Furusaki S. Gas holdup and volumetric oxygen transfer coefficient in a three-phase fluidized bed bioreactor. J Chem Eng Jpn. 1988;21(1):15–20. doi: 10.1252/jcej.21.15. [DOI] [Google Scholar]
- 94.Shen Y, Brown R, Wen Z. Syngas fermentation of Clostridium carboxidivorans P7 in a hollow fiber membrane biofilm reactor: evaluating the mass transfer coefficient and ethanol production performance. Biochem Eng J. 2014;85:21–29. doi: 10.1016/j.bej.2014.01.010. [DOI] [Google Scholar]
- 95.Shen Y, Brown RC, Wen Z. Syngas fermentation by Clostridium carboxidivorans P7 in a horizontal rotating packed bed biofilm reactor with enhanced ethanol production. Appl Energy. 2017;187:585–594. doi: 10.1016/j.apenergy.2016.11.084. [DOI] [Google Scholar]
- 96.van den Heuvel JC, Vredenbregt LHJ, Portegies-Zwart I, Ottengraf SPP. Acceleration of mass transfer in methane-producing loop reactors. Antonie Van Leeuwenhoek. 1995;67(1):125–130. doi: 10.1007/BF00872200. [DOI] [PubMed] [Google Scholar]
- 97.Tian L, Cervenka ND, Low AM, Olson DG, Lynd LR. A mutation in the AdhE alcohol dehydrogenase of Clostridium thermocellum increases tolerance to several primary alcohols, including isobutanol, n-butanol and ethanol. Sci Rep. 2019;9(1):1736. doi: 10.1038/s41598-018-37979-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary information about the equations used for the construction of the model and supporting information about examples given within the main document.
Additional file 2. MatLab codes for the simulation of CO fermentation.
Additional file 3. MatLab codes for the simulation of H2/CO2 fermentation.
Data Availability Statement
The MatLab codes created and used for the current study have been provided as additional files and are also available in the Zenodo repository.