Abstract
Interferon-inducible transmembrane proteins (IFITMs) restrict infection by several viruses, such as influenza A virus, West Nile virus and dengue virus. It has not been determined whether porcine IFITMs (pIFITMs) inhibit infection by pseudorabies virus (PRV), an enveloped, double-stranded DNA virus, which is the etiological agent of Aujeszky’s disease in pigs. Here, we report that PRV infection elicited pIFITM1 expression in PK15 porcine kidney epithelial cells and 3D4/21 alveolar macrophages. pIFITM2 and pIFITM3 expression was only elevated in PK15 cells during PRV infection. Depletion of pIFITM1 using RNA interference, either in PK15 or in 3D4/21 cells, enhanced PRV infection while overexpression of pIFITM1 had the opposite effect. Knockdown of pIFITM2 and pIFITM3 did not influence PRV infection, suggesting that pIFITM2 and pIFITM3 are independent of PRV infection. PRV-induced pIFITM1 expression was dependent on the cGAS/STING/TBK1/IRF3 innate immune pathway and interferon-alpha receptor-1, suggesting that pIFITM1 is up-regulated by the type I interferon signaling pathway. The anti-PRV role of pIFITM1 was inhibited upon PRV entry. Our data demonstrate that pIFITM1 is a host restriction factor that inhibits PRV entry that may shed light on a strategy for prevention of PRV infection.
Keywords: PRV, IFITMs, GAS/STING/TBK1/IRF3 innate immune pathway, Viral entry
1. Introduction
Aujeszky’s disease, a notifiable infectious disease of pigs, causes economic losses worldwide in the pig industry. The causative pathogen is pseudorabies virus (PRV), which is a member of the subfamily Alphaherpesvirinae of the family Herpesviridae [1]. The genome of PRV is approximately 143 kb and encodes at least 70 open reading frames [2]. Although pigs are the natural host of PRV, other mammals, such as ruminants, carnivores and rodents, are susceptible to PRV infection [3]. PRV infection causes fatal fever and encephalomyelitis in pigs and susceptible animals [4]. PRV Bartha-K61 vaccines lacking virulence-determining genes are effective for prevention of morbidity of PRV infection in pigs. However, since 2011, this vaccine has exhibited less protective efficacy against virulent PRV variants in China. Therefore, new applications are urgently needed to control PRV spread.
The innate immune system has evolved as a first line of defense against virus infection. Previous studies have demonstrated that PRV infection can induce an innate immune response through cytosolic pattern recognition receptors that recognize PRV genomic DNA. Knockdown of DNA-dependent activator of interferon (IFN)-regulatory factors (DAI) inhibits IFN-β expression and overexpression of DAI enhances IFN-β expression in response to PRV infection [5]. Later, DEAD (Asp–Glu–Ala–Asp) box polypeptide (DDX)41 was shown to participate in PRV infection [6]. DDX41 recognizes PRV genomic DNA and activates the type I IFN signaling pathway. Our previous study suggests that porcine cyclic GMP–AMP (cGAMP) synthase (cGAS) is also responsible for PRV-induced type I IFN activation [7]. The enzymatic activity of cGAS is activated by the binding between cGAS and PRV DNA. cGAS thereafter catalyzes the generation of cGAMP, an intrinsic second messenger that activates stimulator of interferon genes protein (STING) to promote IFN expression dependent on the cGAS/STING/TBK1/IRF3 innate immune pathway. Contrary to cGAMP production by cGAS, ectonucleotide pyrophosphatase phosphodiesterase 1 can hydrolyze cGAMP to maintain homeostasis of the cGAMP reservoir and prevent IFN overactivation during PRV infection [8]. PRV can also evade IFN-mediated innate immune response to facilitate virus infection [9], suggesting that interaction of PRV and its host is complicated and obscure.
Upon activation by the innate immune pathways, IFNs bind to cell surface receptors and initiate a signaling cascade through the Janus kinase signal transducer and activator of transcription (JAK–STAT) pathway, leading to the transcriptional regulation of hundreds of IFN-stimulated genes (ISGs) that are highly effective at resisting and controlling pathogens [10], [11]. We previously found that porcine cholesterol 25-hydroxylase (CH25H) is an ISG induced by either IFN or PRV infection [12]. CH25H catalyzes 25-hydroxycholesterol production to inhibit PRV entry. In addition, IFN-inducible transmembrane proteins (IFITMs) are a group of antiviral restriction factors that act against a broad range of viruses [13], such as influenza A virus [14], human immunodeficiency virus (HIV)-1 [15], West Nile virus and dengue virus [16], vesicular stomatitis virus [17], severe acute respiratory syndrome coronavirus and Marburg virus [18], classical swine fever virus [19] and African swine fever virus [20]. The mechanisms by which IFITMs influence virus infection are restriction of viral entry, viral assembly or viral protein synthesis [21]. For example, IFITM1, IFITM2 and IFITM3 inhibit HIV-1 replication partially through interfering with virus entry [15]. However, which IFITM member is involved in PRV infection and how IFITMs influence PRV replication have not been documented.
Here, we examine the effect of porcine IFITMs (pIFITMs) on PRV infection. Our data indicated that PRV-induced pIFITM1, pIFITM2 and pIFITM3 expression was different in PK15 porcine kidney epithelial cells and 3D4/21 alveolar macrophages. We further identified that pIFITM1, but not pIFITM2 or pIFITM3, was responsible for PRV infection, and that it interfered with PRV infection predominantly by inhibition of viral entry. This study uncovers the function of pIFITM1 during PRV infection.
2. Materials and methods
2.1. Cells, virus and tissues
PK15, 3D4/21, Vero and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco) at 37 °C in 5% CO2. PRV-QXX (virulent strain) and PRV-GFP (recombinant strain) were used as previously described [8].
2.2. Establishment of pIFITM1-Flag stable cell line
The coding sequence of pIFITM1 with Flag tag fused to the COOH terminus was amplified from the cDNA of PK15 cells with the primers shown in Table 1 . The PCR product was cloned into pLVX-IRES-Puro lentiviral vector (Clontech, Mountain View, CA, USA) to generate pLVX-pIFITM1-Flag. On day 0, human HEK293T cells were seeded in 10-cm dishes at 4 × 106 per dish. On day 1, cells were transfected with 2 μg/dish pLVX-pIFITM1-Flag, 1.5 μg/dish psPAX2 (packaging plasmid), and 0.5 μg/dish pMD2.G (envelope plasmid) using Lipofectamine 3000 (Invitrogen, Grand Island, NY, USA). On day 3, the culture media that contained lentiviruses were collected and used to infect PK15 cells that were then selected with puromycin (4 μg/ml) for 7 days. Expression of pIFITM1-Flag was determined by immunoblotting analysis.
Table 1.
Primers used for gene cloning and RT-qPCR analysis.
| Name | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| Sus IFITM1 | GACGATAAGACCGGTTCTAGAATGATGATCAAGAGCCAGCACGAGA | 375 |
| GAAGTTTGTTGCGCCGGATCCGTAGCCTCTGTTACTCTTTGC | ||
| Q-Sus β-actin | CTGAACCCCAAAGCCAACCGT | 317 |
| TTCTCCTTGATGTCCCGCACG | ||
| Q-Sus IFITM1 | ATTTGTTCCACCCTCCCAGG | 146 |
| TTGATGCAGAGACGGAGCAG | ||
| Q-Sus IFITM2 | CATTCTGACCATCGGAGCCA | 75 |
| TTTGCGCGCTCTAACATCTG | ||
| Q-Sus IFITM3 | TGCGTTCATCATCGTTTGCAC | 64 |
| TATGAGCTGCAGAACTGCTTGG | ||
| Q-Sus IFN-β | CTCTAGCACTGGCTGGAATGAA | 237 |
| CCGGAGGTAATCTGTAAGTCTGTT | ||
| Q-Sus ISG15 | ATGCCCCCTTGCCCTCTCCAGTG | 235 |
| TCCGATGCCATCATGCAGTCCCT | ||
| Q-Sus IL-1β | CCTGTACCCCAACTGGTA | 59 |
| CCAGGAAGACGGGCTTTTG | ||
| Q-PRV gB | GGCATCGCCAACTTCTTCC | 289 |
| CCTCGTCCACGTCGTCCTC | ||
| Q-PRV TK | GGCGTACTGGCGCACTCTG | 279 |
| ATGTCCCCGACGATGAAGC | ||
| Q-PRV gH | CTCGCCATCGTCAGCAA | 187 |
| GCTGCTCCTCCATGTCCTT | ||
2.3. RT-qPCR
Total RNA was isolated by using Trizol Reagent (TaKaRa, Shiga, Japan) and subjected to cDNA synthesis with the PrimeScript™ RT Reagent Kit (TaKaRa). RT-qPCR was performed in triplicate by using SYBR Premix Ex Taq (TaKaRa), and data were normalized by the level of β-actin expression in each individual sample. Melting curve analysis indicated formation of a single product in all cases. The 2−ΔΔCt method was used to calculate relative expression changes. For quantification of PRV genome copy number, PCR product of 187 bp from the gene of PRV glycoprotein H (gH) was cloned into pGEM-T vector. Serial 10-fold dilutions of this plasmid were used to construct a standard curve. The total number of PRV genomic equivalents was determined by comparison with the standard curve. Primers used for RT-qPCR are presented in Table 1.
2.4. Immunoblotting analysis
Whole-cell lysates were extracted with lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mM MgCl2) supplemented with protease and phosphatase inhibitors (Roche, Mannheim, Germany). The protein concentrations in the lysates were quantified with a BCA Protein Assay Kit (DingGuo, Beijing, China), detected with a microplate reader (Awareness Technology Inc., Palm City, FL, USA). Protein samples (50 μg) were separated by SDS-PAGE, transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA), and incubated in 5% nonfat milk (Sangon, Shanghai, China) for 1 h at room temperature. The membranes were incubated with primary antibody overnight at 4 °C and then with a horseradish-peroxidase-conjugated, donkey anti-mouse IgG antibody (diluted 1:5000; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 1 h at room temperature. Primary antibodies used were anti-Flag M2 mouse monoclonal antibody (1:500, Sigma, St. Louis, MO, USA) and anti-actin mouse monoclonal antibody (1:10000, Sigma). Antisera against PRV gE (1:500) was generated by immunizing mice with purified recombinant gE. Immunoblotting results were visualized using Luminata Crescendo Western HRP Substrate (Millipore) on GE AI600 imaging system (Boston, MA, USA).
2.5. Immunofluorescence assay
Cells grown on glass coverslips (Thermo, Waltham, MA, USA) were incubated with PRV-QXX for 1 h at 4 °C and fixed with 4% paraformaldehyde for 20 min. After washing with phosphate-buffered saline (PBS), PRV glycoprotein E (gE) was detected by incubating cells in PBS/10% FBS containing antisera against PRV gE (1:500) for 1 h. After washing with PBS, the cells were further incubated with fluorescent secondary antibodies (1:500; Invitrogen) in PBS/10% FBS for 1 h. Immunofluorescence microscopy was conducted using a Zeiss LSM 800 microscope (Oberkochen, Germany).
2.6. Cell proliferation assay
Cells were seeded at 104 per well in 96-well plates and viability was determined using CCK-8 cell counting assay (DingGuo).
2.7. Fluorescence-activated cell sorting (FACS)
Cells were infected with PRV-GFP (MOI 0.01) for 36 h and digested with trypsin–EDTA (Gibco). Cells were collected by centrifugation at 1000 g for 5 min and suspended in PBS. The percentage of GFP-positive cells was measured by flow cytometry on CytoFLEX (Beckman, Atlanta, GA, USA). Data were analyzed using FlowJo software.
2.8. RNA interference (RNAi)
On day 0, human HEK293T cells were seeded in 10-cm dishes at 4 × 106 per dish. On day 1, the cells were transfected with 2 μg/dish shRNA plasmid (Table 2 ), 1.5 μg/dish psPAX2 (packaging plasmid), and 0.5 μg/dish pMD2.G (envelope plasmid) using Lipofectamine 2000. On day 3, the culture media that contained the viruses were collected, filtered through a 0.45-μm membrane, and stored at −80 °C. For cell infection, PK15 or 3D4/21 cells were cultured in T25 flasks. When the cells had reached 70–80% confluence, they were infected with 1 ml lentivirus-containing medium mixed with 2 ml fresh medium. After 16 h, the medium was changed and the cells were cultured for a further 48 h. The cells were selected in culture medium containing puromycin (4 μg/ml) for 7 days.
Table 2.
Sequences of shRNA used for gene knockdown.
| Name | Sequence (5′-3′) |
|---|---|
| sh-Sus IFITM1-1 | GCCACTGTTCTTCTGGTGTTT |
| sh-Sus IFITM1-2 | GCAAAGAGTAACAGAGGCTAC |
| sh-Sus IFITM2-1 | GATGTTAGAGCGCGCAAAGAG |
| sh-Sus IFITM2-2 | GCGCAAAGAGTAACAGAGGCT |
| sh-Sus IFITM2/3 | GGAAGATGGTGGGAGACATCA |
2.9. Virus titration and infection
The tissue culture infective dose assay (TCID50) was performed to assess virus titration and infectivity. On day 0, Vero cells were seeded in a 96-well plate at 104 per well. On day 1, the cells were inoculated with serially diluted viruses (10−1–10−12 fold) for 1 h at 37 °C. The excess virus inoculum was removed by washing with PBS. Then, 200 μl maintenance medium (DMEM/2% FBS) was added to each well and the cells were cultured for 3–5 days. The cells demonstrating the expected cytopathic effect were observed daily and the TCID50 value was calculated by the Reed–Muench method.
2.10. Generation of gene knockout cell lines via CRISPR/Cas9
Small guide RNAs (sgRNAs) targeting porcine cGAS, STING, TBK1, IRF3 and IFNAR1 (Table 3 ) were synthesized and cloned into the lentiCRISPR v2 vector Addgene, Watertown, MA, USA). Lentiviral production was the same as indicated in the method of RNA interference. PK15 cells were infected with lentiviruses and then selected with puromycin (4 μg/ml) for 7 days. Single clonal knockout cells were obtained by serial dilution and verified by Sanger sequencing.
Table 3.
sgRNA and primers used for DNA sequencing.
| Name | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| cGAS sgRNA | CACCGGAAGCCGCAGGTACGCACG | 24 |
| AAACCGTGCGTACCTGCGGCTTCC | ||
| STING sgRNA | CACCGGAATACACGCTCCGGTGGC | 24 |
| AAACGCCACCGGAGCGTGTATTCC | ||
| TBK1 sgRNA | CACCGCTCGTAGACTTTGAGGCGG | 24 |
| AAACCCGCCTCAAAGTCTACGAGC | ||
| IRF3 sgRNA | CACCGCCCTTGGAAGCACGGCTTG | 24 |
| AAACCAAGCCGTGCTTCCAAGGGC | ||
| IFNAR1 sgRNA | CACCGCTGGTCGCTGGGGCTCCGT | 24 |
| AAACACGGAGCCCCAGCGACCAGC | ||
| cGAS sequencing | TGCGAGCCCTACTGCTG | 709 |
| CTTCACTCGCTCATAGTAGCTC | ||
| STING sequencing | GAGTGTCCGGTGGGTGGT | 720 |
| AGCCCTCCAGTAGCTGCTC | ||
| TBK1 sequencing | CTGAGGAGTGAGTCTAAGCGG | 794 |
| GACCCAAGATCCAAAGCAG | ||
| IRF3 sequencing | GCCCATGGGAACTCAGAA | 595 |
| AAATCCCCCTTACCTCCAC | ||
| IFNAR1 sequencing | ATGAGCGTGGGGCGGGG | 515 |
| CTGGCCGAGGTCTCCCATG | ||
2.11. Statistical analysis
All data were analyzed using the Prism 7 software (GradphPad Software, La Jolla, CA, USA). All data were analyzed with two-tailed Student’s t-test. P < 0.05 was considered statistically significant.
3. Results
3.1. Phylogenetic tree and multiple sequence alignment of IFITMs
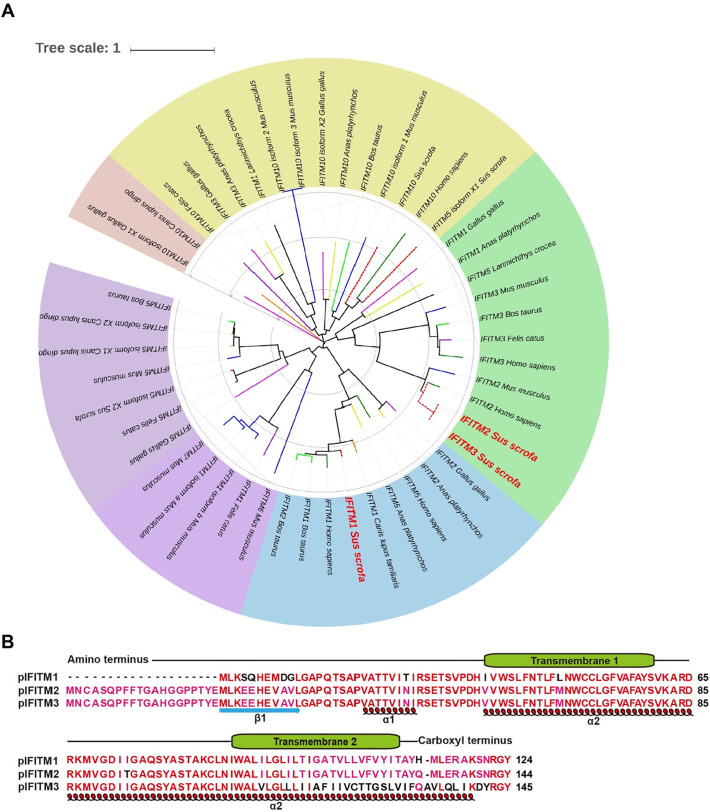
To determine the role of IFITMs in PRV infection, we first analyzed the phylogenetic relationships of IFITMs in different species. We constructed the phylogenetic tree of IFITMs based on amino acid sequences with MEGA 6 software. We analyzed 47 IFITMs from different species. The phylogenetic analysis indicated that pIFITM1 was clustered with that from humans, cows and dogs, and pIFITM2, pIFITM3 and IFITM2 from humans and mice clustered together (Fig. 1 A), suggesting that pIFITM1–3 had an evolutionarily conserved relationship with that from humans.
Fig. 1.
Phylogenetic tree and multiple sequence alignment of IFITM1–3. A Phylogenetic tree of IFITMs from different species was constructed with MEGA 6 software, using the neighbor-joining method. B Multiple sequence alignment and secondary structure prediction of pIFITM1–3. The transmembrane domains of pIFITM1–3 are shown above the amino acid sequence. The α-helices and β-sheets of pIFITM1–3 are shown under the amino acid sequence.
pIFITM1 contained 124 amino acids, whereas pIFITM2 and pIFITM3 harbored 144 and 145 amino acids respectively. pIFITM1 shared 77.08% and 61.38% amino acid identity with pIFITM2 and pIFITM3, and pIFITM2 shared 82.76% amino acid identity with pIFITM3. In addition, we predicted the secondary structure of pIFITM1–3 using Protein Homology/analogY Recognition Engine version 2.0. pIFITM1–3 were composed of 2 transmembrane domains, 2 α-helices and 1 β-strand (Fig. 1B).
3.2. Transcription of pIFITM1, pIFITM2 and pIFITM3 in PRV-infected cells
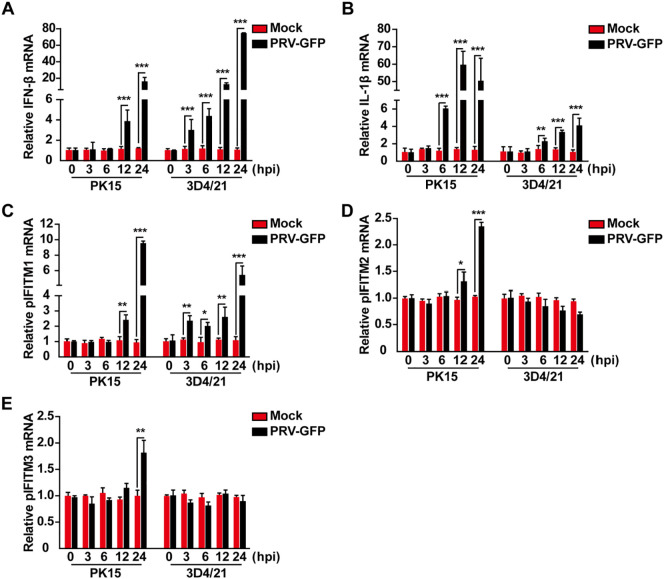
We then infected PK15 porcine kidney epithelial cells and 3D4/21 alveolar macrophages with PRV-GFP to detect whether mRNA expression of pIFITM1, pIFITM2 and pIFITM3 was induced. We examined the effect of PRV infection on IFN-β and interleukin 1β (IL-1β) mRNA expression. PK15 and 3D4/21 cells were infected with PRV-GFP (MOI 0.1) for 0, 3, 6, 12 or 24 h. Total RNA was isolated and reverse transcribed to cDNA for RT-qPCR analysis. PRV infection resulted in activation of IFN-β and IL-1β mRNA expression in PK15 and 3D4/21 cells in a time-dependent manner (Fig. 2 A and 2B), which is consistent with our previous findings that PRV can activate IFN and proinflammatory cytokine expression [7]. We next detected if PRV infection induced IFITM expression. RT-qPCR indicated that PRV-GFP infection promoted pIFITM1 expression in PK15 and 3D4/21 cells (Fig. 2C). However, pIFITM2 and pIFITM3 expression was only enhanced in PK15 cells, but not in 3D4/21 cells during PRV-GFP infection (Fig. 2D and 2E). These results suggest that PRV-induced IFITM expression differs among cell types.
Fig. 2.
Transcription of pIFITM1–3 in response to PRV-GFP infection. A–E RT-qPCR analysis of IFN-β (B), IL-1β (C), pIFITM1 (D), pIFITM2 (E) and pIFITM3 (F) mRNA expression in mock-infected or PRV-GFP (MOI 0.1) infected PK15 and 3D4/21 cells at 0, 3, 6, 12 or 24 h post-infection. All data are shown as mean ± SD based on three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test.
3.3. pIFITM1 knockdown enhances PRV-GFP infection
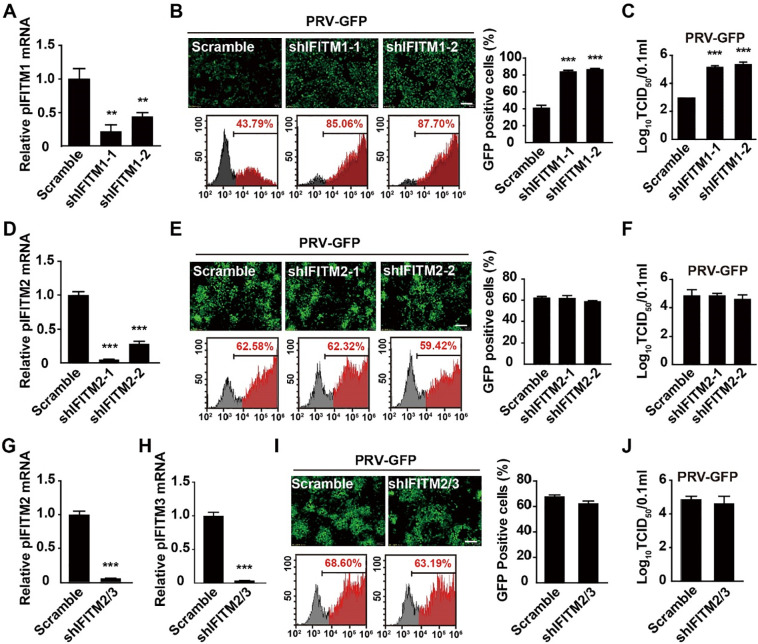
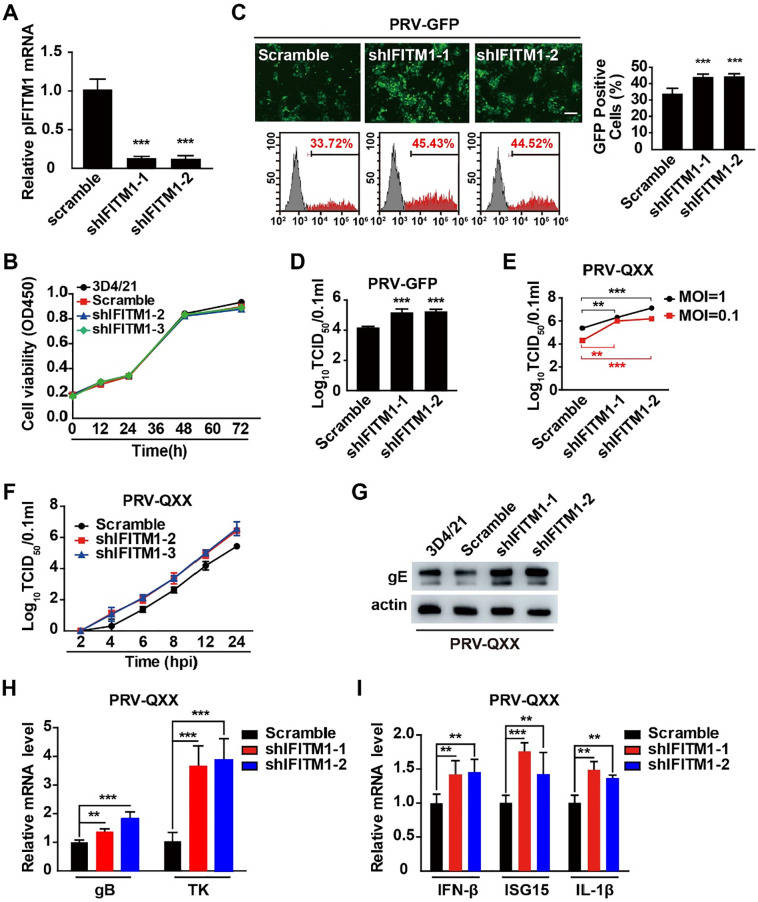
We investigated whether pIFITM1–3 were involved in PRV infection. Lentivirus-delivered shRNAs targeting IFITM mRNA were used to stably knockdown IFITMs in PK15 cells. Two independent shRNAs specifically targeting pIFITM1 (shIFITM1-1 and shIFITM1-2) showed significant knockdown efficiency, proven by RT-qPCR (Fig. 3 A). We infected Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells with PRV-GFP and performed fluorescent microscopy and viral titration analysis that showed that knockdown of pIFITM1 promoted PRV-GFP infection (Fig. 3B and 3C). We also established PK15 cells expressing low level of pIFITM2 (shIFITM2-1 and shIFITM2-2) (Fig. 3D). PRV-GFP replication, detected by fluorescent microscopy or viral titration, was not altered in shIFITM2-1 and shIFITM2-2 compared to that in Scramble PK15 cells, suggesting that pIFITM2 was not involved in PRV infection (Fig. 3E and 3F).
Fig. 3.
pIFITM1 knockdown enhances PRV-GFP infection. A RT-qPCR analysis of pIFITM1 mRNA in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells. B Fluorescent microscopy and FACS analysis of PRV-GFP (MOI 0.01) proliferation in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells for 36 h. Scale bar, 400 μm. C TCID50 assay in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells infected with PRV-GFP (MOI 0.1) for 24 h. D RT-qPCR analysis of pIFITM2 mRNA in Scramble, shIFITM2-1 and shIFITM2-2 PK15 cells. E Fluorescent microscopy and FACS analysis of PRV-GFP (MOI 0.01) proliferation in Scramble, shIFITM2-1 and shIFITM2-2 PK15 cells for 36 h. Scale bar, 400 μm. F TCID50 assay in Scramble, shIFITM2-1 and shIFITM2-2 PK15 cells infected with PRV-GFP (MOI 0.01) for 24 h. G, H RT-qPCR analysis of pIFITM2 and pIFITM3 mRNA in Scramble, shIFITM2/3 PK15 cells. I Fluorescent microscopy and FACS analysis of PRV-GFP (MOI 0.01) proliferation in Scramble and shIFITM2/3 PK15 cells for 36 h. Scale bar, 400 μm. J TCID50 assay in Scramble and shIFITM2/3 PK15 cells infected with PRV-GFP (MOI 0.01) for 24 h. All the data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by two-tailed Student’s t-test.
The nucleic acid sequences of pIFITM2 and pIFITM3 are identical; therefore, we could not design a shRNA specifically targeting pIFITM3. To circumvent this problem, we simultaneously knocked down pIFITM2 and pIFITM3 in PK15 cells (shIFITM2/3). RT-qPCR indicated that mRNA expression of pIFITM2 and pIFITM3 was significantly down-regulated in shIFITM2/3 PK15 cells (Fig. 3G and 3H). Fluorescent microscopy and viral titration showed that PRV-GFP replication in shIFITM2/3 PK15 cells was almost the same as that in control cells (Fig. 3I and 3J). All the results above demonstrate that pIFITM1, but not pIFITM2 and pIFITM3, participates in PRV infection.
3.4. PRV-induced pIFITM1 expression is dependent on cGAS, STING, TBK1, IRF3 and IFNAR1
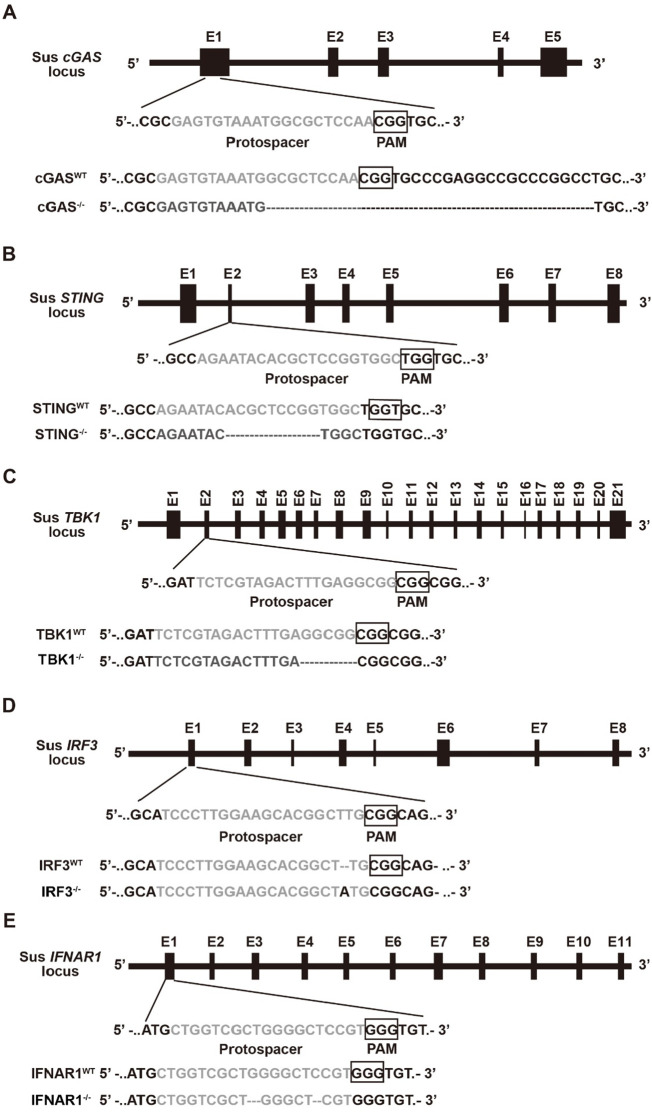
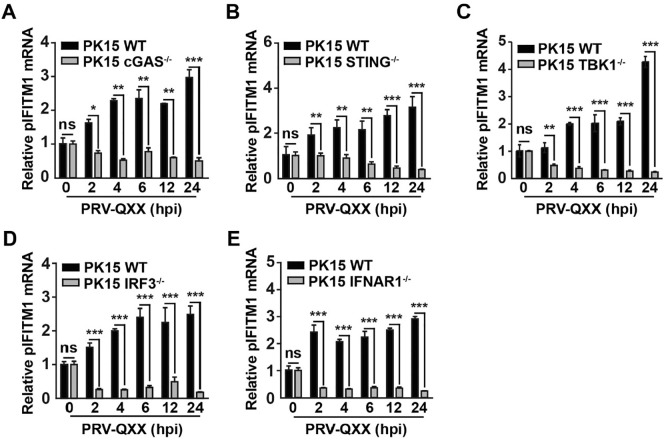
We showed that knockdown of IFITM1 promoted PRV-GFP infection; therefore, we investigated the role of IFITM1 in PRV infection. IFITM1 is regulated by IFN signaling [22] and our previous study indicated that PRV infection induces cGAS/STING/TBK1/IRF3-dependent IFN expression [7]. Thus, we attempted to identify whether PRV-induced IFITM1 expression relied on this innate immune pathway. CRISPR/Cas9 was used to generate cGAS−/−, STING−/−, TBK1−/−, IRF3−/− and IFNAR1−/− knockout PK15 cells (Fig. 4 ). We infected cells with the virulent PRV strain PRV-QXX for 0, 2, 4, 6, 12 or 24 h. Total RNA was isolated and reverse-transcribed to cDNA for RT-qPCR. PRV-QXX infection induced pIFITM1 expression in a time-dependent manner in PK15 cells (Fig. 5 A–5E). However, induction of pIFITM1 expression was abolished in cGAS−/−, STING−/−, TBK1−/−, IRF3−/− or IFNAR1−/− PK15 cells (Fig. 5A–5E). These results indicate that PRV-induced pIFITM1 expression relies on cGAS/STING/TBK1/IRF3 innate immune and IFN signaling pathways.
Fig. 4.
Schematic showing the design of sgRNAs that target porcine cGAS, STING, TBK1, IRF3 and IFNAR1. A–E Schematic representation of sus cGAS, STING, TBK1, IRF3 and IFNAR1 genomic structure and DNA sequencing results of cGAS−/− (A), STING−/− (B), TBK1−/− (C), IRF3−/− (D) and IFNAR1−/− (E) PK15 cells. Protospacer sequences are grey. The PAM sequence is framed by a black box.
Fig. 5.
PRV-induced pIFITM1 expression is dependent on cGAS, STING, TBK1, IRF3 and IFNAR1. A–E RT-qPCR analysis of pIFITM1 mRNA in WT, cGAS−/− (A), STING−/− (B), TBK1−/− (C), IRF3−/− (D) and IFNAR1−/− (E) PK15 cells infected with PRV-QXX (MOI = 1) at 0, 2, 4, 6, 12 or 24 h post-infection. Data are shown as mean ± SD based on three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test. ns, no significance.
3.5. pIFITM1 knockdown enhances PRV-QXX infection in PK15 cells
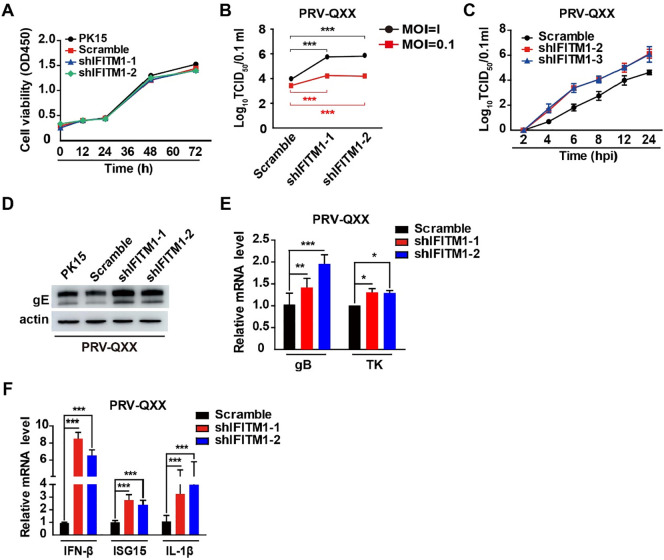
To further validate the role of IFITM1 in PRV infection, we detected PRV-QXX replication in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells. We performed a CCK-8 cell counting assay to identify whether pIFITM1 knockdown influenced cell proliferation. The proliferation of shIFITM1-1 and shIFITM1-2 cells was almost the same as PK15 and Scramble cells (Fig. 6 A). We infected cells with PRV-QXX and performed a viral titration assay. In accordance with PRV-GFP, pIFITM1 knockdown enhanced PRV-QXX replication (Fig. 6B). To gain insight into the effect of pIFITM1 on PRV infection, we detected the growth kinetics of PRV-QXX in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells using a one-step growth curve. As indicated in Fig. 6C, viral yield was higher in shIFITM1-1 and shIFITM1-2 than in Scramble PK15 cells from 2 to 24 h post-infection. We examined PRV-QXX replication by immunoblotting of PRV gE, RT-qPCR of transcription of PRV glycoprotein B (gB), thymidine kinase (TK), and RT-qPCR of PRV-induced IFN-β, ISG15 and IL-1β expression in PK15 cells with pIFITM1 knockdown. PRV gE expression was higher in shIFITM1-1 and shIFITM1-2 than in PK15 and Scramble cells (Fig. 6D). Transcription of PRV gB and TK was enhanced in cells due to pIFITM1 knockdown (Fig. 6E). PRV-induced IFN-β, ISG15 and IL-1β expression was also increased in PK15 cells with pIFITM1 knockdown (Fig. 6F). These data demonstrate that knockdown of pIFITM1 promoted PRV-QXX infection in PK15 cells.
Fig. 6.
pIFITM1 knockdown enhances PRV-QXX infection in PK15 cells. A Cell proliferation assay of PK15, Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells for 0–72 h. B TCID50 assay in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells infected with PRV-QXX (MOI 0.1 or 1) for 24 h. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by two-tailed Student’s t-test. C One-step growth curve of PRV-QXX (MOI 0.1) in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells at 2, 4, 6, 8, 12 or 24 h post-infection. Viral titer was determined by TCID50 assay. D Immunoblotting analysis of whole cell extracts from PK15, Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells infected with PRV-QXX (MOI 0.1) for 24 h with antibodies against PRV gE. Actin served as a loading control. E RT-qPCR analysis of PRV gB and TK mRNA in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells infected with PRV-QXX (MOI 0.1) for 24 h. Data are shown as mean ± SD based on three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 determined by two-tailed Student’s t-test. F RT-qPCR analysis of IFN-β, ISG15 and IL-1β mRNA expression in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells treated as in E. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test.
3.6. pIFITM1 knockdown enhances PRV infection in 3D4/21 cells
To determine whether the role of pIFITM1 in PRV infection was limited to a specific cell type, we further depleted pIFITM1 by RNA interference in 3D4/21 cells infected with PRV-GFP and PRV-QXX. shIFITM1-1 and shIFITM1-2 showed significant knockdown efficiency of pIFITM1 in 3D4/21 cells, as demonstrated by RT-qPCR (Fig. 7 A). A CCK-8 cell counting assay showed that proliferation of 3D4/21 cells was not altered by pIFITM1 knockdown (Fig. 7B). Subsequently, we infected Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells with PRV-GFP and examined viral replication by fluorescent microscopy, FACS and viral titer assay. The percentage of GFP-positive cells was 33.72% in Scramble, and this rate significantly increased to 45.43% and 44.52% in shIFITM1-1 and shIFITM1-2 3D4/21 cells infected with PRV-GFP, respectively (Fig. 7C). Viral titer assay showed that PRV-GFP infection was enhanced in shIFITM1-1 and shIFITM1-2 compared with in Scramble 3D4/21 cells (Fig. 7D). We infected cells with PRV-QXX and found that viral titer was also higher in shIFITM1-1 and shIFITM1-2 than that in Scramble 3D4/21 cells (Fig. 7E). Additionally, we tested the growth kinetics of PRV-QXX in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells by one-step growth curve. Viral titration was higher in shIFITM1-1 and shIFITM1-2 than in Scramble 3D4/21 cells from 2 to 24 h post-infection, which was consistent with that in PK15 cells (Fig. 7F). Moreover, pIFITM1 knockdown enhanced PRV gE expression, PRV gB and TK transcription and PRV-induced IFN-β, ISG15 and IL-1β expression (Fig. 7G–7I). All the data illustrate that knockdown of pIFITM1, either in PK15 or in 3D4/21 cells, promotes PRV infection.
Fig. 7.
pIFITM1 knockdown enhances PRV infection in 3D4/21 cells. A RT-qPCR analysis of pIFITM1 mRNA in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. B Cell proliferation assay of 3D4/21, Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells for 0–72 h. C Fluorescent microscopy and FACS analysis of PRV-GFP (MOI 0.01) proliferation in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells for 36 h. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. D TCID50 assay in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells infected with PRV-GFP (MOI 0.1) for 24 h. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by two-tailed Student’s t-test. E TCID50 assay in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells infected with PRV-QXX (MOI 0.1 or 1) for 24 h. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test. F One-step growth curve of PRV-QXX (MOI 0.1) in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells at 2, 4, 6, 8, 12 or 24 h post-infection. Viral titer was determined by TCID50 assay. G Immunoblotting analysis of whole cell extracts from 3D4/21, Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells infected with PRV-QXX (MOI 0.1) for 24 h with antibodies against PRV gE. Actin served as a loading control. H RT-qPCR analysis of PRV gB and TK mRNA in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells infected with PRV-QXX (MOI 0.1) for 24 h. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test. I RT-qPCR analysis of IFN-β, ISG15 and IL-1β mRNA expression in Scramble, shIFITM1-1 and shIFITM1-2 3D4/21 cells treated as in H. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test.
3.7. Overexpression of pIFITM1 inhibits PRV infection
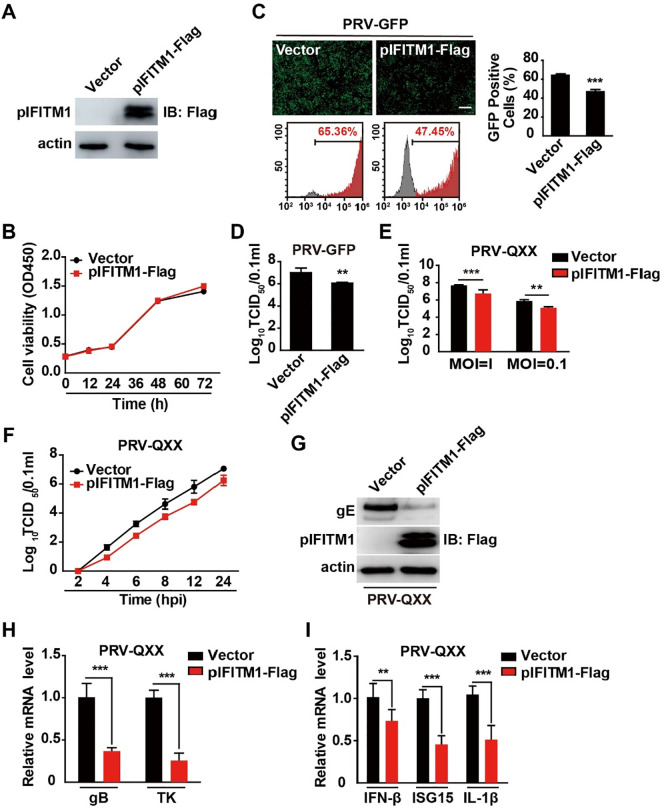
To confirm the role of pIFITM1 in PRV infection, we used lentivirus-mediated delivery to generate PK15 cells overexpressing pIFITM1-Flag (Fig. 8 A). Cell proliferation by CCK-8 cell counting assay indicated that ectopic expression of pIFITM1 did not inhibit PK15 cell proliferation (Fig. 8B). We infected control (vector) and pIFITM1-overexpressing cells with PRV-GFP and fluorescent microscopy and FACS assay showed that GFP intensity was lower in pIFITM1-overexpressing cells than in control cells, suggesting that pIFITM1 overexpression inhibited PRV-GFP infection (Fig. 8C). The viral titer assay indicated that overexpression of pIFITM1 reduced viral production (Fig. 8D). In addition to inhibiting PRV-GFP infection, pIFITM1 overexpression also prevented viral infection in PK15 cells infected with PRV-QXX (Fig. 8E). We then detected the growth kinetics of PRV-QXX in control and pIFITM1-Flag-overexpressing PK15 cells by one-step growth curve. Viral titration was lower in pIFITM1-overexpressing cells than in control cells from 2 to 24 h post-infection (Fig. 8F).
Fig. 8.
Overexpression of pIFITM1 inhibits PRV infection. A Immunoblotting analysis of whole cell extracts from PK15 cells stably expressing vector or pIFITM1-Flag with antibodies against Flag. Actin served as a loading control. B Cell proliferation assay of PK15 cells stably expressing vector or pIFITM1-Flag for 0–72 h. C Fluorescent microscopy and FACS analysis of PRV-GFP (MOI 0.01) proliferation in PK15 cells stably expressing vector or pIFITM1-Flag for 36 h. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. D TCID50 assay in PK15 cells stably expressing vector or pIFITM1-Flag infected with PRV-GFP (MOI 0.1) for 24 h. Data are shown as mean ± SD based on three independent experiments. **P < 0.01 determined by two-tailed Student’s t-test. E TCID50 assay in PK15 cells stably expressing vector or pIFITM1-Flag infected with PRV-QXX (MOI 0.1 or 1) for 24 h. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test. F One-step growth curve of PRV-QXX (MOI 0.1) in K15 cells stably expressing vector or pIFITM1-Flag at 2, 4, 6, 8, 12 or 24 h post-infection. Viral titer was determined by TCID50 assay. G Immunoblotting analysis of whole cell extracts from PK15 cells stably expressing vector or pIFITM1-Flag infected with PRV-QXX (MOI 0.1) for 24 h with antibodies against PRV gE and Flag. Actin served as a loading control. H RT-qPCR analysis of PRV gB and TK mRNA expression in K15 cells stably expressing vector or pIFITM1-Flag infected with PRV-QXX (MOI 0.1) for 24 h. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. I RT-qPCR analysis of IFN-β, ISG15 and IL-1β mRNA expression in PK15 cells stably expressing vector or pIFITM1-Flag treated as in H. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test.
Meanwhile, we examined the effect of pIFITM1 overexpression on PRV-QXX gE expression by immunoblotting analysis. As shown in Fig. 8G, gE expression was decreased due to pIFITM1-Flag expression. PK15 cells expressing pIFITM1-Flag generated less PRV gB and TK mRNA than control cells did (Fig. 8H), and PRV-QXX-induced IFN-β, ISG15 and IL-1β expression was decreased in response to pIFITM1 overexpression (Fig. 8I). These results demonstrate that overexpression of pIFITM influences PRV infection.
3.8. pIFITM1 restricts PRV entry
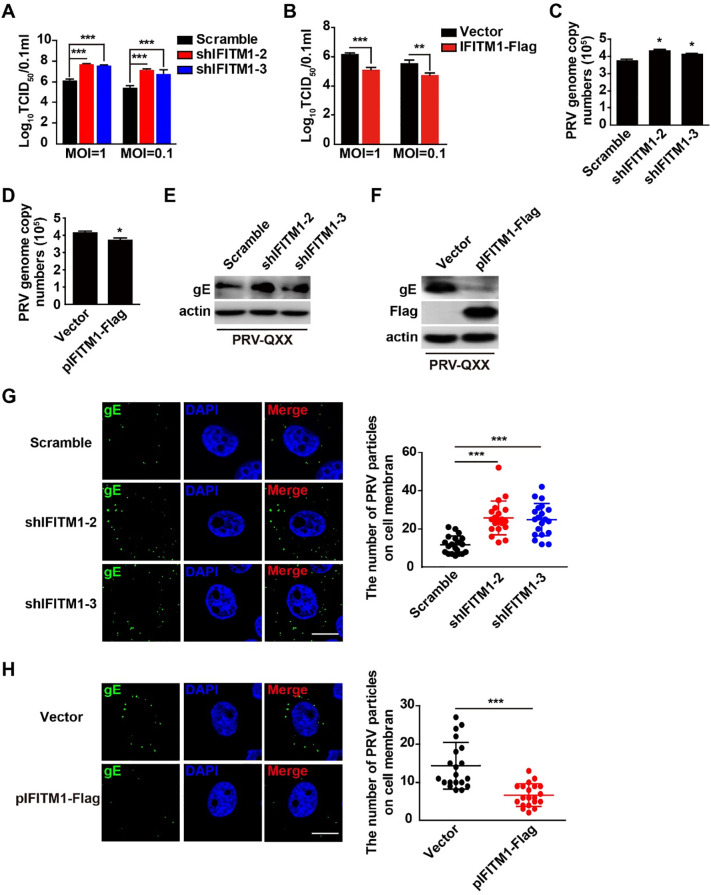
We investigated whether pIFITM1 inhibited PRV entry. Cells were incubated with PRV-QXX for 1 h at 4 °C and then washed twice with ice-cold PBS, and viral titer was determined at 24 h post-infection. Knockdown of pIFITM1 accelerated PRV replication and overexpression of pIFITM1 dampened PRV infection (Fig. 9 A and 9B). We incubated cells with PRV-QXX for 1 h at 4 °C and then washed twice with ice-cold PBS, and examined PRV entry by RT-qPCR, immunoblotting and immunofluorescence analysis. We found more PRV genome copy numbers in shIFITM1-1 and shIFITM1-2 PK15 cells than in Scramble cells (Fig. 9C). On the contrary, PRV genome copy numbers were decreased on cells overexpressing pIFITM1 (Fig. 9D). Immunoblotting analysis showed that more PRV gE was detected on cells with pIFITM1 knockdown and less PRV gE was detected on pIFITM1-PK15 cells (Fig. 9E and 9F). Immunofluorescence analysis indicated that knockdown of pIFITM1 promoted PRV attachment to the cell membrane, and overexpression of pIFITM1 impaired PRV adherence to the cell membrane (Fig. 9G and 9H). All the results above indicate that pIFITM1 inhibits PRV entry during PRV infection.
Fig. 9.
pIFITM1 restricts PRV entry. A TCID50 assay in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells incubated with PRV-QXX (MOI 0.1 or 1) for 1 h at 4 °C. Cells were washed twice with ice-cold PBS and cultured for 24 h at 37 °C. Data are shown as mean ± SD based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. B TCID50 assay of control (Vector) and pIFITM1-Flag PK15 cells treated as in A. Data are shown as mean ± SD based on three independent experiments. **P < 0.01, ***P < 0.001 determined by a two-tailed Student’s t-test. C RT-qPCR analysis of PRV genome copy numbers in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells incubated with PRV-QXX (MOI 1) for 1 h at 4 °C and washed twice with ice-cold PBS. Data are shown as mean ± SD based on three independent experiments. *P < 0.05 determined by a two-tailed Student’s t-test. D RT-qPCR analysis of PRV genome copy numbers from control (Vector) and pIFITM1-Flag PK15 cells treated as in C. Data are shown as mean ± SD based on three independent experiments. *P < 0.05 determined by two-tailed Student’s t-test. E Immunoblotting analysis of whole cell extracts from Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells treated as in C. F Immunoblotting analysis of whole cell extracts from control (Vector) and pIFITM1-Flag PK15 cells treated as in C. G Immunofluorescence of PRV gE in Scramble, shIFITM1-1 and shIFITM1-2 PK15 cells treated as in C. Data are shown as mean ± SD of n = 22 cells based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test. H Immunofluorescence of PRV gE in control (Vector) and pIFITM1-Flag PK15 cells treated as in C. Data are shown as mean ± SD of n = 20 cells based on three independent experiments. ***P < 0.001 determined by a two-tailed Student’s t-test.
4. Discussion
IFITMs are activated in response to IFN and represent a new class of cellular restriction factors that block the replication and pathogenesis of pathogens [23]. Our results showed that PRV infection activated IFN-β expression in PK15 porcine kidney epithelial cells and 3D4/21 alveolar macrophages. We demonstrate that pIFITM1 can be induced in PK15 and 3D4/21 cells, while pIFITM2 and pIFITM3 can be only induced in PK15 cells during PRV infection. The reason why PRV-induced pIFITM expression differs in PK15 and 3D4/21 cells needs further investigation. Lu et al. have examined the expression profile of IFITM in 32 rat tissues and show that IFITM1–3 were ubiquitously expressed [24]. IFITM1 and IFITM3 were also ubiquitously expressed in all the tissues examined in BALB/c mice. However, mice challenged with H9N2 avian influenza virus showed that the expression patterns of IFITM1 and IFITM3 differ among tissues [25]. Further elucidation of the expression profile of IFITMs in porcine tissues will help us to understand the roles of IFITMs in pig infectious diseases.
The detection of cytosolic DNA as a ‘stranger’ and a ‘danger’ signal by DNA sensors has emerged as a key event in the innate immune response to virus infection [26]. A couple of DNA sensors have been identified to participate in innate immune response. The endoplasmic-reticulum-resident adaptor protein STING integrates DNA sensor-transduced signaling and subsequently activates the expression of type I IFN and pro-inflammatory cytokines [27]. DAI was the first putative DNA sensor to be shown to bind DNA, and to be required for the response to viral DNA [28], [29]. Using an RNAi screen of 59 members of the DExD/H helicase family, Zhang and colleagues identified DDX41 as a putative DNA sensor [30]. RNA polymerase III, IFN-γ-inducible protein 16, the high mobility group box proteins, and cGAS all play roles in sensing cytosolic DNA [31], [32], [33], [34]. We showed that PRV-induced pIFITM1 expression relied on the cGAS/STING/TBK1/IRF3 innate immune and IFN signaling pathways. Except for the activation of IFN, cGAMP also has a noncanonical function in inflammasome activation in human and mouse cells [35]. This may arm cells in another beneficial way to fight against PRV infection. DAI and DDX41 have been reported to respond to PRV infection [5], [6]; thus, it is possible that they are involved in PRV-induced pIFITM1 expression. Other DNA sensors may also be responsible for pIFITM1 expression when cells are infected with PRV. The predominant DNA sensor that is important for PRV-induced innate immune response may be determined by its expression profile in different cells and tissues.
We showed that pIFITM1 restricted PRV entry into host cells. The mechanism of how pIFITM1 restricts PRV entry may be due to the subcellular localization of IFITM1. IFITM1 mostly concentrates on the lipid raft and caveola in the plasma membrane and interacts with some membrane proteins such as CD19 and CD81 [36], [37], [38]. IFITM2 and IFITM3 are mainly in the intracellular compartments and co-localize with Rab7, CD63 and lysosomal-associated membrane protein 1 [39]. The C-terminal region of IFITM1 that controls protein subcellular localization is important for modulating the antiviral function against HIV entry [40]. Further studies have illustrated that the C-terminal KRXX motif of IFITM1 that interacts with adaptor protein complex 3 is responsible for its antiviral activity against jaagsiekte sheep retrovirus and 10A1 amphotropic murine leukemia virus [41]. It is also reported that IFITM1 inhibits viral entry and this antiviral function is associated with its cell surface localization [42]. IFITM1 may reduce membrane fluidity and increase spontaneous positive curvature in the outer leaflet of membranes [43], which may influence membrane fusion between PRV envelope and plasma membrane. Another mechanism by which pIFITM1 restricts PRV entry may be clathrin-mediated phagocytosis, because many viruses invade cells through a clathrin-dependent pathway [44]. Treatment of ifitm1 −/− cells with IFN-β reduces the association of clathrin with membrane compartments, suggesting a molecular interaction between clathrin and IFITM1 [45]. Our multiple sequence alignment indicated that pIFITM1 lacks 20 amino acids in the NH2 terminus of pIFITM2 and pIFITM3. Jian et al. have shown that there is a sorting motif YEML, amino acids 20–23, for binding to the μ2 subunit of the AP-2 complex in IFITM3 [46]. Although pIFITM1 lacks this sorting motif, we speculate that there may be other sorting motifs that mediate clathrin-dependent endocytosis of IFITM1.
Acknowledgements
This work was supported by a grant for Ministry of Agriculture and Rural Affairs of China (2016ZX08006001-006), Outstanding Talents of Henan Agricultural University (30600773) and Advantage and Characteristic Discipline of Henan Province (203/18xk0102).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2019.10.162.
Contributor Information
Guo-Yu Yang, Email: haubiochem@163.com.
Bei-Bei Chu, Email: chubeibei_hau@hotmail.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Mettenleiter T.C. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis–state of the art, June 1999. Vet. Res. 2000;31(1):99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 2.Pomeranz L.E., Reynolds A.E., Hengartner C.J. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005;69(3):462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fonseca A.A., Jr., Camargos M.F., de Oliveira A.M., Ciacci-Zanella J.R., Patricio M.A., Braga A.C., Cunha E.S., D'Ambros R., Heinemann M.B., Leite R.C., dos Reis J.K. Molecular epidemiology of Brazilian pseudorabies viral isolates. Vet. Microbiol. 2010;141(3–4):238–245. doi: 10.1016/j.vetmic.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Sun Y., Luo Y., Wang C.H., Yuan J., Li N., Song K., Qiu H.J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016;183:119–124. doi: 10.1016/j.vetmic.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Xie L., Fang L., Wang D., Luo R., Cai K., Chen H., Xiao S. Molecular cloning and functional characterization of porcine DNA-dependent activator of IFN-regulatory factors (DAI) Dev. Comp. Immunol. 2010;34(3):293–299. doi: 10.1016/j.dci.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhu X., Wang D., Zhang H., Zhou Y., Luo R., Chen H., Xiao S., Fang L. Molecular cloning and functional characterization of porcine DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (DDX41) Dev. Comp. Immunol. 2014;47(2):191–196. doi: 10.1016/j.dci.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Chu B., Du L., Han Y., Zhang X., Fan S., Wang Y., Yang G. Molecular cloning and functional characterization of porcine cyclic GMP-AMP synthase. Mol. Immunol. 2015;65(2):436–445. doi: 10.1016/j.molimm.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Lu S.F., Wan B., Ming S.L., Li G.L., Su B.Q., Liu J.Y., Wei Y.S., Yang G.Y., Chu B.B. Maintenance of cyclic GMP-AMP homeostasis by ENPP1 is involved in pseudorabies virus infection. Mol. Immunol. 2018;95:56–63. doi: 10.1016/j.molimm.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Brukman A., Enquist L.W. Suppression of the interferon-mediated innate immune response by pseudorabies virus. J. Virol. 2006;80(13):6345–6356. doi: 10.1128/JVI.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark G.R., Darnell J.E., Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Zeng L., Zhang L., Guo Z.Z., Lu S.F., Ming S.L., Li G.L., Wan B., Tian K.G., Yang G.Y., Chu B.B. Cholesterol 25-hydroxylase acts as a host restriction factor on pseudorabies virus replication. J. Gen. Virol. 2017;98(6):1467–1476. doi: 10.1099/jgv.0.000797. [DOI] [PubMed] [Google Scholar]
- 13.Lewin A.R., Reid L.E., McMahon M., Stark G.R., Kerr I.M. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 1991;199(2):417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 14.Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J., Brass A.L. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J., Pan Q., Rong L., He W., Liu S.L., Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85(5):2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang D., Weidner J.M., Qing M., Pan X.B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.Y., Block T.M., Guo J.T. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 2010;84(16):8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weidner J.M., Jiang D., Pan X.B., Chang J., Block T.M., Guo J.T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010;84(24):12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C., Zheng H., Wang Y., Dong W., Liu Y., Zhang L., Zhang Y. Antiviral role of IFITM proteins in classical swine fever virus infection. Viruses. 2019;11(2) doi: 10.3390/v11020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz-Moreno R., Cuesta-Geijo M.A., Martinez-Romero C., Barrado-Gil L., Galindo I., Garcia-Sastre A., Alonso C. Antiviral role of IFITM proteins in african swine fever virus infection. PLoS ONE. 2016;11(4):e0154366. doi: 10.1371/journal.pone.0154366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao Y., Goraya M.U., Yuan X., Zhang B., Chiu S.H., Chen J.L. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front. Microbiol. 2019;10:1097. doi: 10.3389/fmicb.2019.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatano H., Kudo Y., Ogawa I., Tsunematsu T., Kikuchi A., Abiko Y., Takata T. IFN-induced transmembrane protein 1 promotes invasion at early stage of head and neck cancer progression. Clin. Cancer Res. 2008;14(19):6097–6105. doi: 10.1158/1078-0432.CCR-07-4761. [DOI] [PubMed] [Google Scholar]
- 23.Siegrist F., Ebeling M., Certa U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 2011;31(1):183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y., Zuo Q., Zhang Y., Wang Y., Li T., Han J. The expression profile of IFITM family gene in rats. Intractable Rare Dis. Res. 2017;6(4):274–280. doi: 10.5582/irdr.2017.01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu M., Qi W., Huang Z., Zhang K., Ye J., Liu R., Wang H., Ma Y., Liao M., Ning Z. Expression profile and histological distribution of IFITM1 and IFITM3 during H9N2 avian influenza virus infection in BALB/c mice. Med. Microbiol. Immunol. 2015;204(4):505–514. doi: 10.1007/s00430-014-0361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating S.E., Baran M., Bowie A.G. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32(12):574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Choi M.K., Ban T., Yanai H., Negishi H., Lu Y., Tamura T., Takaoka A., Nishikura K., Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. U S A. 2008;105(14):5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiu Y.H., Macmillan J.B., Chen Z.J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S., Fitzgerald K.A., Paludan S.R., Bowie A.G. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanai H., Ban T., Wang Z., Choi M.K., Kawamura T., Negishi H., Nakasato M., Lu Y., Hangai S., Koshiba R., Savitsky D., Ronfani L., Akira S., Bianchi M.E., Honda K., Tamura T., Kodama T., Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462(7269):99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 34.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson K.V., Junkins R.D., Kurkjian C.J., Holley-Guthrie E., Pendse A.A., El Morabiti R., Petrucelli A., Barber G.N., Benedict C.A., Ting J.P. A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med. 2017;214(12):3611–3626. doi: 10.1084/jem.20171749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith R.A., Young J., Weis J.J., Weis J.H. Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 2006;7(2):113–121. doi: 10.1038/sj.gene.6364278. [DOI] [PubMed] [Google Scholar]
- 37.Weston S., Czieso S., White I.J., Smith S.E., Kellam P., Marsh M. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS ONE. 2014;9(8):e104341. doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y., Yang G., Hu G. Binding of IFITM1 enhances the inhibiting effect of caveolin-1 on ERK activation. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41(6):488–494. doi: 10.1093/abbs/gmp034. [DOI] [PubMed] [Google Scholar]
- 39.Yount J.S., Karssemeijer R.A., Hang H.C. S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287(23):19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia R., Ding S., Pan Q., Liu S.L., Qiao W., Liang C. The C-terminal sequence of IFITM1 regulates its anti-HIV-1 activity. PLoS ONE. 2015;10(3):e0118794. doi: 10.1371/journal.pone.0118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li K., Jia R., Li M., Zheng Y.M., Miao C., Yao Y., Ji H.L., Geng Y., Qiao W., Albritton L.M., Liang C., Liu S.L. A sorting signal suppresses IFITM1 restriction of viral entry. J. Biol. Chem. 2015;290(7):4248–4259. doi: 10.1074/jbc.M114.630780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith S.E., Busse D.C., Binter S., Weston S., Diaz Soria C., Laksono B.M., Clare S., Van Nieuwkoop S., Van den Hoogen B.G., Clement M., Marsden M., Humphreys I.R., Marsh M., de Swart R.L., Wash R.S., Tregoning J.S., Kellam P. Interferon-induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019;93(6) doi: 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., Gratton E., Cohen F.S., Liu S.L. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9(1):e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercer J., Schelhaas M., Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 45.Wee Y.S., Roundy K.M., Weis J.J., Weis J.H. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 2012;18(6):834–845. doi: 10.1177/1753425912443392. [DOI] [PubMed] [Google Scholar]
- 46.Jia R., Xu F., Qian J., Yao Y., Miao C., Zheng Y.M., Liu S.L., Guo F., Geng Y., Qiao W., Liang C. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol. 2014;16(7):1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.