Abstract
The Mediterranean hot spot includes numerous endemic and socio-economically important plant species seriously threatened by climate change and habitat loss. In this study, the genetic diversity of five populations of Cicer graecum, an endangered endemic species from northern Peloponnisos, Greece and a wild relative of the cultivated Cicer arietinum, was investigated using inter-simple sequence repeats (ISSRs) and amplified fragment length polymorphism (AFLP) markers in order to determine levels and structure of genetic variability. Nei’s gene diversity by ISSR and AFLP markers indicated medium to high genetic diversity at the population level. Moreover, AMOVA results suggest that most of the variation exists within (93 % for AFLPs and 65 % for ISSRs), rather than among populations. Furthermore, Principal Component Analysis based on ISSRs positively correlated the genetic differentiation among the populations to the geographic distances, suggesting that the gene flow among distant populations is limited. The ecological adaptation of C. graecum populations was also investigated by correlation of their genetic diversity with certain environmental variables. Aridity arose as the dominant factor positively affecting the genetic diversity of C. graecum populations. We modelled the realized climatic niche of C. graecum in an ensemble forecasting scheme under three different global circulation models and two climate change scenarios. In all cases, a severe range contraction for C. graecum is projected, highlighting the high extinction risk that is probably going to face during the coming decades. These results could be a valuable tool towards the implementation of an integrated in situ and ex situ conservation scheme approach for activating management programmes for this endemic and threatened species.
Keywords: Climate change, conservation, extinction risk, genetic diversity, genetic resources
The genetic diversity of five populations of Cicer graecum, an endangered endemic species from Northern Peloponnisos (Greece), was investigated using inter-simple sequence repeat (ISSR) and amplified fragment length polymorphism (AFLP) markers. Population genetic diversity was correlated with certain environmental variables and the realized climatic niche of C. graecum according to different climate change scenarios was modelled. The results indicated medium to high population genetic diversity, which was most strongly affected by aridity. A severe range contraction and high extinction risk for C. graecum is projected in the near future. The results constitute a valuable tool towards the implementation of effective in situ and ex situ conservation schemes.
Introduction
Growing concern regarding predicted disastrous impacts of climate change on global biodiversity and food security has set crop wild relatives (CWRs) at the core of active conservation efforts globally (Dempewolf et al. 2014). Crop wild relatives often contain traits of great importance for agriculture, and recent cultivars of most major crops already contain genes from them (Heywood 2008). Domesticated crops have been experiencing domestication bottlenecks, resulting in lower allelic diversity than their wild progenitors, exacerbated by the increasing demand for high crop productivity and uniformity (van Heerwaarden et al. 2011; Dempewolf et al. 2017).
Crop wild relatives, however, having natural populations in environmentally diverse areas, usually maintain high genetic diversity through population differentiation and local adaptation. The genetic structure of natural populations is shaped by the influence of several abiotic and biotic variables. Life history traits, e.g. breeding system, dispersal mode, life form, geographical range and ecogeographical characteristics, shape patterns of genetic diversity within and among plant populations (Hamrick and Godt 1996). Understanding their genetic and adaptive diversity patterns across their distribution range is crucial for their effective use in breeding programmes (Ganopoulos et al. 2011; Egan et al. 2018) and for ensuring their long-term survival, as numerous CWRs are currently threatened by habitat loss and climate change (Redden et al. 2015). Despite the urgent need for new, stress-tolerant crop varieties and conservation of genetically important traits of wild populations, the intra-population diversity of most CWRs remains largely unknown. Data deficiency in legume wild relatives (WRs), in particular, is especially high as only a few species have been studied to date (e.g. Kouam et al. 2012; Smýkal et al. 2018).
Cicer (Fabaceae, Cicereae) consists of 42 species of annual and perennial herbs or small shrubs mainly distributed in Central and Southwest Asia, with a scattered distribution in the Mediterranean region and East Africa (Javadi et al. 2007). Among these, Cicer arietinum (chickpea), which is a self-pollinated species, has a high economic value as a food crop and assigns particular importance to this genus. Chickpea is one of the earliest crops cultivated by man (Zohary and Hopf 2000), and continues to play an important role in modern agricultural systems, ranking third behind Phaseolus vulgaris and Pisum sativum in terms of world grain legume production (Croser et al. 2003).
In general, chickpea is characterized by narrow genetic variability (Andeden et al. 2013). The low level of genetic diversity is a limiting factor in the development of new cultivars with resistance to biotic and abiotic stresses. The WRs of the species are an important alternative genetic resource for its breeding (Toker et al. 2014; Srivastava et al. 2016). However, there are few studies on the genetic diversity of annual WRs by using DNA-based molecular markers, while the corresponding research on perennials is even more limited (Ahmad et al. 2005). More specifically, the genetic diversity of annual WRs has been studied by amplified fragment length polymorphisms (AFLPs) (Nguyen et al. 2004; Sudupak 2004), inter-simple sequence repeats (ISSRs) (Rajesh et al. 2003; Sudupak 2004; Amirmoradi et al. 2012), RAPDs (Sudupak et al. 2003; Talebi et al. 2009), ScoT, DAMD-PCR and ISSRs (Amirmoradi et al. 2012), iPBS-Retrotransposons (Andeden et al. 2013), while that of perennial WRs by AFLPs (Nguyen et al. 2004; Sudupak 2004) and ISSRs (Rajesh et al. 2003). Recently, von Wettberg et al. (2018) studied the population genomics of the annuals C. echinospermum and C. reticulatum in combination with traits of potential agronomic importance.
Cicer graecum is a rare perennial species, endemic to northern Peloponnisos (Greece) (Dimopoulos et al. 1996, 2016). It is restricted to the northern slopes of Mts. Kyllini and Chelmos, where it grows at moderate altitudes, from ca. 400 to 1400 m a.s.l. Together with the Anatolian endemics C. floribundum and C. isauricum, and the Balkan-Anatolian C. montbretii form a well-supported monophyletic group (Javadi et al. 2007).
Our primary goal in this study was to reveal the genetic diversity of the narrow endemic C. graecum populations in northern Peloponnisos. Additionally, the relation of genetic diversity with population size was also detected. Several studies have demonstrated that DNA-based molecular markers are valuable tools to unravel the genetic diversity of endemic natural populations (Breinholt et al. 2009; Rameshkumar et al. 2019). For that purpose, a combination of ISSR and AFLP markers was employed to 97 individuals belonging to five populations. The possible genetic structure of C. graecum and its correlation to isolation by distance were also investigated. Furthermore, we used species distribution modelling in order to predict present and future habitat suitability for the species, using distribution data and several environmental variables, in order to estimate its potential extinction risk due to climate change and to develop an effective management and conservation scheme.
Materials and Methods
Plant sampling and DNA extraction
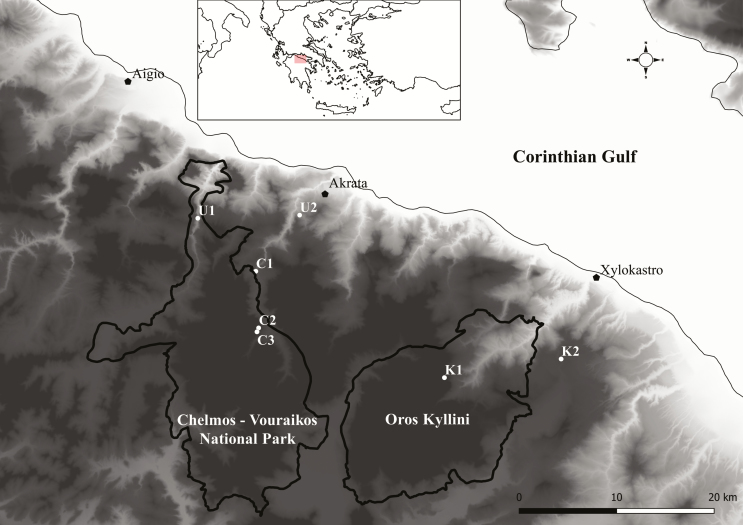
We visited all known localities at the entire distribution range of C. graecum, based on the available herbarium specimens and the results of previous field work. We were able to locate five of the seven known populations (Table 1; Fig. 1) and leaves of individual plants (~20 from each population) were collected. Populations from Mt. Kyllini have the code K1 and K2 while populations from Mt. Chelmos have the code C1, C2 and C3. Isolation of DNA from leaves was performed with the CTAB method (Doyle 1991). The DNA concentration was estimated by standard spectrophotometric method at 260 nm UV lengths by an Eppendorf BioPhotometer and the integrity by gel electrophoresis in a 0.8 % agarose gel. Samples were then diluted to 20 ng μL−1 work concentrations.
Table 1.
Sampled populations of Cicer graecum.
| Population | Abbreviation | Latitude | Longitude | Elevation (m) | Habitat |
|---|---|---|---|---|---|
| Limni Tsivlou | C1 | 38°04′36″ | 22°14′08″ | 740 | Platanus orientalis woodland |
| Peristera 1 | C2 | 38°01′28″ | 22°14′24″ | 1280 | Rocky limestone slope |
| Peristera 2 | C3 | 38°01′15″ | 22°14′18″ | 1230 | Rocky limestone slope |
| Trikala | K1 | 37°58′53″ | 22°27′29″ | 1260 | Abies cephalonica–Pinus nigra woodland |
| Zemeno | K2 | 38°00′02″ | 22°35′38″ | 830 | Quercus coccifera scrub |
Figure 1.
Locations of the populations of C. graecum on Mt. Chelmos (C1–C3) and Mt. Kyllini (K1–K2), and the borders of the protected areas on these mountains. U1 and U2 correspond to known localities where the species was not found.
ISSR procedure
PCR for ISSR analysis was performed in a total volume of 25 μL, containing 20 ng total genomic DNA, 200 mM of each dNTP, 2 mM MgCl2, 40 pmol of primers, 2.5 μL Taq DNA polymerase buffer and 1 U Kapa Taq polymerase (Kapa Biosystems, Wilmington, MA, USA).
To study ISSRs, seven oligonucleotide primers complementary to simple sequence repeats (UBC807, UBC811, UBC826, UBC818, UBC841, UBC850, UBC880) were used for PCR amplification. Binary data points denote the presence/absence of each distinguishable band across all samples for the same primer in both replicate sets of amplifications. PCR amplifications were performed in an Eppendorf Mastercycler EP Thermal Cycler Range as follows: an initial step of 5 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, for denaturation, 90 s at 45–60 °C (depending on the primer used) for annealing and 90 s at 72 °C, for elongation. A 5-min step at 72 °C was programmed as the final extension. Amplification products were separated by electrophoresis on 1.5 % agarose gel and stained with ethidium bromide. A 2-log DNA ladder (New England Biolabs) was used as a size marker. Gel images were placed in a UVItecTransilluminator (UVItec Limited, Cambridge, UK) and analysed using UVIDoc software UVIDocMw version 99.04 (UVItec Limited).
AFLP procedure
For the AFLP procedure total genomic DNA (200 ng) was digested with 4 U of EcoRI and MseI for 3 h at 37 °C. Digested DNA fragments and EcoRI and MseI adapters were ligated with T4 DNA ligase (New England Biolabs) for 3 h at 26 °C. The resulting DNA was used as the primary template DNA in the AFLP analysis. A primer pair based on the sequences of the EcoRI and MseI adapters (Table 2) with one additional selective nucleotide at the 3′ end (EcoRI+A and MseI+C) was used for the first PCR step (pre-amplification). Pre-amplification PCR was performed in a total volume of 20 μL containing 1× Kapa Taq Buffer, 0.2 mM of each dNTP, 2.5 mM MgCl2, 30 ng of each primer EcoRI+A, MseI+C, 1 U Taq DNA polymerase (Kapa Biosystems) and 5 μL of diluted fragments (from the digestion and ligation reaction). Cycling was carried out on a BioRad thermocycler with a 95 °C hold for 30 s followed by 32 cycles of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 1 min and subsequently followed by a final hold at 72 °C for 10 min. A 5-μL aliquot of the reaction was electrophoresed on agarose to verify amplification; the remaining 15 μL was diluted 5-fold with Tris-EDTA. Selective amplifications were carried out in 10-μL total volumes consisting of 3 μL of diluted preselective template and using the same reaction conditions as for preselective amplification, but using 30 ng of an MseI primer and 5 ng of an EcoRI primer per reaction. Selective amplification cycling was performed on a BioRad thermocycler with the following programme: an initial cycle of 95 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min; then 12 cycles of 95 °C for 30 s with an annealing temp starting at 65 °C for 30 s, but decreasing by 0.75 °C each cycle, 72 °C for 1 min; finally, 23 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min, with a final hold at 72 °C for 30 min. Preselective and selective primers for the AFLP procedure are described in Table 2. Replicate analyses were conducted once employing the same DNA extractions.
Table 2.
Sequences of adapters and primers for AFLP analysis.
| 5′→3′ Sequence | |
|---|---|
| Adapters | |
| EcoRI adapter | CTCGTAGACTGCGTACC |
| CTGACGCATGGTTAA | |
| MseI adapter | GACGATGAGTCCTGAG |
| TACTCAGGACTCAT | |
| Preselective primers | |
| EcoRI+A | GACTGCGTACCAATTC-A |
| MseI+C | GATGAGTCCTGAGTAA-C |
| AFLP selective primers-unlabelled | |
| MseI-CAG | GATGAGTCCTGAGTAA-CAG |
| AFLP selective primers-labelled | |
| EcoRI-ACT (fluorescent dye:Hex) | GACTGCGTACCAATTC-ACT |
| EcoRI-AAC (fluorescent dye:Rox) | GACTGCGTACCAATTC-AAC |
| EcoRI-ATG (fluorescent dye:Fam) | GACTGCGTACCAATTC-ATG |
| EcoRI-AAG fluorescent dye:Tamra) | GACTGCGTACCAATTC-AAG |
Fluorescent amplified fragment length polymorphism product mixtures were denatured in formamide at 94 °C for 2 min and electrophoretically separated using an ABI Prism 3730xl automated fluorescence sequencer (Applied Biosystems). Four primer combinations were screened for AFLP analysis; genotypes were scored for the presence or absence of given fragments. The size of detected fragments was determined by the Genemapper v4.0 program using an internal standard (GS 500 LIZ, Applied Biosystems). Fragments ranging from 150 to 500 bases in size were counted and further analysed in order to reduce the impact of potential size homoplasy (Vekemans et al. 2002).
Structure analysis
For ISSR and AFLP markers, possible population structure was investigated using a model-based Bayesian procedure implemented in the software STRUCTURE v2.3.4 (Pritchard et al. 2000). The analysis was carried out using a burning period of 100 000 iterations and a run length of 100 000 Markov chain Monte Carlo replications. We tested a continuous series of K, from 1 to 8, in five independent runs. We did not introduce prior knowledge about the population of origin, and assumed correlated allele frequencies and admixture (Falush et al. 2003). For selecting the optimal value of K, DK values (Evanno et al. 2005) were calculated using STRUCTURE HARVESTER (Earl,and vonHoldt 2012).
Data analysis
Inter-simple sequence repeats and AFLPs are dominant markers, each band representing the phenotype at a single biallelic locus. Only bands that could be unambiguously scored were used in the analysis. Inter-simple sequence repeat- and AFLP-amplified bands were scored for band presence (1) or absence (0), and two binary qualitative data matrices were formed.
Analysis of the ISSR and AFLP data was done with POPGENE version 1.32 (Yeh et al. 2000), using the option for dominant diploid markers. GenAlEx 6.502b (Peakall and Smouse 2006) was used to determine genetic structure at hierarchical levels (AMOVA), to reveal band patterns and frequencies, and to plot grouping of individuals in the principal coordinates analysis (PCoA).
A cluster analysis using an unweighted pair-group method with arithmetic averaging (UPGMA) (Sneath et al. 1975) was performed using the POPGENE 1.32 software (Yeh et al. 2000). The individual contributions of aspect and sites on genotypic variability were assessed under the AMOVA framework using GenALEx ver.6.5b5 (Peakall and Smouse 2006), with sites nested within aspect. Tests for statistical significance were based on 9999 random permutations, followed by sequential Bonferroni correction.
We generated a Moran’s spatial correlogram with ‘fossil’ 0.3.7 (Vavrek 2010) and ‘vegan’ 2.5.2 (Oksanen et al. 2018) packages, to check whether there is any spatial correlation between the genetic distances of the population, based on both ISSR and AFLP markers and on their geographical distance.
Species occurrence data
In order to avoid pseudo-replication and associated spatial sampling biases, we selected occurrences with a minimum distance from each other (250 m). We removed the fewest records necessary to substantially reduce the effects of sampling bias, while simultaneously retaining the greatest amount of useful information. This subsampling reduces the spatial aggregation of records to prevent that SDMs reflect a possible over-representation of environmental conditions associated with regions of higher sampling, which hinder interpretation and application of models (Elith et al. 2010; Aiello-Lammens et al. 2015). This data cleaning and organizing procedure followed the protocols as set out in Robertson et al. (2016) and we used the ‘biogeo’ 1.0 (Robertson et al. 2016) and ‘spThin’ 0.1.0 (Aiello-Lammens et al. 2015) packages. We also evaluated whether any geographical sampling bias existed in our species occurrence data by comparing the statistical distance distribution observed in our data set to a simulated distribution expected under random sampling via the ‘sampbias’ 0.1.1 (https://github.com/azizka/sampling_analyses2.0/tree/master) package.
Environmental data
We obtained current climatic data (19 bioclimatic variables) from the WorldClim database (Hijmans et al. 2005) at a 30-s resolution. Soil variables were obtained from the SoilGrids 250 m database (Hengl et al. 2017) (https://www.soilgrids.org). We used 56 soil variables that provide predicted values for the surface soil layer at varying depths (0–200 cm, 250-m resolution). Elevation data were derived from the CGIAR-CSI data portal (http://srtm.csi.cgiar.org; Jarvis et al. 2008). All environmental variables were statistically downscaled using functions from the ‘raster’ 2.6.7 (Hijmans et al. 2005) and ‘automap’ 1.0.14 (Hiemstra et al. 2009) packages, so as to match the resolution of soil variables.
Regarding future climate, we obtained current climatic data for 2050 and 2070 for three global circulation models (GCMs) that are rendered more suitable and realistic for the study area’s future climate per (McSweeney et al. 2015) and two different Intergovernmental Panel on Climate Change (IPCC) scenarios from the Representative Concentration Pathways (RCPs) family: RCP 2.6 (mild scenario) and RCP 8.5 (severe scenario) from the WorldClim database (Hijmans et al. 2005) at a 30-s resolution. We also constructed 16 additional climatic variables at the same resolution via the ‘envirem’ 1.1 (Title and Bemmels 2018) package based on the 19 bioclimatic variables from WorldClim for current and future climate conditions.
From this initial set of 92 predictors, only eight (temperature seasonality, temperature annual range, precipitation of the wettest month, cation exchange soil capacity, weight percentage of the clay and silt particles, volumetric percentage of coarse fragments and soil organic carbon content) were not highly correlated (Spearman rank correlation < 0.7 and VIF < 5; Dormann et al. 2013). Multicollinearity assessment was performed with the ‘usdm’ 1.1.18 (Naimi et al. 2014) package.
Species Distribution Models and Risk Assessment
Model parameterization and evaluation.
We modelled he realized climatic niche of C. graecum by combining the available occurrence data with current environmental predictors with the ‘biomod2’ 3.3.7 (Thuiller et al. 2009) and ‘ecospat’ 2.2.0 (Di Cola et al. 2017) packages. We used three different modelling algorithms for our study species: Random Forest (RF), Classification Tree Analysis (CTA) and Artificial Neural Networks (ANN) in an ensemble modelling scheme, as ensemble forecasting integrates the results of multiple SDM algorithms into a single geographical projection for each time period, reducing the uncertainties associated with the use of a single model algorithm (Araújo and New 2007; Araújo et al. 2019). Since C. graecum is a rare species with very few known localities, we followed the ensemble of small models (ESMs) framework (Breiner et al. 2015), which is suitable for modelling rare species (Breiner et al. 2015, 2017, 2018). As the algorithms we used require presence/absence (PA) data, we generated PAs following the recommendations of Barbet-Massin et al. (2012) and pseudo-absences were generated at a minimum distance of 5.7 km from present locations to reduce the probability of false absences. We chose that minimum distance due to the median autocorrelation of 5.6 km among the non-collinear environmental variables, which we computed with ‘blockCV’ 1.0.0 (Valavi et al. 2019) package. Presence/absence generation and model calibration were repeated 100 times to ensure that the selected pseudo-absences did not bias the final predictions. We calibrated ESMs by fitting numerous bivariate models, which were then averaged into an ensemble model using weights based on model performances. For all models, the weighted sum of presences was equal to that of the PAs. The predictive performance of the models was evaluated via the true skill statistic (TSS; Allouche et al. 2006), based on a repeated (10 times) split-sampling approach in which models were calibrated with 80 % of the data and evaluated over the remaining 20 %. We used null model significance testing (Raes and ter Steege 2007) to evaluate the performance of our model and estimated the probability that each model performed better than 100 null models. Our model outperformed the null expectation at P < 0.001.
Model projections.
Calibrated models were used to project the suitable area for our species in the study area under current and future conditions through an ensemble forecast approach (Araújo and New 2007). The contribution of each model to the ensemble forecast was weighted according to its TSS score. To avoid working with poorly calibrated models those with a TSS score < 0.8 were excluded from building projections. Note that while model evaluation was carried out using the data-splitting procedure mentioned above, the final models used for spatial projections were calibrated using 100 % of the data, thus allowing taking advantage of all available data. Binary transformations were carried out using the value maximizing the TSS score as the threshold for distinguishing presence and absence predictions. As a conservative approach, the suitability of all cells showing variable values not experienced during the model training (values greater than zero in the clamping mask) was set to zero (Elith et al. 2010). We subsequently applied a mask representing urban and suburban areas to avoid projections at locations that are unsuitable regardless of the prevailing environmental conditions.
Area range change.
To assess whether C. graecum will experience range contraction or expansion under future conditions, we used functions from the ‘biomod’ 3.3.7 (Thuiller et al. 2009) package. Species were not assumed to have unlimited dispersal capability, since this assumption could be overoptimistic.
Correlation of genetic diversity with environmental factors.
In order to explore the effect of key environmental variables on the genetic diversity of our study species, we applied a best subset regression analysis with H index (ISSRs) as the response variable. We used Akaike’s Information Criterion (AIC) to identify the minimum adequate models. This process also allowed calculating the relative importance of each explanatory variable, which captured the percentage of variation explained by each factor when the other factors were held constant.
The predictor variables that were not collinear (Spearman’s rank correlation < 0.7) and thus included in the analysis were temperature seasonality, aridity, soil organic carbon content and weight percentage of the clay particles. Temperature seasonality was obtained from the WorldClim database (Hijmans et al. 2005) at a 30-s resolution, while aridity was constructed at the same resolution via the ‘envirem’ 1.1 (Title and Bemmels 2018) package. Soil variables were obtained from the SoilGrids 250 m database (Hengl et al. 2017; https://www.soilgrids.org).
Most of the predictor variables had frequency distributions that were strongly positively skewed. Hence, the variables were log10-transformed to normalize their distribution so that they could be compared with bivariate and multivariate regression methods without heteroscedastic biases and improve the linearity of the relationships in the regression models. A goodness-of-fit test (Shapiro–Wilk and Kolmogorov–Smirnov with 95 % level of confidence) to a normal distribution was used to confirm that each transformed variable was successfully transformed to an approximately normal distribution.
By fitting the full model, the total adjusted coefficient of multiple determinations (R2adj) was assessed. All analyses were carried out in the R 3.4.2 (R Development Core Team 2017) using core functions and functions from the ‘leaps’ 3.0 (Lumley 2017) package.
IUCN measures.
We calculated the standard IUCN measures EOO and AOO and we assigned C. graecum to a preliminary IUCN threat category according to Criterion B under current and future conditions by applying the ‘ConR’ 1.1.1 (Dauby et al. 2017) package.
Results
Genetic diversity based on ISSRs
The population in the present study was identified and collected at the Mt. Kyllini (K) and Mt. Chelmos (C) in Peloponissos based on previous field work. We scored 145 loci of which 100 % were polymorphic at the species level with a mean of 64.28 % at the population level and a range of 59.31 % in C. graecum population K1 and 68.97 % in C1 (Table 3). There was one private allele found in C2, C3 and K1 populations, six found only in C1 population and seven found only in K2 population. The value of Shannon’s information index (I) showed similar trends and ranged from 0.287 to 0.345 in populations K1 and C1, respectively (mean I = 0.288). Nei’s (1978) gene diversity at the population level ranged from 0.188 (population K1) to 0.228 (population K2) with an average value of 0.205. Results showed that the gene diversity of C. graecum from the C1 population was the richest among the five populations. The K1 population presented the poorest gene diversity. The gene diversity of the total sample was relatively high (P = 100 %, I = 0.313, GD = 0.205; Table 3).
Table 3.
Genetic diversity of five populations of Cicer graecum using ISSR and AFLP markers. N = numbers of individuals; NPB = numbers of polymorphic bands; NPrB = numbers of private bands; PPB (%) = percentage of polymorphic bands; I = Shannon’s information index; GD = Nei’s gene diversity.
| ISSR | AFLP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | NPB | NPrB | PPB (%) | I | GD | NPB | NPrB | PPB (%) | I | GD |
| C1 | 21 | 100 | 6 | 68.97 | 0.345 | 0.227 | 260 | 33 | 42.30 | 0.191 | 0.130 |
| C2 | 20 | 96 | 1 | 66.21 | 0.300 | 0.192 | 240 | 13 | 38.85 | 0.176 | 0.120 |
| C3 | 20 | 97 | 1 | 62.76 | 0.294 | 0.191 | 283 | 66 | 46.23 | 0.188 | 0.126 |
| K1 | 20 | 88 | 1 | 59.31 | 0.287 | 0.188 | 254 | 22 | 40.66 | 0.195 | 0.136 |
| K2 | 16 | 97 | 7 | 64.14 | 0.340 | 0.228 | 294 | 50 | 48.03 | 0.217 | 0.148 |
| Mean | – | 64.28 | 0.313 | 0.205 | 43.21 | 0.193 | 0.132 |
According to the inter-population AMOVA analysis of the ISSR markers, there were highly significant (P < 0.001) genetic differences among the five C. graecum populations. The 35 % of the total gene diversity was attributed to among-population differentiation. Thus, AMOVA (Φ st = 0.354) also supports the results of Nei’s (1978) gene diversity statistics and Shannon’s information index, indicating genetic differentiation among populations (Table 4).
Table 4.
Analysis of molecular variance results for five Cicer graecum populations based on ISSR markers and AFLP markers. df = degree of freedom; SS = sum of squared differences; MS = mean square; Est. Var. = estimated variance; Φ st = Wright’s Fst.
| Loci | Source | df | SS | MS | Est. Var. | % | Φ st | P-value |
|---|---|---|---|---|---|---|---|---|
| ISSR | Among | 4 | 726.410 | 181.603 | 8.572 | 35 % | 0.354 | P < 0.001 |
| Within | 92 | 1438.208 | 15.633 | 1.633 | 65 % | |||
| Total | 96 | 2164.619 | 24.205 | 100 % | ||||
| AFLP | Among | 4 | 407.024 | 101.756 | 3.178 | 7 % | 0.073 | P < 0.001 |
| Within | 92 | 3700.234 | 40.220 | 40.220 | 93 % | |||
| Total | 96 | 4107.258 | 43.398 | 100 % |
Relatedness among populations was illustrated by a dendrogram using the UPGMA algorithm based on Nei’s (1978) distance (Fig. 2A). An ISSR data-based dendrogram grouped the five populations into two main clusters (Fig. 2). The C1, C2, C3 and K1 populations formed a cluster and K2 another one. However, in the first cluster, two subclusters were formed with C1, C2 and C3 population in one and K1 in the other. Furthermore, PCoA (Fig. 3A) results illustrated two major clusters, which was congruent to the UPGMA dendrogram (Fig. 2A) and correlated with the geographical origin of the populations (Table 1). The first two principal coordinates accounted for 63.47 % of the total variation (Fig. 3A).
Figure 2.
UPGMA dendrogram generated with (Α) ISSR and (Β) AFLP data and based on a Nei’s genetic distance matrix.
Figure 3.
Principal coordinates analysis (PCoA) of five populations of C. graecum based on (A) 145 ISSR loci and (B) 610 AFLP loci.
The Mantel test for the five populations studied revealed a significant correlation between the geographic and the genetic distance (r = 0.798, P < 0.001).
Genetic diversity based on AFLPs
With four primer combinations, the AFLP analysis yielded a total of 610 AFLP loci among the five C. graecum populations. Of these loci 263 (43.21 %) were polymorphic and used for subsequent analysis. Number of private bands ranged from 13 (C2) to 66 (C3). Mean Shannon’s information index (I) had an average value of 0.193 and unbiased genetic diversity ranged from 0.120 for population C1 to 0.148 for population K2 (Table 3). Furthermore, results from AMOVA (Φ st = 0.073) showed that the majority of the variation was found within populations (93 %), rather than among populations (7 %) (Table 4). From PCoA results, the first two axes accounted for >75 % cumulatively of the total amount of variation and all populations grouped separately (Fig. 3B).
An UPGMA tree was constructed and grouped populations based on Nei’s (1978) distance (Fig. 2B) differently from ISSR results. The dendrogram contained two subclusters one with C1 and C2 and one with C3 and K1 and a separate cluster for K2.
The Mantel test revealed non-significant correlation between the geographic and the genetic distance based on AFLPs.
Population structure based on ISSRs and AFLPs
STRUCTURE analysis with ISSR markers was performed without prior information on the geographic origin of samples, and the highest likelihood of the data was obtained for K = 5. The data set was partitioned into clusters or gene pools corresponding roughly to geography: individuals from C1, C2, C3, K1 and K2 tended to be classified in separate clusters. Low levels of admixture were observed in four populations. A moderate level of admixture was detected in the C2 population (Fig. 4A). Contrariwise, when STRUCTURE analysis performed with AFLP markers, the highest likelihood of the data was obtained for K = 2. High levels of admixture were observed in five populations (Fig. 4B).
Figure 4.
STRUCTURE analysis for (A) ISSR data with K = 5 clusters and five populations of C. graecum. Each vertical bar represents a single plant (n = 97) and the probability of its membership in five clusters. (B) AFLP data with K = 2 and five populations of C. graecum. Each vertical bar represents a single plant (n = 97) and the probability of its membership in two clusters. Estimation of the number of populations for K ranging from 1 to 8 was conducted by calculating delta K values.
Environmental factors affecting genetic diversity
We investigated whether certain environmental factors might affect the ecological adaptation of C. graecum populations. Our predictive model explains 99 % (P < 0.001) of C. graecum population diversity (Table 5). Aridity arises as the most important environmental variable, positively correlated with genetic diversity. Temperature seasonality and weight percentage of the clay particles were also retained in the optimal model for the H index (Table 5), with aridity being the most important variable (Table 5). Soil organic carbon content did not emerge as a significant factor in this type of analysis.
Table 5.
Summary statistics for the predictive model of H index with aridity, temperature seasonality (TS) and weight percentage of clay particles (WPC) as proposed by the best subset regression. All predictor variables were log10-transformed. The adjusted R2 (total variance explained by each model), the P-value, Akaike’s Information Criterion (AIC), the estimate (Est.), the significance (P), the Bonferroni-corrected P-values (PBon), as well as the relative importance (rw) of each significant predictor of the best subset regression are reported.
| Est. | t | P | P Bon | AIC | R 2 adj | r w | F | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| H | −83.00 | 0.99 | – | 223 | <0.001 | ||||
| Intercept | −1.89 | −179.00 | – | – | – | ||||
| Aridity | 1.34 | 672.20 | 0.00 | 0.03 | – | 85.9 | |||
| TS | −0.16 | −24.30 | 0.03 | 0.08 | 8.1 | ||||
| WPC | −0.12 | −44.4 | 0.01 | 0.04 | – | 6.0 |
Species distribution modelling and risk assessment
The ESMs framework predictions were very good, with sufficient predictive power (TSS ≥ 0.99 for all algorithms and the ensemble prediction). Among the eight studied variables, soil organic carbon content had the highest contribution among the response variables, followed by the temperature annual range and precipitation of the wettest month. The resulting habitat suitability map (Fig. 5A) was converted into a binary map and then compared to the binary maps obtained for each GCM, RCP scenario and time period. Since the trends for the future potential distribution of C. graecum were largely identical across GCMs, RCPs and time periods, we present only the area-range change for 2050 time period and the CCSM4 GCM and the RCP 2.6 scenario. Our results indicate that by 2050 C. graecum will experience a severe range contraction, with no variation across GCMs, scenarios and time periods (100.0 %; Fig. 5B).
Figure 5.
(A) Habitat suitability map of C. graecum based on present-day environmental predictors, (B) Predicted potential distribution map for 2050 and the CCSM4 GCM and the RCP 2.6 scenario.
According to the IUCN Criterion B, C. graecum is considered as Endangered, with an Area of Occupancy equalling to 16 km2 and an Extent of Occurrence of 143.3 km2. The risk assessment status of C. graecum could be greatly deteriorated, since under any GCM and RCP included in our analyses, it is projected to become extinct.
Discussion
Genetic diversity of Cicer graecum
In the present study, the genetic diversity of C. graecum, a perennial, endemic, endangered species with narrow geographical distribution was assessed by using the dominant molecular markers AFLPs and ISSRs. According to our results, a similar overall trend for the genetic diversity and population structure was detected by the two marker systems used. However, higher genetic diversity and genetic differentiation were detected by ISSRs compared to AFLPs. Furthermore, the ISSRs clearly clustered the populations from Chelmos together, which is more reliable than the outcome of AFLP clusters. The aforementioned discrepancies that were found by analysing the genetic diversity of the five populations with two marker systems are suggesting that marker specificity affects the manner of polymorphism. Similar results have been also reported by other studies when multiple markers were used to detect genetic structure (Zhao et al. 2012; Roy et al. 2015). Nevertheless, different techniques that calculate the intra- and inter-population variation give comparable results in general (Constandinou et al. 2018). Many studies, which compared the use of various marker systems in the genetic study of locally restricted endemic species, highlighted the informativeness and efficiency of ISSRs (Ci et al. 2008; El-Bakatoushi and Ahmed 2018).
The genetic diversity of C. graecum populations based on Nei’s indices was lower than expected in outcrossing species (h = 0.27), but higher than the ones obtained from self-pollinating species (h = 0.12) (Nybom 2004). In this respect, C. graecum is closer to self-pollinating species based on AFLPs (h = 0.13), but closer to mixed reproductive (h = 0.18) and endemic species (h = 0.20) (Nybom 2004) based on ISSRs (h = 0.21). There is no information on the mating system of C. graecum. Generally, the Cicer spp. are predominantly self-pollinated, but outcrossing by insects also occurs (Sharma et al. 2013).
As a general rule, narrow distributed endemic species typically display lower genetic variation compared to widespread taxa, due to their small range size, genetic drift and limited gene flow (Spielman et al. 2004). However, some studies have pointed out high genetic diversity of rare or endemic species (Qian et al. 2013). Moreover, the genetic diversity of endemic species in comparison with their widespread congeners has been investigated in various studies with contradictory results (Dettori et al. 2016). In some cases, the endemic taxa had lower genetic diversity compared to their congeners and in others higher or similar (Dettori et al. 2016). In the present study, C. graecum had higher percentage of polymorphism and genetic diversity at species level (Table 6) compared to other annual (such as, C. reticulatum, C. echinospermum, C. bijugum, C. juidaicum) and perennial (such as, C. anatolicum, C. macranthum, C. oxyodon) Cicer species with broader geographical distribution based on AFLPs and ISSRs (Nguyen et al. 2004; Sudupak 2004; Andeden et al. 2013). This moderate to high genetic diversity may suggest that its current populations have originated from larger and more contiguous populations that existed in the past (Colling et al. 2010; Jiménez et al. 2017). Furthermore, its perennial life cycle could enable it to accumulate mutants in different populations due to environmental processes.
Table 6.
The genetic diversity of annual and perennial Cicer species based on ISSRs and AFLPs compared to Cicer graecum.
| GD | |||
|---|---|---|---|
| ISSR | AFLP | ||
| Cicer graecum | 0.205 | 0.132 | |
| Annual | Sudupak (2004) | Andeden et al. (2013) | Nguyen et al. (2004) |
| Cicer reticulatum | 0.085 | 0.045 | 0.104 |
| Cicer echinospermum | 0.150 | 0.075 | 0.038 |
| Cicer bijugum | 0.034 | 0.080 | |
| Cicer juidaicum | 0.005 | 0.119 | |
| Perennial | |||
| Cicer anatolicum | 0.027 | 0.023 | |
| Cicer macranthum | 0.015 | ||
| Cicer oxyodon | 0.023 |
According to the AMOVA, most of the variation was found within (93 % for AFLPs and 65 % for ISSRs), rather than among populations. The high percentage of total variation within the populations indicates that C. graecum is probably not exclusively self-pollinated. Genetic differentiation among the studied populations was detected by both marker systems. However, this differentiation was much higher based on ISSRs compared to AFLPs. The Φ st value of C. graecum based on the ISSRs (0.35) is close to the Φ st values for short-lived perennials (0.39) and narrowly distributed species (0.34), and higher than those for species with mixed mating system (0.27), as reported by Nybom and Bartixh (2000) based on a review of RAPD analysis on plant species. The intra-inter population genetic diversity is shaped by the equilibrium among multiple factors, such as genetic drift, gene flow, mutation and selection (Loveless and Hamrick 1984). This equilibrium depends on biological factors, such as the mating system and life form of the plant species (Hamrick and Godt 1996) but also is affected by both abiotic and biotic environmental factors (Odat et al. 2004).
Furthermore, the genetic differentiation among the populations was positively correlated to the geographic distances based on ISSRs, indicating that the gene flow among distant populations is limited. This is probably the result of geographic isolation, which prevents the gene flow via pollen and/or seeds. In this respect, the results of UPGMA and PCoA reveal clear geographic trends among the populations. In particular, the populations from Mt. Chelmos are clearly grouped together while the K1 population from Mt. Kyllini seems closer to Mt. Chelmos’ populations than to K2. However, the clustering was slightly different when conducting an AFLP data clustering method. While PCoA formed a group for all populations from Chelmos and one group for all populations from Kyllini, results from UPGMA analysis were slightly different and inconclusive. Differences in clustering of the population studied based on different marker system are common in literature (Muminović et al. 2005; Sarwat et al. 2008), as each marker system is designed to sample different region of the genome (Mueller and Wolfenbanger 1999). In spite of this, in future studies more AFLP primer combinations should be used in order to have more available fragments for subsequent analysis.
Possible admixture in these populations was assessed by STRUCTURE, which revealed that each population studied corresponded to a unique gene cluster. STRUCTURE results further verified UPGMA and PCoA results and showed that most individuals clustered into distinct population-specific groups. Overall, STRUCTURE suggested a low level of population admixture in these populations.
Among the populations studied, C1 and K2 had the higher genetic diversity compared to the others based on both markers. Furthermore, it is noteworthy that C1 and K2 populations showed the highest numbers of private bands (6 and 7 respectively). This result suggests that C1 and K2 populations did not face recent bottlenecks (Breinholt et al. 2009). Both of them grew at woodland openings, at lower elevation than the others and finally they are the most isolated by the distance (Fig. 1; Table 1). Interestingly, C1 and K2 form the largest (>200 individuals) and the smallest (16 individuals) population, respectively. The general assumption is that the genetic diversity can be reduced in small populations due to bottlenecks, genetic drift and/or founder effects (Young et al. 1996; Lowe et al. 2004). It seems that the population size did not influence the genetic diversity of C. graecum in the present study. These results are in agreement with other studies in which there was no correlation between population size and genetic diversity (Tero et al. 2003; Kang et al. 2005; Ci et al. 2008). On the other hand, the C2 and C3 which were present in a highly disturbed habitat had the lowest genetic diversity compared to the others. The low diversity probably suggests that the C2 and C3 populations have been established by a recent founder event, a similar response to environmental stress and/or the survival of the most resilient individuals, which is further supported by the occurrence of a single private band.
Patterns of population genetic structure, even in small spatial scales, are often due to environmental factors (Huang et al. 2016). Stressful abiotic conditions, in particular, can promote adaptive changes, thus increasing genetic diversity (Hoffmann and Hercus 2000). In the present study, aridity was determined as the dominant environmental variable that positively affects population diversity of C. graecum. Its effect is also reflected in the increased genetic diversity of lowland populations of C. graecum. As the dry areas of Central and Southwest Asia are the diversity centres of the genus Cicer (van der Maessen 1972), phylogenetic niche conservatism has probably influenced environmental adaptation and distribution of Cicer spp. also in peripheral regions like the Mediterranean. In this respect, the high genetic diversity and the increased number of private bands observed in the populations of C. graecum at low altitudes indicate their long-lasting presence at the low elevation intervals of Mts. Chelmos and Kyllini. Our results also indicate that populations at higher altitudes might be the outcome of an upslope range shift, due to niche tracking as a response to climate change, a phenomenon already reported for several other plant species (Crimmins et al. 2009; Urban 2018).
Distribution under climate change
Our results highlight the role of soil and climate variables in shaping the distribution of C. graecum and forecast an especially uncertain future for this rare endemic. It is likely that C. graecum is going to experience a severe range contraction, as suggested by the 100 % reduction of suitable habitats across all GCMs, scenarios and time periods. This result for a locally restricted endemic species constitutes an especially high extinction risk. Climate change has already affected plant species distribution in many parts of the world, and Mediterranean mountains have been identified as the most sensitive regions in Europe with a projected species loss of 62 % by 2080 (Thuiller et al. 2009). As Mediterranean mountains host a large number of endemic plants, it is likely that we are going to face a wave of mass extinction of mountain plant species over the coming decades. Whether the high genetic diversity of certain endemic species, like C. graecum, may ultimately favour adaptability and persistence remains largely uncertain. Species survival will depend on factors like genetic make-up, fitness, habitat fragmentation and dispersal ability. Seeds of C. graecum lack any obvious adaptation for long-distance dispersal and barochory is likely the major dispersal mode. However, seed dispersal by grazing mammals that consume the seeds along with the foliage cannot be ruled out (Janzen 1984), although the viability of C. graecum seeds during passage through livestock is unknown. The projected lack of suitable habitats within the distribution range of C. graecum by 2050 together with its low dispersal rates that probably could not keep pace with fast rates of environmental change (Engler et al. 2009), make the long-term survival of this species in its natural habitat especially uncertain.
Conservation implications
Cicer graecum has been considered an endangered (EN) species by several studies (Economou and Maxted 2011; Bilz et al. 2011) along with the present study. Furthermore, it is projected to experience a severe range contraction (~100 %) by 2050, while there is no seed accession from this species deposited in any Greek or international germplasm collections. A combination of both in situ and ex situ conservation actions is necessary to safeguard species survival. Our results on the genetic diversity allocation across the natural populations of C. graecum could be a valuable tool towards the implementation of an effective in situ conservation scheme for this species. Furthermore, they could effectively guide future germplasm collection efforts to maximize the efficacy of the ex situ conservation actions. C1 and K2 populations should have priority in future conservation plans, since they maintain a high genetic diversity (Table 3) and belong to different clusters (Fig. 2). The ex situ conservation of C. graecum is strongly advised, as it is the only conservation measure able to effectively safeguard the long-term survival of the species. Creating a stock of preserved propagules representing the entire genetic diversity of C. graecum would have multiple advantages, as it would ensure its use in future chickpea breeding programmes and allow future reintroduction or even relocation projects to be developed.
Data
A link to a permanent, open-access data depository: https://github.com/kostask84/Cicer_ESM_ISSR
Acknowledgements
We would like to thank Joseph Pentheroudakis for carefully editing our manuscript.
Contributions by the Authors
Conceptualization, E.T., E.A. and P.T.; data curation, E.S., K.K., Ev.A. and I.G.; formal analysis, Ev.A., P.T., K.K. and I.G.; funding acquisition, E.T.; methodology, E.S., K.K., Ev.A. and I.G; resources, P.T. and K.K.; supervision, E.T.; validation, E.T.; writing – original draft, all; writing – review & editing, all.
Conflict of Interest
None declared.
Literature Cited
- Ahmad F, Gaur PM, Croser J. 2005. Chickpea (Cicer arietinum L.). In: Singh RJ, Jauhar PP, eds. Genetic resources, chromosome engineering and crop improvement-grain legumes. vol 1. Boca Raton: CRC Press, 187–217. [Google Scholar]
- Aiello‐Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP. 2015. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38:541–545. [Google Scholar]
- Allouche O, Tsoar A, Kadmon R. 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). Journal of Applied Ecology 43:1223–1232. [Google Scholar]
- Amirmoradi B, Talebi R, Karami E. 2012. Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Systematics and Evolution 298:1679–1688. [Google Scholar]
- Andeden EE, Baloch FS, Derya M, Kilian B, Özkan H. 2013. iPBS-Retrotransposons-based genetic diversity and relationship among wild annual Cicer species. Journal of Plant Biochemistry and Biotechnology 22:453–466. [Google Scholar]
- Araújo MB, Anderson RP, Márcia Barbosa A, Beale CM, Dormann CF, Early R, Garcia RA, Guisan A, Maiorano L, Naimi B, O’Hara RB, Zimmermann NE, Rahbek C. 2019. Standards for distribution models in biodiversity assessments. Science Advances 5:eaat4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo MB, New M. 2007. Ensemble forecasting of species distributions. Trends in Ecology & Evolution 22:42–47. [DOI] [PubMed] [Google Scholar]
- Barbet‐Massin M, Jiguet F, Albert CH, Thuiller W. 2012. Selecting pseudo-absences for species distribution models: how, where and how many? Methods in Ecology and Evolution 3:327–338. [Google Scholar]
- Bilz M, Kell SP, Maxted N, Lansdown RV. 2011. European red list of vascular plants. Luxembourg: Publications Office of the European Union. [Google Scholar]
- Breiner FT, Guisan A, Bergamini A, Nobis MP. 2015. Overcoming limitations of modelling rare species by using ensembles of small models. Methods in Ecology and Evolution 6:1210–1218. [Google Scholar]
- Breiner FT, Guisan A, Nobis MP, Bergamini A. 2017. Including environmental niche information to improve IUCN Red List assessments. Diversity and Distributions 23:484–495. [Google Scholar]
- Breiner FT, Nobis MP, Bergamini A, Guisan A. 2018. Optimizing ensembles of small models for predicting the distribution of species with few occurrences. Methods in Ecology and Evolution 9:802–808. [Google Scholar]
- Breinholt JW, Van Buren R, Kopp OR, Stephen CL. 2009. Population genetic structure of an endangered Utah endemic, Astragalus ampullarioides (Fabaceae). American Journal of Botany 96:661–667. [DOI] [PubMed] [Google Scholar]
- Ci X, Chen J, Li Q. 2008. AFLP and ISSR analysis reveals high genetic variation and inter-population differentiation in fragmented populations of the endangered Litsea szemaois (Lauraceae) from Southwest China. Plant Systematics and Evolution 273:237–246. [Google Scholar]
- Colling G, Hemmer P, Bonniot A, Hermant S, Matthies D. 2010. Population genetic structure of wild daffodils (Narcissus pseudonarcissus L.) at different spatial scales. Plant Systematics and Evolution 287:99–111. [Google Scholar]
- Constandinou S, Nikoloudakis N, Kyratzis AC, Katsiotis A. 2018. Genetic diversity of Avena ventricosa populations along an ecogeographical transect in Cyprus is correlated to environmental variables. PLoS One 13:e0193885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins TM, Crimmins MA, Bertelsen CD. 2009. Flowering range changes across an elevation gradient in response to warming summer temperatures. Global Change Biology 15:1141–1152. [Google Scholar]
- Croser JS, Ahmad F, Clarke HJ, Siddique KHM. 2003. Utilisation of wild Cicer in chickpea improvement—progress, constraints, and prospects. Australian Journal of Agricultural Research 54:429–444. [Google Scholar]
- Dauby G, Stévart T, Droissart V, Cosiaux A, Deblauwe V, Simo-Droissart M, Sosef MSM, Lowry PP 2nd, Schatz GE, Gereau RE, Couvreur TLP. 2017. ConR: an R package to assist large-scale multispecies preliminary conservation assessments using distribution data. Ecology and Evolution 7:11292–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L. 2017. Past and future use of wild relatives in crop breeding. Crop Science 57:1070–1082. [Google Scholar]
- Dempewolf H, Eastwood RJ, Guarino L, Khoury CK, Müller JV, Toll J. 2014. Adapting agriculture to climate change: a global initiative to collect, conserve, and use crop wild relatives. Agroecology and Sustainable Food Systems 38:369–77. [Google Scholar]
- Dettori CA, Loi MC, Brullo S, Fraga i Arguimbau P, Tamburini E, Bacchetta G. 2016. The genetic diversity and structure of the Ferula communis L. complex (Apiaceae) in the Tyrrhenian area. Flora 223:138–146. [Google Scholar]
- Di Cola V, Broennimann O, Petitpierre B, Breiner FT, D’Amen M, Randin C, Engler R, Pottier J, Pio D, Dubuis A, Pellissier L, Mateo RG, Hordijk W, Salamin N, Guisan A. 2017. ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography 40:774–787. [Google Scholar]
- Dimopoulos P, Georgiadis T, Sykora K. 1996. Phytosociological research on the montane coniferous forests of Greece: Mount Kyllini (NE Peloponnisos—S Greece). Folia Geobotanica & Phytotaxonomica 31:169–195. [Google Scholar]
- Dimopoulos P, Raus T, Bergmeier E, Constantinidis T, Iatrou G, Kokkini S, Strid A, Tzanoudakis D. 2016. Vascular plants of Greece: an annotated checklist. Supplement. Willdenowia 46:301–347. [Google Scholar]
- Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, Marquéz JRG, Gruber B, Lafourcade B, Leitão PJ, Münkemüller T. 2013. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36:27–46. [Google Scholar]
- Doyle J. 1991. DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW, eds. Molecular techniques in taxonomy. NATO ASI Series (Series H: Cell Biology). Berlin, Heidelberg: Springer, 283–293. [Google Scholar]
- Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. [Google Scholar]
- Economou G, Maxted N. 2011. Cicer graecum. The IUCN Red List of Threatened Species. e.T176608A7275603. 10.2305/IUCN.UK.2011-1.RLTS.T176608A7275603 (14 March 2020) [DOI] [Google Scholar]
- Egan PA, Muola A, Stenberg JA. 2018. Capturing genetic variation in crop wild relatives: an evolutionary approach. Evolutionary Applications 11:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Bakatoushi R, Ahmed DGA. 2018. Evaluation of genetic diversity in wild populations of Peganum harmala L., a medicinal plant. Journal, Genetic Engineering & Biotechnology 16:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elith J, Kearney M. Phillips S. 2010. The art of modelling range-shifting species. Methods in Ecology and Evolution 1:330–342. [Google Scholar]
- Engler R, Randin CF, Vittoz P, Czáka T, Beniston M, Zimmermann NE, Guisan A. 2009. Predicting future distributions of mountain plants under climate change: does dispersal capacity matter?’ Ecography 32:34–45. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganopoulos IV, Aravanopoulos FA, Argiriou A, Kalivas A, Tsaftaris AS. 2011. Is the genetic diversity of small scattered forest tree populations at the southern limits of their range more prone to stochastic events? A wild cherry case study by microsatellite-based markers. Tree Genetics & Genomes 7:1299–1313. [Google Scholar]
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 351:1291–1298. [Google Scholar]
- Hengl T, Mendes de Jesus J, Heuvelink GB, Ruiperez Gonzalez M, Kilibarda M, Blagotić A, Shangguan W, Wright MN, Geng X, Bauer-Marschallinger B, Guevara MA, Vargas R, MacMillan RA, Batjes NH, Leenaars JG, Ribeiro E, Wheeler I, Mantel S, Kempen B. 2017. SoilGrids250m: global gridded soil information based on machine learning. PLoS One 12:e0169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood VH. 2008. Challenges of in situ conservation of crop wild relatives. Turkish Journal of Botany 32:421–432. [Google Scholar]
- Hiemstra PH, Pebesma EJ, Twenhöfel CJW, Heuvelink GBM. 2009. Real-time automatic interpolation of ambient gamma dose rates from the Dutch radioactivity monitoring network. Computers & Geosciences 35:1711–1721. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25:1965–1978. [Google Scholar]
- Hoffmann AA, Hercus MJ. 2000. Environmental stress as an evolutionary force. Bioscience 50:217–226. [Google Scholar]
- Huang W, Zhao X, Zhao X, Li Y, Lian J. 2016. Effects of environmental factors on genetic diversity of Caragana microphylla in Horqin Sandy Land, northeast China. Ecology and Evolution 6:8256–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DH. 1984. Dispersal of small seeds by big herbivores: foliage is the fruit. The American Naturalist 123:338–353. [Google Scholar]
- Jarvis A, Reuter HI, Nelson A, Guevara E. 2008. Hole-filled SRTM for the globe version 4. Available from the CGIAR-CSI SRTM 90m database. http://srtm.csi.cgiar.org (14 March 2020), 15, 25–54. [Google Scholar]
- Javadi F, Wojciechowski MF, Yamaguchi H. 2007. Geographical diversification of the genus Cicer (Leguminosae: Papilionoideae) inferred from molecular phylogenetic analyses of chloroplast and nuclear DNA sequences. Botanical Journal of the Linnean Society 154:175–186. [Google Scholar]
- Jiménez JF, López-Romero C, Rosselló JA, Sánchez-Gómez P. 2017. Genetic diversity of Narcissus tortifolius, an endangered endemic species from Southeastern Spain. Plant Biosystems 151:117–25. [Google Scholar]
- Kang M, Ye Q, Huang H. 2005. Genetic consequence of restricted habitat and population decline in endangered Isoetes sinensis (Isoetaceae). Annals of Botany 96:1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouam EB, Pasquet RS, Campagne P, Tignegre JB, Thoen K, Gaudin R, Ouedraogo JT, Salifu AB, Muluvi GM, Gepts P. 2012. Genetic structure and mating system of wild cowpea populations in West Africa. BMC Plant Biology 12:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless MD, Hamrick JL. 1984. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15:65–95. [Google Scholar]
- Lowe A, Harris S, Ashton P. 2004. Ecological genetics: design, analysis, and application. Oxford, UK: Blackwell Publishing. [Google Scholar]
- Lumley T. 2017. leaps: regression subset selection. R package version 3.0 (based on Fortran code by Alan Miller). [Google Scholar]
- McSweeney CF, Jones RG, Lee RW, Rowell DP. 2015. Selecting CMIP5 GCMs for downscaling over multiple regions. Climate Dynamics 44:3237–3260. [Google Scholar]
- Mueller UG, Wolfenbanger LL. 1999. AFLP genotypes and fingerprinting. Trend in Ecology and Evolution 14:389–394. [DOI] [PubMed] [Google Scholar]
- Muminović J, Merz A, Melchinger AE, Lübberstedt T. 2005. Genetic structure and diversity among radish varieties as inferred from AFLP and ISSR analyses. Journal of the American Society for Horticultural Science 130:79–87. [Google Scholar]
- Naimi B, Hamm NA, Groen TA, Skidmore AK, Toxopeus AG. 2014. Where is positional uncertainty a problem for species distribution modelling? Ecography 37:191–203. [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Taylor PWJ, Redden RJ. 2004. Genetic diversity estimates in Cicer using AFLP analysis. Plant Breeding 123:173–179. [Google Scholar]
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13:1143–1155. [DOI] [PubMed] [Google Scholar]
- Nybom H, Bartish I. 2000. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3:93–144. [Google Scholar]
- Odat N, Jetschke G, Hellwig FH. 2004. Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Molecular Ecology 13:1251–1257. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H. 2018. vegan: community ecology package. R package version 2.5-2. [Google Scholar]
- Peakall ROD, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Wang CX, Tian M. 2013. Genetic diversity and population differentiation of Calanthe tsoongiana, a rare and endemic orchid in China. International Journal of Molecular Sciences 14:20399–20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes N, ter Steege H. 2007. A null-model for significance testing of presence-only species distribution models. Ecography 30:727–736. [Google Scholar]
- Rajesh PN, Sant V, Gupta V, Muehlbauer FJ, Ranjekar PK. 2003. Genetic relationships among annual and perennial wild species of Cicer using inter simple sequence repeat (ISSR) polymorphism. Euphytica 129:15–23. [Google Scholar]
- Rameshkumar R, Pandian S, Rathinapriya P, Selvi CT, Satish L, Gowrishankar S, Leung DWM, Ramesh M. 2019. Genetic diversity and phylogenetic relationship of Nilgirianthus ciliatus populations using ISSR and RAPD markers: implications for conservation of an endemic and vulnerable medicinal plant. Biocatalysis and Agricultural Biotechnology 18:101072. [Google Scholar]
- R Development Core Team 2017. “R: A language and environment for statistical computing (Version 3.4. 2).” Vienna, Austria: R foundation for statistical computing. [Google Scholar]
- Redden R, Yadav SS, Maxted N, Dulloo ME, Guarino L, Smith P. 2015. Crop wild relatives and climate change. Plant genetic resources and climate change, 1st edn. Oxford: Wiley‐Blackwell. [Google Scholar]
- Robertson MP, Visser V, Hui C. 2016. Biogeo: an R package for assessing and improving data quality of occurrence record datasets. Ecography 39:394–401. [Google Scholar]
- Roy S, Banerjee A, Mawkhlieng B, Misra AK, Pattanayak A, Harish GD, Singh SK, Ngachan SV, Bansal KC. 2015. Genetic diversity and population structure in aromatic and quality rice (Oryza sativa L.) landraces from North-Eastern India. PLoS One 10:e0129607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwat M, Das S, Srivastava PS. 2008. Analysis of genetic diversity through AFLP, SAMPL, ISSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Reports 27:519–528. [DOI] [PubMed] [Google Scholar]
- Sharma S, Yadav N, Singh A. 2013. Nutritional and antinutritional profile of newly developed chickpea (Cicer arietinum L.) varieties. International Food Research Journal 20:805–810. [Google Scholar]
- Smýkal P, Trněný O, Brus J, Hanáček P, Rathore A, Roma RD, Pechanec V, Duchoslav M, Bhattacharyya D, Bariotakis M, Pirintsos S, Berger J, Toker C. 2018. Correction: genetic structure of wild pea (Pisum sativum subsp. elatius) populations in the northern part of the Fertile Crescent reflects moderate cross-pollination and strong effect of geographic but not environmental distance. PLoS One 13:e0196376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR, Freeman WH. 1975. Numerical taxonomy. The principles and practice of numerical classification. Systematic Zoology 24:263–268. [Google Scholar]
- Spielman D, Brook BW, Frankham R. 2004. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences of the United States of America 101:15261–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Bajaj D, Malik A, Singh M, Parida SK. 2016. Transcriptome landscape of perennial wild Cicer microphyllum uncovers functionally relevant molecular tags regulating agronomic traits in chickpea. Scientific Reports 6:33616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudupak MA. 2004. Inter and intra-species Inter Simple Sequence Repeat (ISSR) variations in the genus Cicer. Euphytica 135:229–238. [Google Scholar]
- Sudupak M, Akkaya M, Kence A. 2003. Analysis of genetic relationships among perennial and annual Cicer species growing in Turkey using RAPD markers. Theoretical and Applied Genetics 105:1220–1228. [DOI] [PubMed] [Google Scholar]
- Talebi R, Jelodar NB, Mardi M, Fayaz F, Furman BJ, Bagheri N. 2009. Phylogenetic diversity and relationship among annual Cicer species using random amplified polymorphic DNA markers. General and Applied Plant Physiology 35:3–12. [Google Scholar]
- Tero N, Aspi J, Siikamäki P, Jäkäläniemi A, Tuomi J. 2003. Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Molecular Ecology 12:2073–2085. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lafourcade B, Engler R, Araújo MB. 2009. BIOMOD – a platform for ensemble forecasting of species distributions. Ecography 32:369–373. [Google Scholar]
- Title PO, Bemmels JB. 2018. ENVIREM: an expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 41:291–307. [Google Scholar]
- Toker C, Uzun B, Ceylan FO, Ikten C. 2014. Chickpea. In: Pratap A, Kumar J, eds. Alien gene transfer in crop plants. New York, NY: Springer, 121–151. [Google Scholar]
- Urban MC. 2018. Escalator to extinction. Proceedings of the National Academy of Sciences of the United States of America 115:11871–11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valavi R, Elith J, Lahoz-Monfort JJ, Guillera-Arroita G. 2019. blockCV: an R package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models, bioRxiv, 357798. Methods of Ecology and Evolution 10:225–232. [Google Scholar]
- Van der Maessen LJG. 1972. Cicer L., a monograph of the genus, with special reference to the chickpea (Cicer arietinum L.), its ecology and cultivation. PhD Thesis, University of Wageningen, The Netherlands. [Google Scholar]
- van Heerwaarden J, Doebley J, Briggs WH, Glaubitz JC, Goodman MM, de Jesus Sanchez Gonzalez J, Ross-Ibarra J. 2011. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proceedings of the National Academy of Sciences of the United States of America 108:1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavrek MJ. 2010. Fossil: palaeoecological and palaeogeographical analysis tools. Palaeontologia Electronica 14:16. [Google Scholar]
- Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I. 2002. Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molecular Ecology 11:139–151. [DOI] [PubMed] [Google Scholar]
- von Wettberg EJB, Chang PL, Başdemir F, Carrasquila-Garcia N, Korbu LB, Moenga SM, Bedada G, Greenlon A, Moriuchi KS, Singh V, Cordeiro MA, Noujdina NV, Dinegde KN, Shah Sani SGA, Getahun T, Vance L, Bergmann E, Lindsay D, Mamo BE, Warschefsky EJ, Dacosta-Calheiros E, Marques E, Yilmaz MA, Cakmak A, Rose J, Migneault A, Krieg CP, Saylak S, Temel H, Friesen ML, Siler E, Akhmetov Z, Ozcelik H, Kholova J, Can C, Gaur P, Yildirim M, Sharma H, Vadez V, Tesfaye K, Woldemedhin AF, Tar’an B, Aydogan A, Bukun B, Penmetsa RV, Berger J, Kahraman A, Nuzhdin SV, Cook DR. 2018. Ecology and genomics of an important crop wild relative as a prelude to agricultural innovation. Nature Communications 9:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle TBJ, Ye Z, Xiyan JM, Yang R, Boyle TJ. 2000. PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. [Google Scholar]
- Young A, Boyle T, Brown T. 1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology & Evolution 11:413–418. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ma Y, Sun W, Wen X, Milne R. 2012. High genetic diversity and low differentiation of Michelia coriacea (Magnoliaceae), a critically endangered endemic in Southeast Yunnan, China. International Journal of Molecular Sciences 13:4396–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D., Hopf M. 2000. Domestication of plants in the Old World: the origin and spread of cultivated plants in West Asia, Europe and the Nile Valley, 3rd edn. Oxford: Oxford University Press. [Google Scholar]