Abstract
Objective
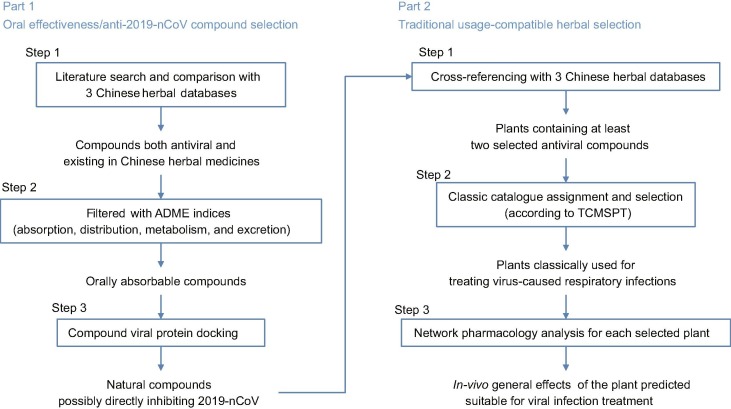
In this study we execute a rational screen to identify Chinese medical herbs that are commonly used in treating viral respiratory infections and also contain compounds that might directly inhibit 2019 novel coronavirus (2019-nCoV), an ongoing novel coronavirus that causes pneumonia.
Methods
There were two main steps in the screening process. In the first step we conducted a literature search for natural compounds that had been biologically confirmed as against sever acute respiratory syndrome coronavirus or Middle East respiratory syndrome coronavirus. Resulting compounds were cross-checked for listing in the Traditional Chinese Medicine Systems Pharmacology Database. Compounds meeting both requirements were subjected to absorption, distribution, metabolism and excretion (ADME) evaluation to verify that oral administration would be effective. Next, a docking analysis was used to test whether the compound had the potential for direct 2019-nCoV protein interaction. In the second step we searched Chinese herbal databases to identify plants containing the selected compounds. Plants containing 2 or more of the compounds identified in our screen were then checked against the catalogue for classic herbal usage. Finally, network pharmacology analysis was used to predict the general in vivo effects of each selected herb.
Results
Of the natural compounds screened, 13 that exist in traditional Chinese medicines were also found to have potential anti-2019-nCoV activity. Further, 125 Chinese herbs were found to contain 2 or more of these 13 compounds. Of these 125 herbs, 26 are classically catalogued as treating viral respiratory infections. Network pharmacology analysis predicted that the general in vivo roles of these 26 herbal plants were related to regulating viral infection, immune/inflammation reactions and hypoxia response.
Conclusion
Chinese herbal treatments classically used for treating viral respiratory infection might contain direct anti-2019-nCoV compounds.
Keywords: 2019-nCoV, Wuhan coronavirus, Drugs, Chinese herbal, Pneumonia, Natural compounds, Molecular docking, Network pharmacology
1. Introduction
Toward the end of December 2019, a novel coronavirus (2019-nCoV) with human-to-human transmission and severe human infection, originating in Wuhan, China, was identified [1]. This virus has affected many persons in China and spread to other countries in a very short time. On January 30, 2020, the Director-General of the World Health Organization declared that the outbreak of 2019-nCoV constitutes a public health emergency of international concern and issued temporary recommendations under the International Health Regulations [2]. According to the Daily Report of China National Health Commission, as of this writing on February 2, 2020, 14,488 cases, including 304 deaths, have been confirmed in China; 146 cases, including 1 death, have also been reported among 23 other countries. This pandemic is still ongoing, so it is urgent to find new preventive and therapeutic agents as soon as possible. In addition, commensurate with the risk, strong measures for early detection, isolation and treatment of cases, as well as minimization of transmission through social interaction must be implemented.
While specific vaccines and antiviral agents are the most effective methods to prevent and treat viral infection, there are not yet effective treatments that target the 2019-nCoV. Development of these treatments may require months or years, meaning that a more immediate treatment or control mechanism should be found if possible. Herbs used in traditional Chinese medicine present a potentially valuable resource to this end. The effectiveness of herbal treatment to control contagious disease was demonstrated during the 2003 severe acute respiratory syndrome (SARS) outbreak [3]. As such, the Chinese government is encouraging the use of herbal plants in fighting this new viral pneumonia. However, the application of herbal treatment is mainly guided by the type of herb (based on the catalogue of classic literature on herbs) and the patient’s symptoms or signs. There is often not enough information to predetermine whether the herbs in question can directly target the viral cause, in other words, herbal usage is generally not guided by viral pathology. We think more detailed knowledge about the direct antiviral effects of different plants would be greatly helpful to the doctors selecting them.
In fact, after the outbreak of SARS, many groups dedicated themselves to finding anti-coronavirus agents, including some natural compounds that exist in traditional Chinese herbal medicines [4], [5], [6], [7], [8], [9], [10], [11], [12]. The coronavirus encodes more than one dozen proteins, some of which are essential to viral entry and replication. Among these proteins, the most well-studied are papain-like protease (PLpro), 3C-like protease (3CLpro) and spike protein. Coronavirus PLpro not only processes the viral polypeptide onto functional proteins but is also a deubiquitinating enzyme that can dampen host anti-viral response by hijacking the ubiquitin (Ub) system. For example, SARS PLpro cleaves ISG15, a two-domain Ub-like protein, and Lys48-linked polyUb chains, releasing diUbLys48 products [13], [14]. SARS-3CLpro is a cysteine protease indispensable to the viral life cycle [15]. Coronavirus spike protein uses angiotensin-converting enzyme 2 as a receptor to help the virus enter cells [16]. These three proteins make attractive targets for drug development.
Through in silico and biological processing, a series of small molecules, including those from natural compounds, have been screened and confirmed to directly inhibit these important proteins in SARS or Middle East respiratory syndrome (MERS) coronavirus [17], [18], [19], [20], [21], [22], [23]. The gene sequence of 2019-nCoV has been released, which suggests high similarities between the main proteins in this virus and those previously identified in SARS-Cov or MERS-Cov [24], [25]. In this sense, previously reported anti-SARS-Cov or anti-MERS-Cov natural compounds may become a valuable guide to finding anti-coronavirus (2019-nCoV) herbal plants among the traditional Chinese herbs used to treat viral pneumonia.
It is a challenge to screen out the herbs containing anti-coronavirus (2019-nCoV) compounds from the large number of those possibly being used for patients infected with this pathogen, especially in very short time. Here, we propose two principles to guide such work: oral effectiveness and traditional usage compatibility. The first principle refers to the fact that most Chinese herbal plants are orally ingested after boiling with water, meaning that the anti-coronavirus (2019-nCoV) ingredients in selected plants should be absorbable via oral preparation. The second principle recognizes that candidate plants should be consistent with the type classifications for traditional herbal usage, since type-guided applications are integral to herbal use, as mentioned above. Following these two principles, we used a 6-step selection process (3 for each principle), including drug-likeness, evaluation of oral bioavailability, molecular docking, network pharmacology analysis and other methods to identify herbs that have both a high possibility of containing effective anti-coronavirus (2019-nCoV) compounds and are classified as treating virus-caused respiratory infection.
2. Materials and methods
2.1. Literature search and compound selection
PubMed literature concerning natural compounds against SARS or MERS coronavirus activity was selected using the query “coronavirus AND inhibitor AND (SARS OR MERS OR SARS-CoV OR MERS-CoV).” After careful reading of the studies returned by this search, the natural compounds that had biologically confirmed antiviral activities were compared with the Traditional Chinese Medicine Systems Pharmacology database (TCMSP, http://www.tcmspw.com/browse.php?qc=herbs), the Encyclopedia of Traditional Chinese Medicine (ETCM, http://www.nrc.ac.cn:9090/ETCM/) and SymMap (https://www.symmap.org/). Natural compounds both associated with antiviral activity and contained in herbs were examined in the next step of our study.
2.2. ADME screening of natural compounds
Since Chinese herbal treatments are always taken orally after boiling with water, an in silico integrative model of absorption, distribution, metabolism and excretion (ADME) was used to screen for natural compounds that may be bioactive via oral administration. The indices used for the screening include evaluation of oral bioavailability, Caco-2 permeability, drug-like value, and drug half-life. The threshold values indicating effectiveness for these four indices were > 30%, > −0.4, > 0.18 and > 3 h, respectively, as recommended by Hu et al [26]. The values of these four indices can be obtained from the TCMSP database.
2.3. Protein-molecular docking
We used molecular docking software AutoDock 4 to perform protein compound docking analysis, according to the following procedure: (1) We built three-dimensional (3D) structure files of the proteins of interest. We used the online server SWISS-MODEL (https://swissmodel.expasy.org/) to build the 3D structures of the proteins of interest by template-based modeling, these template structures being the reported 3D structures of the corresponding proteins from SARS-CoV. The models built were of Protein Data Bank (PDB) format. (2) To retrieve the required 3D structure files of compounds, the structure data file (SDF) format of compounds were retrieved from the PubChem website and then converted to PDB format by Discovery Studio. (3) AutoDock 4.2 was used to prepare PDBQT format files for target and ligand screening (Target.pdbqt and Ligand.pdbqt) and grid and docking parameter files (a.gpf and a.dpf). (4) Molecular docking was performed using AutoDock in Cygwin and finally the results were analyzed. The process and parameters used were detailed by Rizvi et al [27].
2.4. Plant selection
Herbs were selected through three steps. (1) Primary selection: molecules chosen from the above steps were used as input for the TCMSP, ETCM and SymMap to search for plants containing that input and the plants were filtered by the numbers of antiviral compounds they contain. Those containing 2 or more antiviral compounds were selected for the next step. (2) Classic usage catalogue cross-reference: only herbs traditionally used to treat viral respiratory infection were retained for further study. (3) Predication of general effects in vivo with network pharmacology analysis, which is detailed as follows.
2.5. Network pharmacology analysis
The TCMSP provided the main components of each herb and the protein targets for each component. We identified the reported chemical constituents for each plant in the final analysis and used the ADME indices listed above to find the orally absorbable and drug-like compounds for the plant. The protein targets of these compounds were downloaded from the TCMSP database. All protein targets for each individual plant were used as input for the String online server (https://string-db.org/) to perform protein–protein interaction analysis and pathway enrichment. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched (with P < 0.01) by the input were downloaded.
All data were processed using the statistical language R (3.6.2), unless otherwise specified.
3. Results
3.1. Overlapping of natural compounds biologically confirmed to be anti-SARS or anti-MERS coronavirus in literature and in Chinese herbal database
We received 261 hits from conducting our search in the PubMed database. After careful evaluation of the abstracts from these citations we downloaded and carefully analyzed the full text of 23 highly relevant papers. The natural compounds reported to have biologically confirmed anti-coronavirus activity were identified and then compared to the ingredients listed in three Chinese herbal databases. The result was 115 overlapping ingredients, which we used for further testing (Fig. 1 ).
Fig. 1.
Workflow scheme. The work is divided into two main parts, natural compound selection and herbal plant selection. Each part consists of three steps. As detailed in the text, oral effectiveness is important in compound selection, while in the plant selection portion, the selected herbs should be compatible with the classic usages of herbal treatment in traditional Chinese medicine. TCMSP: Traditional Chinese Medicine Systems Pharmacology.
3.2. Filtration of compounds selected by ADME
The antiviral activities of the 115 natural compounds were reportedly confirmed with enzyme-based (cell-free) or cell-based experimental systems. To be utilized as a Chinese herbal medicine, they must be absorbable via oral prescription. Therefore, we performed ADME screening for the 115 natural compounds, reducing the number of candidates to 13.
3.3. Docking between selected compounds and their reported targets
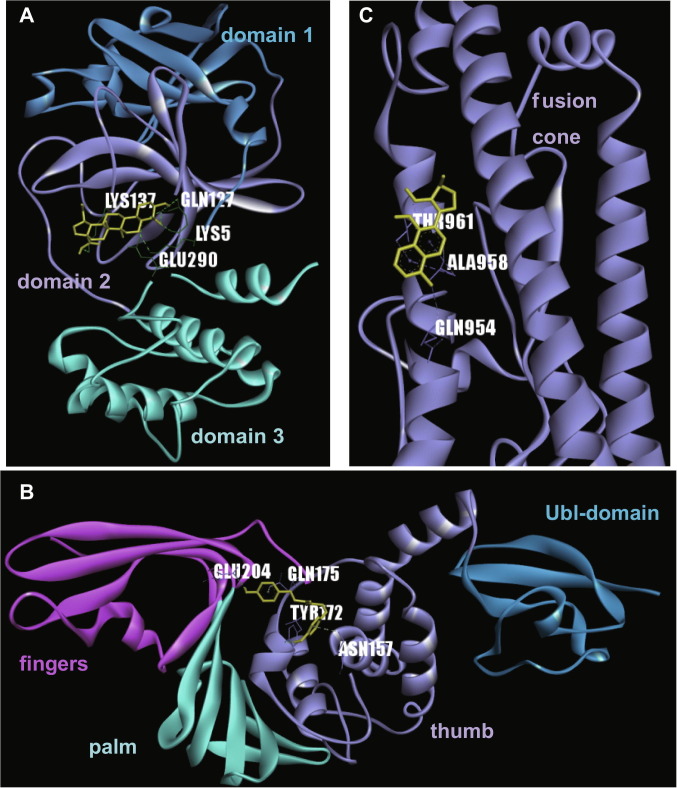
To perform the docking analysis, the 3D structure files of 2019-nCoV PLpro, 3CLpro and spike proteins were built based on the corresponding SARS-CoV templates, i.e., PDB 5e6j, 1uj1 and 6cad, respectively. Then, molecule-protein docking was carried out between the molecules and their reported targets. If the molecules were reported to inhibit viral entry, they were docked with spike proteins (Table 1 ). Each separate analysis returned positive results (Table 1, Fig. 2 and online supplementary Fig. S1), indicating the natural compounds we selected might directly inhibit 2019-nCoV. The molecules selected to target PLpro (M2, M3, M7, M9, M10, M11 and M13) mainly bound in the region between the thumb and palm domains, which might interfere with substrate entering this enzyme’s active sites, located at the bottom of the two domains [29]. The molecules reported to inhibit 3CLpro (M1, M2, M3, M4, M5, M7, M8, M10, M11, M12 and M13) mainly entered the region between domains 2 and 3, and this region is important for 3CLpro to form a dimmer [30]. M6 was reported to inhibit viral entry, accordingly it bound the fusion cone of spike protein; this cone structure is important for viral membrane fusion [31] (Fig. 2 and online supplementary Fig. S1).
Table 1.
The molecules and their docking proteins, binding energy (kcal/mol).
| No. | Molecular name | Targets or inhibition | Reference | Docking (binding energy) |
||
|---|---|---|---|---|---|---|
| PLpro | 3CLpro | Spike | ||||
| M1 | Betulinic acid | Replication, 3CLpro | [16] | Undo | −4.23 | Undo |
| M2 | Coumaroyltyramine | PLpro and 3CLpro | [11], [20] | −3.22 | −4.18 | Undo |
| M3 | Cryptotanshinone | PLpro and 3CLpro | [18] | −5.25 | −6.23 | Undo |
| M4 | Desmethoxyreserpine | Replication, 3CLpro, and entry | [6] | Undo | −3.52 | Undo |
| M5 | Dihomo-γ-linolenic acid | 3CLpro | [7] | Undo | −3.88 | Undo |
| M6 | Dihydrotanshinone Ⅰ | Entry, and spike protein | [28] | Undo | Undo | −5.16 |
| M7 | Kaempferol | PLpro and 3CLpro | [11] | −2.15 | −6.01 | Undo |
| M8 | Lignan | Replication, 3CLpro | [16] | Undo | −4.27 | Undo |
| M9 | Moupinamide | PLpro | [20] | −3.05 | Undo | Undo |
| M10 | N-cis-feruloyltyramine | PLpro and 3CLpro | [11], [20] | −3.11 | −4.31 | Undo |
| M11 | Quercetin | PLpro and 3CLpro | [20] | −4.62 | −6.25 | Undo |
| M12 | Sugiol | Replication, 3CLpro | [16] | Undo | −6.04 | Undo |
| M13 | Tanshinone IIa | PLpro and 3CLpro | [18] | −5.02 | −5.17 | Undo |
3CLpro: 3C-like protease; PLpro: papain-like protease.
Fig. 2.
Docking between selected natural compounds and their reported targets. A. PLpro and natural compound M2; B. 3CL and M1; C. Spike and M6. Docking is performed with AutoDock 4 which is detailed in Materials and Methods. The protein structure files are listed in Table 1. Protein domains are shown in different colors, while natural compounds are shown in dark yellow. The amino acids labeled were those interacting with compound. 3CLpro: 3C-like protease; PLpro: papain-like protease.
3.4. Selection of antiviral herbal plants
The 13 molecules passing the three-round selection process were then compared to the three Chinese herbal databases, and we found 230 herbs containing these molecules (online supplementary file Table S1). We then evaluated these herbs for those containing 2 or more of the 13 natural compounds, leaving 125 results. We cross-referenced the 125 results with the classic categorizations for herbal usage in the TCMSP database, finally choosing 11 types that are traditionally used to treat viral respiratory infections. There are 26 herbal plants within the 11 types. The timeframe during the course of a viral infection that each of these 26 herbal plants (Table 2 ) should be used was also documented by seeking advice from senior practitioners of traditional Chinese medicine. For example, plants catalogued as antipyretic detoxifying drugs, qi-reinforcing drugs, antitussive antiasthmatics, pungent cool diaphoretics and phlegm-resolving medicines may all be used throughout the course of infection, whereas drugs belonging to the interior warming group may be best utilized in prevention.
Table 2.
The 26 Chinese herbals screened and the possible time for usage.
| No. | Herbal name |
Number* | Classic catalogue |
Time to use | ||
|---|---|---|---|---|---|---|
| Latin | Chinese | Latin/English | Chinese | |||
| 1 | Forsythiae fructus | 连翘 | 3 | Antipyretic detoxifying | 清热解毒药 | Full course |
| 2 | Licorice | 甘草 | 3 | Qi-reinforcing | 补气药 | Full course |
| 3 | Mori cortex | 桑白皮 | 3 | Antitussive antiasthmatics | 止咳平喘药 | Full course |
| 4 | Chrysanthemi flos | 菊花 | 2 | Pungent cool diaphoretics | 辛凉解表药 | Full course |
| 5 | Farfarae flos | 款冬花 | 2 | Antitussive antiasthmatics | 止咳平喘药 | Full course |
| 6 | Lonicerae japonicae flos | 金银花 | 2 | Antipyretic-detoxifying drugs | 清热解毒药 | Full course |
| 7 | Mori follum | 桑叶 | 2 | Pungent cool diaphoretics | 辛凉解表药 | Full course |
| 8 | Peucedani radix | 前胡 | 2 | Phlegm-resolving medicine | 化痰药 | Full course |
| 9 | Rhizoma fagopyri cymosi | 金荞麦 | 2 | Antipyretic detoxifying | 清热解毒药 | Full course |
| 10 | Tamaricis cacumen | 西河柳 | 3 | Pungent-warm exterior-releasing medicine | 辛温解表药 | Early |
| 11 | Erigeron breviscapus | 灯盏细辛 | 2 | Pungent-warm exterior-releasing medicine | 辛温解表药 | Early |
| 12 | Radix bupleuri | 柴胡 | 2 | Pungent cool diaphoretics | 辛凉解表药 | Early |
| 13 | Coptidis rhizoma | 黄连 | 2 | Heat-clearing and dampness drying medicine | 清热燥湿药 | Middle |
| 14 | Houttuyniae herba | 鱼腥草 | 2 | Antipyretic-detoxifying | 清热解毒药 | Middle |
| 15 | Hoveniae dulcis semen | 枳椇子 | 2 | Antipyretic-detoxifying | 清热解毒药 | Middle |
| 16 | Inulae flos | 旋覆花 | 2 | Phlegm resolving medicine | 化痰药 | Middle |
| 17 | Eriobotryae folium | 枇杷叶 | 3 | Antitussive antiasthmatics | 止咳平喘药 | Middle and later |
| 18 | Hedysarum multijugum maxim. | 黄芪 | 3 | Qi-reinforcing | 补气药 | Middle and later |
| 19 | Lepidii semen descurainiae semen | 葶苈子 | 3 | Antitussive antiasthmatics | 止咳平喘药 | Middle and later |
| 20 | Ardisiae japonicae herba | 矮地茶 | 2 | Antitussive antiasthmatics | 止咳平喘药 | Middle and later |
| 21 | Asteris radix et rhizoma | 紫菀 | 2 | Antitussive antiasthmatics | 止咳平喘药 | Middle and later |
| 22 | Euphorbiae helioscopiae herba | 泽漆 | 2 | Diuretic dampness-excreting | 利水渗湿药 | Middle and later |
| 23 | Ginkgo semen | 白果 | 2 | Antitussive antiasthmatics | 止咳平喘药 | Middle and later |
| 24 | Anemarrhenae rhizoma | 知母 | 3 | Fire-purging | 清热泻火药 | Later |
| 25 | Epimrdii herba | 淫羊藿 | 2 | Yang-reinforcing | 补阳药 | Later |
| 26 | Fortunes bossfern rhizome | 贯众 | 2 | Warming interior | 温里药 | Prevention |
The number of antiviral natural compounds contained in the plant.
3.5. Network analysis of possible effects or mechanisms
Each of the potentially effective herbal remedies contains many ingredients in addition to the antiviral ones found here. Thus, the general effects of each plant should be examined by combining the effects of all of the orally absorbable and biologically active ingredients in it. To evaluate the possible general in vivo effects of each of our identified herbs, we used the ADME indices listed above to examine each of the orally absorbable and drug-like ingredients recorded in the TCMSP database for each plant. We then extracted the target proteins for each ingredient which had passed the screening process. All proteins belonging to a single plant were combined as input on the online protein–protein interaction analysis server, String, to find the pathway enrichment.
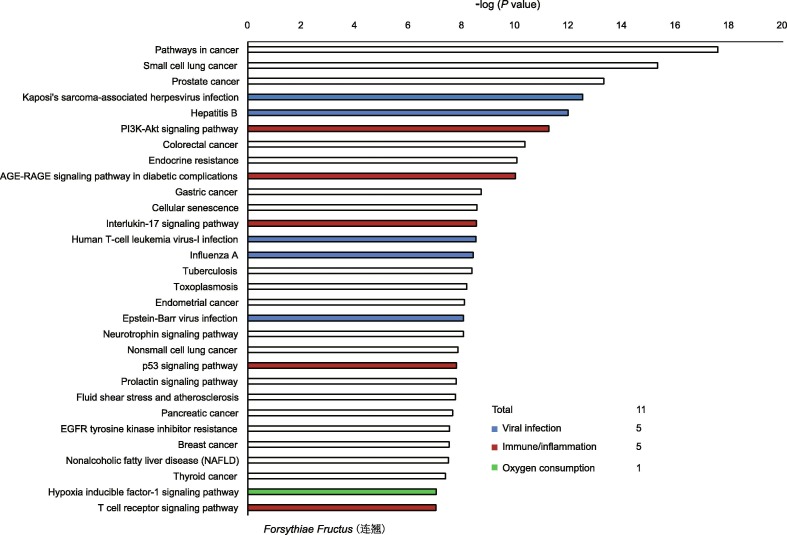
For the 26 herbs, about 1/3 of the top 30 KEGG-enriched pathways (mean = 11) were related to regulating viral infection, immune/inflammatory reactions and hypoxia response, indicating that they are potentially effective treatments for viral respiratory infection (Fig. 3 and online supplementary Fig. S2). Note that some of the herbal plants selected here had been reported to be effective for SARS-CoV infection in 2003 (online supplementary Table S2).
Fig. 3.
Kyoto Encyclopedia of Genes and Genomes pathways enriched for the herb Forsythiae Fructus in network pharmacology analysis. The top 30 pathways are shown. The blue, red and green bars represent the pathways related to viral infection, immune/inflammation response, and hypoxia response, respectively. EGFR: epidermal growth factor receptor; AGE: advanced glycation end product; RAGE: receptor for AGE.
4. Discussion
In this work, we undertook a multiple step selection process and screened out 26 herbal plants with a high probability of directly inhibiting the novel coronavirus (2019-nCoV), possibly providing instant help in the prevention and treatment of the pneumonia that it can cause. While mainly in China at this point, viral spread is ongoing and has affect persons worldwide.
Two principles guided our screening work. The first is that the anti-coronavirus (2019-nCoV) components contained in the source plants should be absorbable via oral prescription. This principle requires that the herbs selected should contain biologically proven anti-coronavirus (2019-nCoV) ingredients, and that these natural compounds should pass the drug-likeness and oral bioavailability evaluations. Therefore, we conducted a three-step screening process. First, we extracted natural compounds verified in PubMed as being effective in treating SARS or MERS coronavirus and then cross-checked these compounds in the Chinese herbal databases. There were 115 overlapping compounds. This method was an expeditious way to identify natural components both pre-existing in Chinese herbal treatment and having a high possibility of anti-coronavirus (2019-nCoV) activity. This is important, as the anti-coronavirus effects of the selected compounds have been biologically confirmed, and the genetic similarities between coronavirus (2019-nCoV) and SARS or MERS coronavirus are high [24], [25].
The anti-coronavirus effects of the natural compounds screened by the above method have been mainly confirmed in vitro by direct loading onto cultured cells, thus it does not guarantee their effectiveness in vivo, especially with oral preparation—the principal way in which Chinese herbals are administered. Therefore, to meet the first principle, we ran ADME filters on the natural compounds selected by 4 indices, as used by Hu et al. [26]. Among the 115 compounds highlighted by our first step, only 13 passed this screening, showing the necessity of such a test.
The novel coronavirus has some mutations when compared to SARS or MERS coronavirus, so the natural compounds effectiveness against the two previous coronaviruses might not be present in the new virus. To reduce this risk, and as the third step of our first principle, we reconstructed the 3D structure of the new coronavirus using the reported structures of SARS and MERS coronavirus proteins as a guide, and then used molecular docking technology to simulate whether the 13 natural compounds selected could combine with the structures we constructed for the new coronavirus proteins. All 13 compounds could bind to the proteins as predicted for the new coronavirus. We believe that the high success rate of our docking screening was due to the high genetic similarity between the new coronavirus and the SARS or MERS virus [24], [25].
Our second principle for screening should also be emphasized and elaborated upon. It states that the selected herbal plants must conform to traditional usages. There are many kinds of Chinese herbs that have been used for thousands of years. Based on this rich history and experience, Chinese herbal medicines are divided into different types, each type dedicated to certain kinds of diseases. Ignoring these grouping guidelines can lead to serious side effects. Therefore, as a further condition for the medicine screened here, we verified that they have been routinely used to treat viral pneumonia. To meet this principle, we conducted another three-step screening process for the herbal plants.
First, we searched the Chinese medicine database for herbs containing the 13 natural compounds identified. Herbs containing at least 2 of these potentially useful compounds were selected, and a total of 125 herbal plants were identified. The second step in targeted plant selection was based on type classification. Of the 125 results, only 26 herbs were found to be routinely used in treating viral respiratory infection.
Finally, network pharmacological analysis was performed to predict the possible therapeutic effects of these 26 plants. Because Chinese herbal medicines contain many ingredients, and multiple absorbable ingredients might exert their effects on the body, the general effects of herbs may be dictated by all of the absorbable ingredients they contain. With this consideration in mind, we extracted the recorded ingredients of each of the plants selected from the Chinese medicine database and screened these ingredients for drug-likeness and oral availability (via ADME filter) [26], [32]. The target proteins of all ingredients passing ADME selection were used for network enrichment to predict the general effects of the herbal plants. For all the plants analyzed, nearly half of the top 30 pathways enriched in KEGG are related to antiviral, immune/inflammatory responses and hypoxia response indicating that these herbs are suitable for anti-viral usage. In fact, some of the herbal plants selected here had been reported effective in against SARS-CoV infection in 2003 (online supplementary Table S2). We thought that the general antiviral and immune/inflammation effects predicted for the 26 plants are correlated with the fact that these plants were selected according to Chinese herbal type classifications.
Of course, it should be pointed out that Chinese herbs that have not been identified through this screening process may still have beneficial effects. Further, considering that the biologically validated natural compounds reported in the literature cannot cover all antiviral natural compounds, and the natural compounds included in the Chinese medicine database are not complete, the process that we have followed may have excluded herbs that would be well suited to this treatment. Nevertheless, the purpose of this screening was to provide a rational approach for selecting Chinese herbal medicines with a high potential efficacy in treating 2019-nCoV and related viruses. The specific dosage and usage of each herb should be determined based on patients’ manifestations. Finally, the key step in this screening was molecular docking. The 3D structures of the proteins used here are based on reported gene sequences. If the virus mutates during transmission, a new screening is recommended.
In conclusion, this work has identified several Chinese medicinal plants classified as antiviral/pneumonia-effective that might directly inhibit the novel coronavirus, 2019-nCoV. Additionally, we propose screening principles and methods which may provide guidance in screening antiviral drugs from other natural drug databases.
Author contributions
DZ conceived the study, participated in its design, coordination, and all the work processes. KW participated in herbal selection. XZ participated in data collection and network pharmacology analysis. SD helped to collect data. BP helped to draft the manuscript.
Funding
This work is supported by Shanghai Leading Talent Grants in Medicine (No. 2019LG26), Shanghai Traditional Chinese Medicine Content Construction Innovation Project (No. ZY3-CCCX-3-7001) and Postdoctoral Funding of Shanghai Gongil Hospital (No. GLBH2017002).
Acknowledgments
Acknowledgement
Thanks to Mr. Steven Schwab for tidying up the English expressions and grammar.
Conflicts of interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.joim.2020.02.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). (2020-01-30) [2020-02-02]. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov).
- 3.Chen Z., Nakamura T. Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytother Res. 2004;18(7):592–594. doi: 10.1002/ptr.1485. [DOI] [PubMed] [Google Scholar]
- 4.Lai L., Han X., Chen H., Wei P., Huang C., Liu S. Quaternary structure, substrate selectivity and inhibitor design for SARS 3C-like proteinase. Cur Pharm Des. 2006;12(35):4555–4564. doi: 10.2174/138161206779010396. [DOI] [PubMed] [Google Scholar]
- 5.Wang S.Q., Du Q.S., Zhao K., Li A.X., Wei D.Q., Chou K.C. Virtual screening for finding natural inhibitor against cathepsin-L for SARS therapy. Amino Acids. 2007;33(1):129–135. doi: 10.1007/s00726-006-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesel A.J. Synthesis of novel test compounds for antiviral chemotherapy of severe acute respiratory syndrome (SARS) Curr Med Chem. 2005;12(18):2095–2162. doi: 10.2174/0929867054637644. [DOI] [PubMed] [Google Scholar]
- 7.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc Natl Acad Sci U S A. 2004;101(27):10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B., Zhou J. SARS-CoV protease inhibitors design using virtual screening method from natural products libraries. J Comput Chem. 2005;26(5):484–490. doi: 10.1002/jcc.20186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem. 2005;48(4):1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 10.Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Li J., Luo C., Liu H., Xu W., Chen G. Binding interaction of quercetin-3-β-galactoside and its synthetic derivatives with SARS-CoV 3CL(pro): structure-activity relationship studies reveal salient pharmacophore features. Bioorg Med Chem. 2006;14(24):8295–8306. doi: 10.1016/j.bmc.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzyme Inhib Med Chem. 2017;32(1):504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458(7237):430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5(6):559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee P., Shah F., Desai P., Avery M. Inhibitors of SARS-3CLpro: virtual screening, biological evaluation, and molecular dynamics simulation studies. J Chem Inf Model. 2011;51(6):1376–1392. doi: 10.1021/ci1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50(17):4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 18.Ryu Y.B., Park S.J., Kim Y.M., Lee J.Y., Seo W.D., Chang J.S. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorg Med Chem Lett. 2010;20(6):1873–1876. doi: 10.1016/j.bmcl.2010.01.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg Med Chem. 2012;20(19):5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J.Y., Kim J.H., Kwon J.M., Kwon H.J., Jeong H.J., Kim Y.M. Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg Med Chem. 2013;21(13):3730–3737. doi: 10.1016/j.bmc.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y.H., Kim D.W., Curtis-Long M.J., Yuk H.J., Wang Y., Zhuang N. Papain-like protease (PLpro) inhibitory effects of cinnamic amides from Tribulus terrestris fruits. Biol Pharm Bull. 2014;37(6):1021–1028. doi: 10.1248/bpb.b14-00026. [DOI] [PubMed] [Google Scholar]
- 22.Park J.Y., Ko J.A., Kim D.W., Kim Y.M., Kwon H.J., Jeong H.J. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J Enzyme Inhib Med Chem. 2016;31(1):23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 23.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol. 2019;93(12):e00023–e119. doi: 10.1128/JVI.00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian H.Y. 2019-nCoV: new challenges from coronavirus. Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:E001. doi: 10.3760/cma.j.issn.0253-9624.2020.0001. [Chinese with abstract in English] [DOI] [PubMed] [Google Scholar]
- 26.Hu W., Fu W., Wei X., Yang Y., Lu C., Liu Z. A network pharmacology study on the active ingredients and potential targets of Tripterygium wilfordii Hook for treatment of rheumatoid arthritis. Evid Based Complement Alternat Med. 2019;2019:5276865. doi: 10.1155/2019/5276865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi S.M., Shakil S., Haneef M. A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J. 2013;12:831–857. [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J.Y., Kim Y.I., Park S.J., Kim I.K., Choi Y.K., Kim S.H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int J Antimicrob Agents. 2018;52(5):730–732. doi: 10.1016/j.ijantimicag.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279(47):49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W., Huai Y., Miao Z., Qian A., Wang Y. Systems pharmacology for investigation of the mechanisms of action of traditional Chinese medicine in drug discovery. Front Pharmacol. 2019;10:743. doi: 10.3389/fphar.2019.00743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.