Abstract
SARS-CoV-2 causes the recent global COVID-19 public health emergency. ACE2 is the receptor for both SARS-CoV-2 and SARS-CoV. To predict the potential host range of SARS-CoV-2, we analyzed the key residues of ACE2 for recognizing S protein. We found that most of the selected mammals including pets (dog and cat), pangolin and Circetidae mammals remained the most of key residues for association with S protein from SARS-CoV and SARS-CoV-2. The interaction interface between cat/dog/pangolin/Chinese hamster ACE2 and SARS-CoV/SARS-CoV-2 S protein was simulated through homology modeling. We identified that N82 in ACE2 showed a closer contact with SARS-CoV-2 S protein than M82 in human ACE2. Our finding will provide important insights into the host range of SARS-CoV-2 and a new strategy to design an optimized ACE2 for SARS-CoV-2 infection.
Keywords: SARS-CoV-2, Spike protein, ACE2, Structure, Host range
Abbreviations: COVID-19, Corona Virus Disease 2019; SARS-CoV-2, Severe Acute Respiratory Syndrome Corona Virus 2; SARSr-CoV, SARS-related coronavirus; S, spike protein; M, membrane protein; E, envelope protein; N, nucleocapsid protein; ACE2, angiotensin-converting enzyme 2; RBM, receptor-binding motif; RBD, receptor-binding domain; JTT, Jones-Taylor-Thornton
Highlights
-
•
Pets (dog and cat), pangolin and Circetidae remained the key residues for association with S from SARS-CoV and SARS-CoV-2.
-
•
The interface of the interaction between cat/dog/pangolin/Chinese hamster ACE2 and SARS-CoV/SARS-CoV-2 RBD was simulated.
-
•
N82 of ACE2 showed a closer contact with SARS-CoV-2 S than M82, suggesting an optimized ACE2 for SARS-CoV-2 infection.
1. Introduction
Corona Virus Disease 2019 (COVID-19), which was reported from Wuhan city, Hubei province of China, has caused over 78,000 human infections and more than 2700 deaths (as of February 25, 2020) [1,2]. Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) was identified as the pathogen of COVID-19 [[1], [2], [3]]. After SARS-CoV and MERS-CoV, SARS-CoV-2 has become the third coronavirus that causes severe respiratory disease and human death [4,5].
Belonging to the subgenus sarbecvirus of Coronaviridae, both SARS-CoV-2 and SARS-CoV are human SARS-related coronavirus (SARSr-CoV). Its genome is a single-stranded RNA composed of about 30 kb nucleotides. SARS-CoV-2 encodes at least four major structural proteins, namely spike protein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N) [6]. S protein, which is a type I glycoprotein, protrudes from the surface of the virus and can contact the host cell earlier. S protein has attracted great attention because of its function in receptor binding.
Angiotensin-converting enzyme 2 (ACE2) binds to the receptor-binding motif (RBM) in the receptor-binding domain (RBD) of SARS-CoV and functions as a receptor for SARS-CoV [7,8]. ACE2 is widely distributed in heart, liver, testis, kidney, intestine and other tissues. It has the physiological functions of regulating heart and kidney function and controlling blood pressure [9]. Recently, it has been found that human ACE2 promoted the entry of SARS-CoV-2 into the cells [3,10]. RBD domain of SARS-CoV-2 interacts with human ACE2. Thus, ACE2 is defined as the receptor for SARS-CoV-2.
The specificity of the interaction between virus and receptor determines the host tropism and host range. The origin of SARS-CoV-2 is presumed to be bat [3]. However, the intermediate host is not clear, and some studies suggest that pangolin is involved in the evolution of SARS-CoV-2 [11,12]. It is not clear which mammals are involved in the evolution of SARS-CoV-2 and which animals may be infected by SARS-CoV-2. By sequence alignment of key amino acids binding to RBD in ACE2, the interaction between RBD of SARS-CoV-2/SARS-CoV and mammalian ACE2 was predicted. Based on the potential interaction between S protein and mammalian ACE2, it was speculated that SARS-CoV-2/SARS-CoV preserved the ability to infect many mammals including cat, dog, pangolin and Chinese hamster. From the structure stimulation, we identified N82 in ACE2 show closer contact with F486 of SARS-CoV-2 S protein compared with M82 of ACE2.
2. Methods
2.1. Sequence analysis of RBM of S from SARS-CoV-2 and SARS-CoV
The S protein sequence of SARS-CoV-2 is YP_009724390.1, and the S protein sequence of SARS-CoV is NP_828851.1. RBM of SARS-CoV is from 424 to 494. RBM of SARS-CoV-2 is from 437 to 508.
2.2. Sequence analysis of mammal ACE2
A total of 42 mammalian ACE2 protein sequences were selected from the wild animal protection lists of Hubei Province and Jiangxi Province, primates, bats, dog, and cat. These sequences are as follows: hACE2: Homo sapiens (BAB40370.1), RhiACE2: Rhinopithecus roxellana (XP_010364367.2), MacmACE2: Macaca mulatta (NP_001129168.1), MuseACE2: Mustela erminea (XP_032187679.1), CamdACE2: Camelus dromedarius (XP_031301717.1), PlACE2: Procyon lotor (XP_031301717.1), PcACE2: Paguma larvata (AAX63775.1), RmACE2: Rhinolophus macrotis (ADN93471.1), RfACE2: Rhinolophus ferrumequinum (BAH02663.1), RsACE2: Rhinolophus sinicus (ADN93472.1), RlACE2: Rousettus leschenaultii (BAF50705.1), SsACE2: Sus scrofa (NP_001116542.1), MpfACE2: Mustela putorius furo (BAE53380.1), RatACE2: Rattus norvegicus (Q5EGZ1), MmACE2: Mus musculus (Q3URC9), ClfACE2: Canis lupus familiaris (J9P7Y2), FcACE2: Felis catus (A0A384DV19), MjACE2: Manis javanica (XP_017505752.1), RpACE2: Rhinolophus pearsonii (ABU54053.1), PvACE2: Pteropus vampyrus (XP_011361275.1), PoaACE2: Pongo abelii (NP_001124604.1), EcACE2: Equus caballus (F6V9L3), BtACE2: Bos taurus (Q58DD0), PtACE2: Pan troglodytes (A0A2J8KU96), OraACE2: Ornithorhynchus anatinus (F7FDA2), OvaACE2: Ovis aries (W5PSB6), PanACE2: Papio Anubis (A0A096N4X9), LaACE2: Loxodonta africana (G3T6Q2), SsdACE2: Sus scrofa domesticus (A0A220QT48), EeACE2: Erinaceus europaeus (A0A1S3APE5), OcACE2: Oryctolagus cuniculus (G1TEF4), NpACE2: Nyctereutes procyonoides (B4XEP4), VvACE2: Vulpes vulpes (A0A3Q7RAT9), PhcACE2: Phodopus campbelli (C7ECU7), MaACE2: Mesocricetus auratus (C7ECV1), CjACE2: Callithrix jacchus (F7CNJ6), SusACE2: Suricata suricatta (XP_029786256.1), HgACE2: Heterocephalus glaber (A0A0N8EUX7), DoACE2: Dipodomys ordii (A0A1S3GHT7), ItACE2: Ictidomys tridecemlineatus (XP_005316051.3), CpACE2: Cavia porcellus (XP_023417808.1), CgACE2: Cricetulus griseus (A0A061HZ66). Based on the known key sites in SARS-CoV S protein interacting with human ACE2, we analyzed whether these sites were conserved on ACE2 from wild mammals and domestic pets. Phylogenetic and molecular evolutionary analyses of ACE2 were conducted using MEGA version X [13]. Phylogenetic tree was generated with Jones-Taylor-Thornton (JTT) evolutionary model using Maximum Likelihood method.
2.3. Structure simulation of ACE2-RBD complex
The interaction interfaces of SARSr-CoV S and ACE2 from cat/dog/pangolin/Chinese hamster were simulated by Chimera software Ver 1.14 [14]. The simulation were based on the structures of hACE2 with SARS-CoV S RBD (PDB: 2AJF) [7] and hACE2 with SARS-CoV-2 S RBD (PDB: 6LZG).
3. Results
3.1. Sequence alignment of RBM from SARS-CoV-2 and SARS-CoV
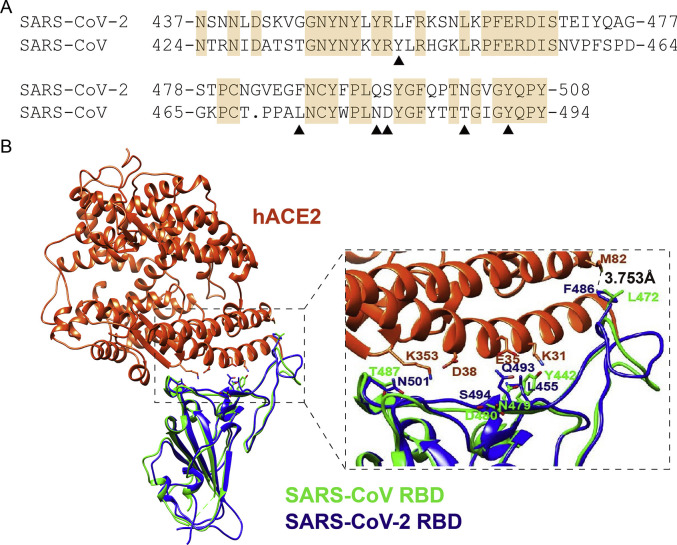
Human ACE2 is the receptor for both SARS-CoV and SARS-CoV-2. According to the literature [15], the key amino acids (AAs) in the S protein of SARS-CoV interacting with human ACE2 are Y442, L472, N479, D480, T487, Y491 [7,15]. We compared the RBM in S protein of SARS-CoV-2 with that of SARS-CoV (Fig. 1 A). These amino acids corresponding to SARS-CoV-2 are L455, F486, Q493, S494, N501 and Y505 (Fig. 1A). Although five of six key AAs in SARS-CoV-2 are changed compared with SARS-CoV, the overall structure of interfaces of ACE2-RBM are similar (Fig. 1B).
Fig. 1.
Alignment of RBM region of S proteins from SARS-CoV-2 and SARS-CoV. (A) Sequence alignment of RBM region of S protein from SARS-CoV-2 and SARS-CoV. ▲ represents the six key amino acids in the S protein interacting with human ACE2. For SARS-CoV, they are Y442, L472, N479, D480, T487 and Y491. The S protein sequence of SARS-CoV-2 comes from YP_009724390.1, and the S protein sequence of SARS-CoV comes from NP_828851.1. (B) Alignment of the structure of ACE2 recognition of RBD from SARS-CoV-2 and SARS-CoV. Human ACE2 (hACE2), SARS-CoV-2 RBD, and SARS-CoV RBD are in orange red, blue, and green, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Sequence alignment of ACE2
The key AAs in hACE2 for interacting with RBM are K31, E35, D38, M82 and K353 [7,15]. Among them, K31 and K353 in hACE2 are most critical residues for RBM recognition. Because the overall structure of interfaces of ACE2-RBM in SARS-CoV-2 and SARS-CoV are similar, we analyzed the key RBD recognizing AAs of ACE2 protein from selected mammals, as shown in Table 1 . We predicted that the mammals whose ACE2 could bind to the S protein of SARS-CoV-2 and SARS-CoV were Homo sapiens, Rhinopithecus roxellana, Macaca mulatta, Mustela erminea, Paguma larvata, Rhinolophus macrotis, Rhinolophus sinicus, Rousettus leschenaultii, Sus scrofa, Mustela putorius furo, Canis lupus familiaris, Felis catus, Manis javanica (pangolin), Rhinolophus pearsonii, Pteropus vampyrus, Pongo abelii, Equus caballus, Bos taurus, Pan troglodytes, Ovis aries, Papio Anubis, Sus scrofa domesticus, Oryctolagus cuniculus, Vulpes vulpes, Phodopus campbelli, Mesocricetus auratus, Callithrix jacchus, Heterocephalus glaber, Ictidomys tridecemlineatus, and Cricetulus griseus (Chinese hamster). The mammals whose ACE2 could not bind to S protein of SARS-CoV-2 and SARS-CoV included Camelus dromedarius, Procyon lotor, Rhinolophus ferrumequinum, Rattus norvegicus, Mus musculus, Ornithorhynchus anatinus, Loxodonta africana, Erinaceus europaeus, Nyctereutes procyonoides, Suricata suricatta, Dipodomys ordii, and Cavia porcellus.
Table 1.
Prediction of the RBD binding capacity of mammalian ACE2.
| ACE2 | AA position |
matched AA | binding capacity | ||||
|---|---|---|---|---|---|---|---|
| 31 | 35 | 38 | 82 | 353 | |||
| hACE2 | K | E | D | M | K | 5/5 | + |
| RhiACE2 | K | E | D | M | K | 5/5 | + |
| MacmACE2 | K | E | D | M | K | 5/5 | + |
| MuseACE2 | K | E | E | T | K | 3/5 | + |
| CamdACE2 | E | E | D | T | K | 3/5 | – |
| PlACE2 | N | E | E | T | K | 2/5 | – |
| PcACE2 | T | E | E | T | K | 2/5 | + |
| RmACE2 | K | K | D | N | K | 3/5 | + |
| RfACE2 | D | E | N | N | K | 2/5 | – |
| RsACE2 | K | K | D | N | K | 3/5 | + |
| RlACE2 | K | E | D | T | K | 4/5 | + |
| SsACE2 | K | E | D | T | K | 4/5 | + |
| MpfACE2 | K | E | E | T | K | 3/5 | + |
| RatACE2 | K | E | D | N | H | 3/5 | – |
| MmACE2 | N | E | D | S | H | 2/5 | – |
| ClfACE2 | K | E | E | T | K | 3/5 | + |
| FcACE2 | K | E | E | T | K | 3/5 | + |
| MjACE2 | K | E | E | N | K | 3/5 | + |
| RpACE2 | K | E | D | D | K | 4/5 | + |
| PvACE2 | K | E | D | A | K | 4/5 | + |
| PoaACE2 | K | E | D | M | K | 5/5 | + |
| EcACE2 | K | E | E | T | K | 3/5 | + |
| BtACE2 | K | E | D | T | K | 4/5 | + |
| PtACE2 | K | E | D | M | K | 5/5 | + |
| OraACE2 | Q | Q | D | K | K | 2/5 | – |
| OvaACE2 | K | E | D | T | K | 4/5 | + |
| PanACE2 | K | E | D | M | K | 5/5 | + |
| LaACE2 | T | E | D | D | K | 3/5 | – |
| SsdACE2 | K | E | D | T | K | 4/5 | + |
| EeACE2 | D | Q | N | N | N | 0/5 | – |
| OcACE2 | K | E | D | T | K | 4/5 | + |
| NpACE2 | K | E | E | T | R | 2/5 | – |
| VvACE2 | K | E | E | T | K | 3/5 | + |
| PhcACE2 | K | E | D | N | K | 4/5 | + |
| MaACE2 | K | E | D | N | K | 4/5 | + |
| CjACE2 | K | E | D | T | K | 4/5 | + |
| SusACE2 | Q | E | E | A | K | 2/5 | – |
| HgACE2 | K | E | D | A | K | 4/5 | + |
| DoACE2 | N | E | D | I | K | 3/5 | – |
| ItACE2 | K | E | D | A | K | 4/5 | + |
| CpACE2 | E | K | D | A | K | 2/5 | – |
| CgACE2 | K | E | D | N | K | 4/5 | + |
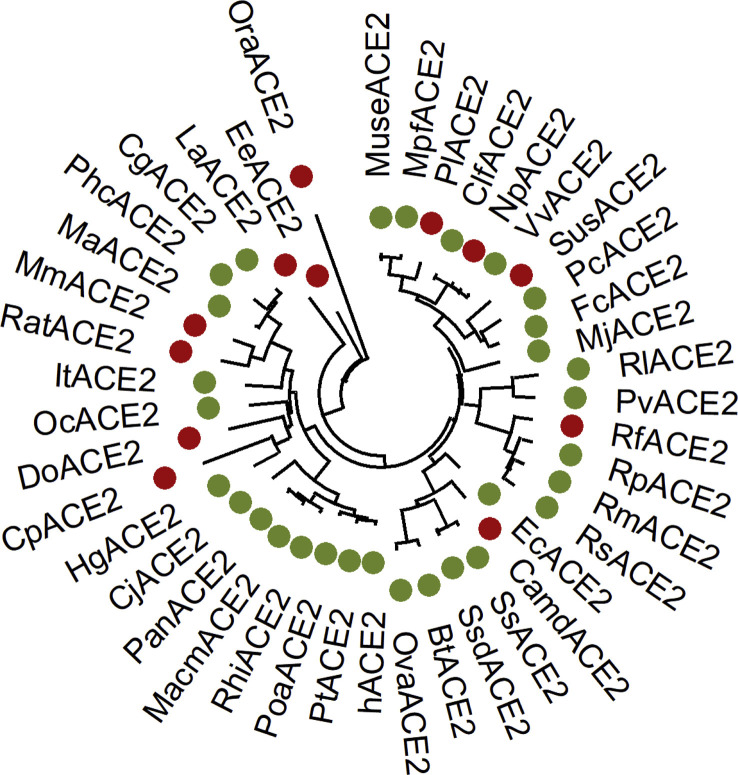
Next, we constructed a phylogenetic tree for mammalian ACE2 proteins. The ACE2 protein sequences of 42 mammalian animals were compared by ClustalW method of MEGA-X software. Then the JTT model of maximum likelihood method was used to construct ACE2 phylogenetic tree. As shown in Fig. 2 , species that cannot bind to S protein are marked in red, and species that can bind to S protein are marked in green. No correlation between genetic distance and the interaction of ACE2/S was found. Some Pets including dog (Canis lupus familiaris) and cat (Felis catus) potentially recognize S protein, indicating the importance to monitor the pets for SARS-CoV-2 infection. We found that four members of Circetidae including Mesocricetus auratus, Phodopus campbelli, Ictidomys tridecemlineatus, and Cricetulus griseus remained the key residues for association with S protein from SARS-CoV and SARS-CoV-2, though two members of Muridae (Rattus norvegicus, Mus musculus) could not bind to S protein. This founding suggested that Circetidae mammals could be developed as SARSr-CoV small animal models.
Fig. 2.
Phylogenetic tree of mammalian ACE2 proteins. ACE2 sequences from a total of 42 mammals were analyzed by MEGA-X and the phylogenetic tree was constructed with JTT evolutionary model using Maximum Likelihood method. The red represents the species whose ACE2 cannot bind to S protein, and the green is the species whose ACE2 associate with S protein. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Structure simulation of the protein complex of SARSr-CoV RBD and cat/dog/pangolin/Chinese hamster ACE2
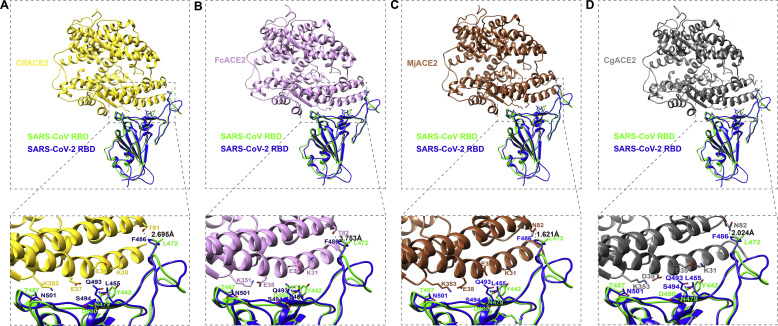
We noticed that ACE2 from dog, cat, pangolin and Chinese hamster potentially associated with S protein (Table 1). Next, we predicted the structure of cat/dog/pangolin/Chinese hamster ACE2 with SARSr-CoV RBD. The structure of protein complex between RBD region of S protein of SARS-CoV and human ACE2 has been resolved (PDB: 2AJF) [7]. Recently, the structure of SARS-CoV-2 RBD with human ACE2 was also determined [16,17]. We used Chimera software to display homologous model, and obtained the interaction complex structure of RBD region of SARSr-CoV (SARS-CoV-2 and SARS-CoV) and cat/dog/pangolin/Chinese hamster ACE2. Overall, the RBM structures of S protein of SARS-CoV-2 and SARS-CoV are similar (Fig. 3 ). Interaction interface of SARSr-CoV RBD and cat/dog/pangolin/Chinese hamster ACE2 confirmed the potential interaction between SARS-CoV-2 and cat/dog/pangolin/Chinese hamster ACE2, indicating that these ACE2 could support SARS-CoV-2 entry. The AA in 82 position of human ACE2 is M82, while the corresponding AA in cat, dog, pangolin and Chinese hamster ACE2 is T82, T81, N82 and N82, respectively. The distance between F486 of SARS-CoV-2 S protein and the corresponding AA in ACE2 is 3.753Å for human (Fig. 1A), 2.695Å for dog (Fig. 3A), 3.753Å for cat (Fig. 3B), 1.621Å for pangolin (Fig. 3C), and 2.024Å for Chinese hamster (Fig. 3D), respectively. We concluded that N82 in ACE2 showed closer contact with F486 of SARS-CoV-2 S protein than M82 of ACE2.
Fig. 3.
Structure simulation of SARSr-CoV RBD with ACE2 from dog, cat, pangolin and Chinese hamster. (A) Structural simulation of the protein complex of dog ACE2 and SARSr-CoV RBD. Dog ACE2, SARS-CoV-2 RBD, SARS-CoV RBD are in gold, blue, and green, respectively. (B) Structural simulation of the protein complex of cat ACE2 and SARSr-CoV RBD. Cat ACE2, SARS-CoV-2 RBD, and SARS-CoV RBD are in pulm, blue and green, respectively. (C) Structural simulation of the protein complex of pangolin ACE2 and SARSr-CoV RBD. Pangolin ACE2, SARS-CoV-2 RBD, and SARS-CoV RBD are in sandy brown, blue and green, respectively. (D) Structural simulation of the protein complex of Chinese hamster ACE2 and SARSr-CoV RBD. Chinese hamster ACE2, SARS-CoV-2 RBD, and SARS-CoV RBD are in dim gray, blue and green, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
The host tropism of zoonotic coronavirus is hybrid, and it is important to determine the natural host and host range of coronavirus. In the past two decades, SARS-CoV, MERS-CoV and SARS-CoV-2 have caused serious outbreaks of human infectious diseases. All the three human coronaviruses originated from bats, but the intermediate hosts were different. SARS-CoV is believed to come from the Paguma larvata [18], and the intermediate host of MERS-CoV is Camelus dromedaries [19]. The new coronavirus SARS-CoV-2 has recently caused a serious epidemic in China, but its intermediate hosts are not clear.
S protein of SARS-CoV-2 interacts with human ACE2, which promotes the entry of SARS-CoV-2, indicating that human ACE2 is the receptor of SARS-CoV-2 [3]. ACE2 contains at least five key amino acids critical for binding S protein of SARSr-CoV [15]. Based on these five amino acids, we analyzed the corresponding amino acids of different mammals to determine which mammalian ACE2 could interact with S protein of human SARSr-CoV. By analyzing the protein sequence of mammalian ACE2, we found that the ACE2 of Camelus dromedarius, Procyon lotor, Rhinolophus ferrumequinum, Rattus norvegicus, Mus musculus, Ornithorhynchus anatinus, Loxodonta africana, Erinaceus europaeus, Nyctereutes procyonoides, Suricata suricatta, Dipodomys ordii, and Cavia porcellus lose the capability to associate with S protein (Table 1). These mammals could be ruled out from the potential host list for SARS-CoV-2. We found that S protein may bind to ACE2 from some wild mammals, which suggests that we should investigate whether these animals may be intermediate hosts for SARS-CoV-2. It has been reported that the RBM region in S protein of pangolin coronavirus is similar to that of S protein of SARS-CoV-2 [11,12], which may be involved in the recombination of SARS-CoV-2. We identified that N82 of pangolin ACE2 showed closer contact with RBD than human ACE2 (Fig. 3C), indicating that pangolin ACE2 might show better affinity to SARS-CoV-2. This finding further supports the hypothesis that pangolin is involved in SARS-CoV-2 evolution. In current study, only a limited list of wild mammals is covered. In the future, we should select more mammals for study. Although no SARS-CoV-2 has been found in domestic cats and dogs, cat/dog ACE2 may bind to S protein of SARS-CoV-2. In the future, we should pay attention to monitoring whether domestic cats and dogs could be infected by SARS-CoV-2.
Animal model is an important tool in the study of infectious diseases. ACE2 of mice cannot interact with SARS-CoV-2, so it cannot be used as animal model of SARS-CoV-2 directly. Some studies have generated mice transfected with human ACE2 as the models to study SARS-CoV [20], and these mice can also be used as animal models for SARS-CoV-2 infection. Interestingly, we identified that N82 in ACE2 is closer to RBD than M82 (Fig. 3C and D), indicating a novel strategy to design an optimized ACE2 for SARS-CoV-2 infection. We speculated that small peptide based on N82 of ACE2 might show higher affinity to SARS-CoV-2 RBD. We proposed that if M82 in human ACE2 was mutated to N82, the modified human ACE2 will enhance SARS-CoV-2 infection. In the future, those ideas will be tested in cell culture and animal model. We noticed that the ACE2 proteins from Circetidae (Mesocricetus auratus, Phodopus campbelli, Ictidomys tridecemlineatus, and Cricetulus griseus) are capable to recognize RBD. Mesocricetus auratus (golden hamster) and Cricetulus griseus (Chinese hamster) are experimental animals and our finding indicates the possibility to develop small animal models for SARS-CoV-2 infection using Chinese hamster and golden hamster.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by grants from National Key Plan for Research and Development of China [2016YFD0500300], National Natural Science Foundation of China [81871663 and 81672035], Shandong Academy of Medical Sciences Grant [2017-52] and Academic promotion programme of Shandong First Medical University. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. We thank Dr. Jianxun Qi for sharing the structure of 6LZG.
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrc.2020.03.047.
Transparency document
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anguiano L., Riera M., Pascual J., Soler M.J. Circulating ACE2 in cardiovascular and kidney diseases. Curr. Med. Chem. 2017;24:3231–3241. doi: 10.2174/0929867324666170414162841. [DOI] [PubMed] [Google Scholar]
- 10.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronavirus. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong M.C., Cregeen S.J.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020 [Google Scholar]
- 12.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F., Leung G.M., Holmes E.C., Hu Y.-L., Guan Y. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. 2020 2002.2013.945485. [Google Scholar]
- 13.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Mega X. Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 15.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020 doi: 10.1128/JVI.00127-20. pii: JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan R., Zhang Y., Guo Y., Xia L., Zhou Q. Structural basis for the recognition of the 2019-nCoV by human ACE2. bioRxiv. 2020 doi: 10.1126/science.abb2762. 2002.2019.956946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Crystal structure of the 2019-nCoV spike receptor-binding domain bound with the ACE2 receptor. bioRxiv. 2020 doi: 10.1038/s41586-020-2180-5. 2002.2019.956235. [DOI] [PubMed] [Google Scholar]
- 18.Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., Cui B., Xu Y., Zhang E., Wang H., Ye J., Li G., Li M., Cui Z., Qi X., Chen K., Du L., Gao K., Zhao Y.T., Zou X.Z., Feng Y.J., Gao Y.F., Hai R., Yu D., Guan Y., Xu J. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan R.W., Hemida M.G., Kayali G., Chu D.K., Poon L.L., Alnaeem A., Ali M.A., Tao K.P., Ng H.Y., Chan M.C., Guan Y., Nicholls J.M., Peiris J.S. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med. 2014;2:813–822. doi: 10.1016/S2213-2600(14)70158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X.H., Deng W., Tong Z., Liu Y.X., Zhang L.F., Zhu H., Gao H., Huang L., Liu Y.L., Ma C.M., Xu Y.F., Ding M.X., Deng H.K., Qin C. Mice transgenic for human angiotensin-converting enzyme 2 provide a model for SARS coronavirus infection. Comp. Med. 2007;57:450–459. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.