Abstract
SARS-CoV-2 has caused tens of thousands of infections and more than one thousand deaths. There are currently no registered therapies for treating coronavirus infections. Because of time consuming process of new drug development, drug repositioning may be the only solution to the epidemic of sudden infectious diseases. We systematically analyzed all the proteins encoded by SARS-CoV-2 genes, compared them with proteins from other coronaviruses, predicted their structures, and built 19 structures that could be done by homology modeling. By performing target-based virtual ligand screening, a total of 21 targets (including two human targets) were screened against compound libraries including ZINC drug database and our own database of natural products. Structure and screening results of important targets such as 3-chymotrypsin-like protease (3CLpro), Spike, RNA-dependent RNA polymerase (RdRp), and papain like protease (PLpro) were discussed in detail. In addition, a database of 78 commonly used anti-viral drugs including those currently on the market and undergoing clinical trials for SARS-CoV-2 was constructed. Possible targets of these compounds and potential drugs acting on a certain target were predicted. This study will provide new lead compounds and targets for further in vitro and in vivo studies of SARS-CoV-2, new insights for those drugs currently ongoing clinical studies, and also possible new strategies for drug repositioning to treat SARS-CoV-2 infections.
Key words: SARS-CoV-2, Drug repurposing, Molecular docking, Remdesivir, Homology modeling
Abbreviations: 3CLpro, 3-chymotrypsin-like protease; E, envelope; M, membrane protein; N, nucleocapsid protein; Nsp, non-structure protein; ORF, open reading frame; PDB, protein data bank; RdRp, RNA-Dependence RNA polymerase; S, Spike; SUD, SARS unique domain; UB, ubiquitin-like domain
Graphical abstract
Twenty structures including 19 SARS-CoV-2 targets and one human target were built by homology modeling. Library of ZINC drug database, natural products, 78 anti-viral drugs were screened against these targets plus human ACE2. This study provides drug repositioning candidates and targets for further in vitro and in vivo studies of SARS-CoV-2.
1. Introduction
Coronaviruses (CoVs) have caused a major outbreak of human fatal pneumonia since the beginning of the 21st century. Severe acute respiratory syndrome coronavirus (SARS-CoV) broke out and spread to five continents in 2003 with a lethal rate of 10%1,2. The Middle East Respiratory Syndrome Coronavirus (MERS-CoV) broke out in the Arabian Peninsula in 2012 with a fatality rate of 35%3,4. Both SARS-CoV and MERS-CoV are zoonotic viruses, and their hosts are bat/civet and dromedary, respectively5,6. To date, no specific therapeutic drug or vaccine has been approved for the treatment of human coronavirus. Therefore, CoVs are considered to be a kind of viruses, of which the outbreak poses a huge threat to humans. The novel coronavirus found at the end of 2019 was named as 2019 novel coronavirus or “2019-nCoV” by the World Health Organization (WHO) on January 12, 20207,8. Since 2019-nCoV is highly homologous with SARS-CoV, it is considered a close relative of SARS-CoV. The International Virus Classification Commission (ICTV) classified 2019-nCoV as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) on February 11, 2020. At the same time, WHO named the disease caused by 2019-nCoV as COVID-19. Common symptoms of a person infected with coronavirus include respiratory symptoms, fever, cough, shortness of breath, and dyspnea. In more severe cases, infection can cause pneumonia, severe acute respiratory syndrome, kidney failure, and even death. There is currently no specific medicine or treatment for diseases caused by SARS-CoV-29.
CoVs are enveloped viruses with a positive RNA genome, belonging to the Coronaviridae family of the order Nidovirales, which are divided into four genera (α, β, γ, and δ). The SARS-CoV-2 belongs to the β genus. CoVs contain at least four structural proteins: Spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein10. Among them, Spike promotes host attachment and virus–cell membrane fusion during virus infection. Therefore, Spike determines to some extent the host range.
Potential anti-coronavirus therapies can be divided into two categories depending on the target, one is acting on the human immune system or human cells, and the other is on coronavirus itself. In terms of the human immune system, the innate immune system response plays an important role in controlling the replication and infection of coronavirus, and interferon is expected to enhance the immune response11. Blocking the signal pathways of human cells required for virus replication may show a certain anti-viral effect. In addition, viruses often bind to receptor proteins on the surface of cells in order to entering human cells, for example, the SARS virus binds to the angiotensin-converting enzyme 2 (ACE2) receptor12, 13, 14 and the MERS binds to the DPP4 receptor15,16. The therapies acting on the coronavirus itself include preventing the synthesis of viral RNA through acting on the genetic material of the virus, inhibiting virus replication through acting on critical enzymes of virus, and blocking the virus binding to human cell receptors or inhibiting the virus's self-assembly process through acting on some structural proteins.
In the fight against coronavirus, scientists have come up with three strategies for developing new drugs17. The first strategy is to test existing broad-spectrum anti-virals18. Interferons, ribavirin, and cyclophilin inhibitors used to treat coronavirus pneumonia fall into this category. The advantages of these therapies are that their metabolic characteristics, dosages used, potential efficacy and side effects are clear as they have been approved for treating viral infections. But the disadvantage is that these therapies are too “broad-spectrum” and cannot kill coronaviruses in a targeted manner, and their side effects should not be underestimated. The second strategy is to use existing molecular databases to screen for molecules that may have therapeutic effect on coronavirus19,20. High-throughput screening makes this strategy possible, and new functions of many drug molecules can be found through this strategy, for example, the discovery of anti-HIV infection drug lopinavir/ritonavir. The third strategy is directly based on the genomic information and pathological characteristics of different coronaviruses to develop new targeted drugs from scratch. Theoretically, the drugs found through these therapies would exhibit better anti-coronavirus effects, but the research procedure of new drug might cost several years, or even more than 10 years11.
For the development of medicines treating SARS-CoV-2, the fastest way is to find potential molecules from the marketed drugs. Once the efficacy is determined, it can be approved by the Green Channel or approved by the hospital ethics committee for rapid clinical treatment of patients. Herein, bioinformatics analysis on the proteins encoded by the novel coronavirus genes was systematically conducted, and the proteins of SARS-CoV-2 were compared with other coronaviruses, such as SARS-CoV and MERS-CoV. We conducted homology modeling to build all possible protein structures, including viral papain like protease (PLpro), main protease (3CLpro, also named 3-chymotrypsin-like protease), RNA-dependent RNA polymerase (RdRp), helicase, Spike, etc. Further, we used these proteins and human relative proteins [human ACE2 and type-II transmembrane serine protease (TMPRSS2) enzymes] as targets to screen ZINC U. S. Food and Drug Administration (FDA)-approved drug database (ZINC drug database, ZDD), our own database of traditional Chinese medicine and natural products (including reported common anti-viral components from traditional Chinese medicine), and the database of commonly used anti-viral drugs (78 compounds) by virtual ligand screening method. This study predicts a variety of compounds that may inhibit novel coronaviruses and provides scientists with information on compounds that may be effective. Subsequent validation of anti-viral effects in vitro and in vivo will provide useful information for clinical treatment of novel coronavirus pneumonia.
2. Methods and materials
2.1. Homology genome blast and genomes information
The complete genome of SARS-CoV-2/WHU02 (MN988669.1) was downloaded from NCBI nucleotide database. Thenucleotide sequences were aligned with whole database using BLASTn to search for homology viral genomes(Alogorithm parameters, max target sequences: 1000, expect threshold: 10). Accession numbers of 22 sequences in GenBank are listed as follows: BetaCoV/YN2018B(MK211376.1), Human/CoV HKU1 isolate SI17244 (MH940245.1), Human/CoV HKU1 genotype B(DQ415911.1), Rodent/RtTn-CoV (KY370043.1), DUCK/CoV (KM454473.1), Feline/CoV UU5(FJ938056.1), Bat/CoV HKU9-4 (EF065516.1), MERS NL13845 (MG021451.1), SARS-CoV-2/WHU02 (MN988669.1), 2019-nCoV/USA-IL1(MN988713.1), SARS-CoV-2/USA-CA2 (MN994468.1), SARS-CoV-2/HKU-SZ-005b (MN975262.1), SARS-CoV-2/USA-WA1/2020 (MN985325.1), SARS-CoV-2/USA-AZ1/2020 (MN997409.1), SARS-CoV-2/USA-CA1 (MN994467.1).
2.2. Nucleotide and amino acid sequence alignment and analysis
Nucleotide sequence editing was conducted using Bioedit and DNAMAN, and sequence alignment was conducted using DNAMAN and ClustaIW. The evolutionary history was inferred using the Neighbor-Joining method in MEGA seven software package. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test was determined by 500 replicates. Protein sequence management and analysis were carried out by using snap gene view, and sequence alignment was performed using Clustalw method in Jalview software. The second structure of protein sequences were predicted by Jpred4.
The homology model prediction was carried out through searching in PDB1018 database included in Fold and Function Assignment System (ffas.godziklab.org)21. Prediction of transmembrane helices in proteins was carried out in TMHMM Server v. 2.0 online (http://www.cbs.dtu.dk/services/TMHMM/). 3D structure structures are aligned by pymol structure alignment tool.
2.3. Compounds database
Approved drug database was from the subset of ZINC database, ZDD (ZINC drug database) containing 2924 compounds22. Natural products database was constructed by ourselves, containing 1066 chemicals separated from traditional Chinese herbals in own laboratory and naturally occurring potential anti-viral components and derivatives. Anti-viral compounds library contains 78 known anti-viral drugs and reported anti-viral compounds through literature search.
2.4. Homology modeling and molecular docking
Corresponding homology models predicted by Fold and Function Assignment System server for each target protein were downloaded from Protein Data Bank (www.rcsb.org). Alignment of two protein sequences and subsequent homology modeling were performed by bioinformatics module of ICM 3.7.3 modeling software on an Intel i7 4960 processor (MolSoft LLC, San Diego, CA, USA). For the structure-based virtual screening, ligands were continuously resiliently made to dock with the target that was represented in potential energy maps by ICM 3.7.3 software, to identify possible drug candidates. 3D compounds of each database were scored according to the Internal Coordinate Mechanics (Internal Coordinate Mechanics, ICM)23. Based on Monte Carlo method, stochastic global optimization procedure and pseudo-Brownian positional/torsional steps, the position of intrinsic molecular was optimized. By visually inspecting, compounds outside the active site, as well as those weakly fitting to the active site were eliminated. Compounds with scores less than −30 or mfScores less than −100 (generally represents strong interactions) have priority to be selected. Protein–protein docking was followed the manual of ICM software.
3. Results
3.1. Analysis, structure prediction and homology modeling of SARS-CoV-2 encoded proteins
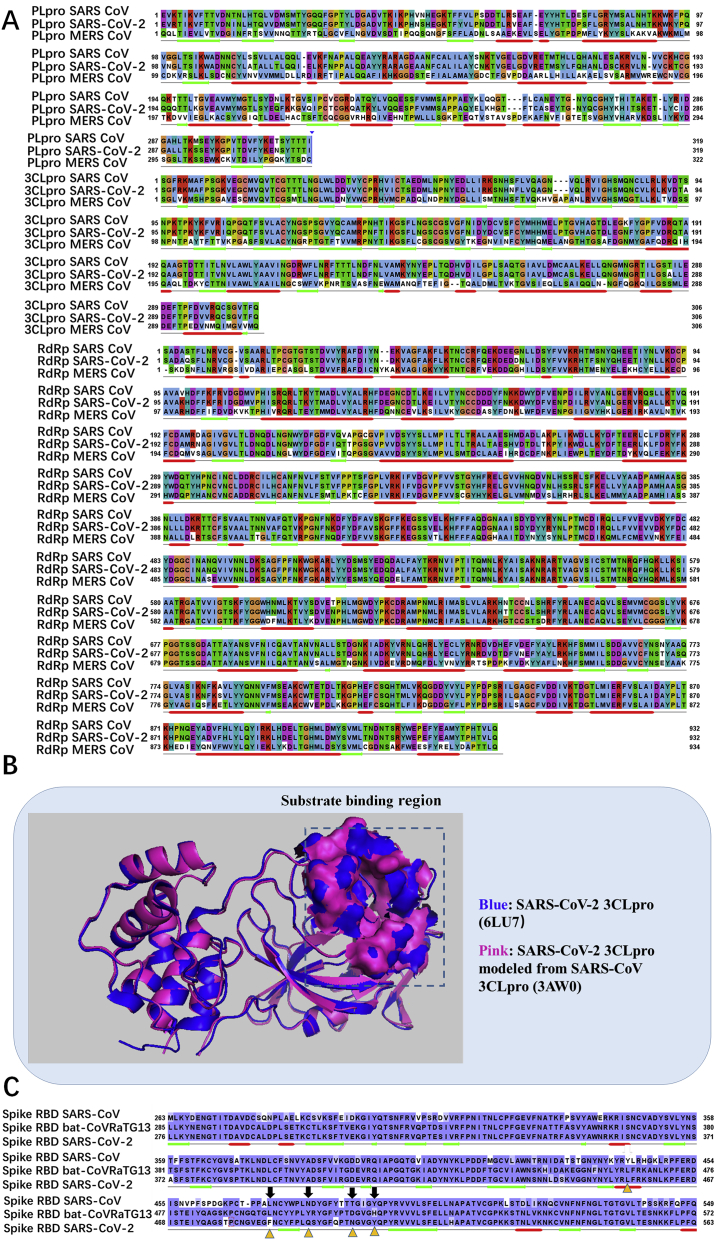
We obtained the SARS-CoV-2 genome from Gene Bank. The genome sequence of SARS-CoV-2/WHU02 was aligned with whole database using BLASTn to search for homology viral genomes. After phylogenetic analysis and sequence alignment of 22 coronaviruses from various species. We found three coronaviruses from bat (96%, 88% and 88% for Bat-Cov RaTG13, bat-SL-CoVZXC12 and bat-SL-CoVZC45, respectively) have the highest genome sequence identity to SARS-CoV-2 (Fig. 1A). Moreover, as shown in Fig. 1B, Bat-Cov RaTG13 exhibited the closest linkage with SARS-CoV-2. Among all coronaviruses from human, SARS-CoV (80%) exhibited the highest genome sequence identity to SARS-CoV-2. And MERS/isolate NL13845 also has 50% identity with SARS-CoV-2. SARS-CoV is the most clearly studied one among all these viruses according to previous literatures. Structure and function of its most genome encoded proteins have been elucidated in recent years. In this study, because of high genome identity between SARS-CoV-2 and SARS-CoV, the structure and function prediction of SARS-CoV-2 genome encoded protein were mainly based on those researches on homology protein in SARS-CoV. SARS-CoV-2 genome has 10 open reading frames (Fig. 2A). ORF1ab encodes replicase polyprotein 1 ab. After cleaved by two proteases, replicase proteins showed multifunction involved in transcription and replication of viral RNAs. ORF2-10 encodes viral structural proteins such as S, M, N, and E proteins, and other auxiliary proteins. The S, M, E proteins are involved in the formation of the viral coat, and the N protein is involved in the packaging of the RNA genome (Fig. 2C). By aligning with the amino acid sequence of SARS PP1ab and analyzing the characteristics of restriction cleavage sites recognized by 3CLpro and PLpro, we speculated 14 specific proteolytic sites of 3CLpro and PLpro in SARS-CoV-2 PP1ab (Fig. 2B). PLpro cleaves three sites at 181–182, 818–819, and 2763–2764 at the N-terminus and 3CLpro cuts at the other 11 sites at the C-terminus, and forming 15 non-structural proteins. Among them, Nsp3 contains multiple domains, including a segment of SARS unique domain and a deubiquitination and proteolytic enzyme PLpro. Nsp5 is 3CLpro, Nsp12 is an RdRp, and Nsp13 is helicase. As a new coronavirus, structure biology study about these proteins still at early stage. Until now, only one crystal structure of 3CLpro has been deposited in PDB (pdb code: 6LU7).
Figure 1.
Phylogenetic analysis and sequence alignment of 22 coronaviruses from different hosts. (A) Homology tree of 22 nucleotide sequences alignment. The homology of each branch is marked in red. (B) The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown above the branches.
Figure 2.
Overall genome and protein analysis of SARS-CoV-2. (A) Genome of SARS-CoV-2. (B) The large replicase polyprotein PP1ab. The purple rectangle represents the replicase polyprotein PP1ab which contains 7096 amino acids. The position of red triangle means cleavage site of PLpro, and position of yellow triangle means the site of 3CLpro, the numbers below the purple rectangle represent the residues that bound the individual proteins or domains. Orange rectangle below the individual proteins or domains means exactly homology 3D structure were detected. (C) Four structure proteins are represented in the cartoon pattern of coronavirus.
In order to acquire more three-dimensional structure information of proteins about these new coronaviruses for subsequent drug screening, we aligned all protein sequences from SARS-CoV-2 with all sequences in PDB1018 database in Fold & Function Assignment System (Supporting Information Figs. S19–S34). Fortunately, most of these proteins have found their high homology proteins that have three-dimensional structure, with homology between 72%–99% (Supporting Information Table S1). Those PDB codes were labeled below the corresponding sequences in Fig. 2B. Unsurprisingly, all these proteins with the highest homology are from SARS. Nevertheless, there are some proteins still have not high homologous in the database. After the prediction of transmembrane helices in these proteins carried out in TMHMM Server, as expected, we found that all these proteins are transmembrane proteins except for Nsp2 (Supporting Information Figs. S35–S41). So far, we have found as much structural information of this viral proteins as possible, which provides the basis for subsequent homology modeling and drug screening. Before homology modeling, all these proteins sequences were aligned with model sequences derived from SARS-CoV and predicted secondary structure as the same time (Fig. 3A and Table S1). Interestingly, as shown in Fig. 3A, important anti-virus drug target protein like 3CLpro, PLpro, and RdRp are highly conserved between those two human coronaviruses, especially in functional region. As shown in Table S1, all these potential drug target proteins have been homologously modeled, and all generated protein models were provided as the PDB files in Supporting Information. All other coordinates of target-screening hit complexes can be provided upon request.
Figure 3.
Sequence alignment of the proteins from SARS-CoV-2 with other CoVs. (A) Sequence alignment of PLpro, 3CLpro, RdRp among SARS-CoV, SARS-CoV-2 and MERS CoV. (B) The computational structure of SARS-CoV-2 3CLpro (shown as pink cartoon) modeled from SARS 3CLpro (3WA0) aligned with crystal structure of SARS-CoV-2 3CLpro (6LU7) (shown in blue cartoon). The computational model of the of SARS-CoV-2 3CLpro showed a Cα RMSD of 0.471 Å on the overall structure compared to the SARS-CoV-2 3CLpro structure, and that showed a Cα RMSD of 0.126 Å on the substrate binding region. (C) Sequence alignment of spike protein receptor binding domain among SARS-CoV, bat-CoVRaTG13 and SARS-CoV-2. “▵” represents residues previously identified as importantly interact with human ACE2 from SARS. Black arrow marked amino acids that are different between SARS-CoV-2 and bat-CoVRaTG13.
In order to verify the accuracy of homologous modeling, we aligned the computational structure of the SARS-CoV-2 3CLpro that modeled from the SARS-CoV 3CLpro with its crystal structure of SARS-CoV-2 3CLpro (6LU7) just solved and released during this manuscript was being prepared. The computational model of SARS-CoV-2 3CLpro showed a Cα RMSD of 0.471 Å on the overall structure compared to the SARS-CoV-2 3CLpro structure (Fig. 3B). What's more, the Cα RMSD between their substrate binding regions is only 0.126 Å, showing very subtle difference.
ACE2 molecule was known as a human entry receptor for Spike which facilitates its cross-species transmission. Sequence alignment results show that the homology of the Spike-receptor binding domain (RBD) sequence between SARS-CoV-2 and SARS-CoV is 76% (Fig. 3C). Recent researches speculated that SARS-CoV-2 could also bind to ACE2, and this was verified by computational docking and ELISA measurement24,25. Moreover, homology of the Spike-RBD sequence between SARS-CoV-2 and Bat-CoV RaTG13 is as high as 95%. Despite the high homology of RaTG13 and SARS-CoV-2 in Spike sequence, our analysis found that four among the five most important amino acids (L465, L495, Y502, D510, and H514) that bind to ACE212 in Bat-CoV RaTG13 differ from SARS-CoV-2 (Fig. 3C). And there is no related research literature about whether Bat-CoV RaTG13 can infect human yet. We also performed homology modeling on the Bat-CoV RaTG13 Spike RBD (Supporting Information Fig. S1). Three Spike RBD structures have been docked with human ACE2. Among them, for the conformations which most resemble the crystal structure of SARS RBD–ACE2 complex26, the binding free energy between SARS-CoV-2 Spike RBD and human ACE2 was −33.72 kcal/mol (Supporting Information Fig. S2), that between SARS-CoV Spike RBD and ACE2 was −49.22 kcal/mol (Supporting Information Fig. S3), and that between Bat-CoV RaTG13 Spike RBD and ACE2 was −31.06 kcal/mol (Supporting Information Fig. S4).
3.2. Virtual ligand screening of potential drug targets of SARS-CoV-2
The therapies that act on the coronavirus can be divided into several categories based on the specific pathways: (1) some acting on enzymes or functional proteins that are critical to virus, preventing the virus RNA synthesis and replication; (2) some acting on structural proteins of virus, blocking virus from binding to human cell receptors, or inhibiting the virus's self-assembly process; (3) some producing virulence factor to restore host's innate immunity; (4) some acting on host's specific receptors or enzymes, preventing virus from entering into host's cells. The related target proteins include Nsp1, Nsp3 (Nsp3b, Nsp3c, PLpro, and Nsp3e), Nsp7_Nsp8 complex, Nsp9–Nsp10, and Nsp14–Nsp16, 3CLpro, E-channel (E protein), ORF7a, Spike, ACE2, C-terminal RNA binding domain (CRBD), N-terminal RNA binding domain (NRBD), helicase, RdRp, and TMPRSSS2.
Based on homology models of the above 18 viral proteins (19 models) and two human targets, we resorted to structure-based virtual ligand screening method using ICM 3.7.3 modeling software (MolSoft LLC) to screen potential small-molecule compounds from a ZINC Drug Database (2924 compounds) and a small in-house database of traditional Chinese medicine and natural products (including reported common anti-viral components from traditional Chinese medicine) and derivatives (1066 compounds). Compounds with lower calculated binding energies (being expressed with scores and mfScores) are considered to have higher binding affinities with the target protein.
3.2.1. Targets preventing the virus RNA synthesis and replication
As significant functional proteins of coronavirus, Nsps are involved in RNA transcription, translation, protein synthesis, processing and modification, virus replication and infection of the host. Among them, 3CLpro, PLpro, RdRp and helicase are the most important targets for the development of small-molecule inhibitors due to their clear biological functions and vital enzyme active site.
3.2.1.1. Papain-like proteinase (PLpro)
PLpro is responsible for the cleavages of N-terminus of the replicase poly-protein to release Nsp1, Nsp2 and Nsp3, which is essential for correcting virus replication27. PLpro was also confirmed to be significant to antagonize the host's innate immunity28, 29, 30. As an indispensable enzyme in the process of coronavirus replication and infection of the host, PLpro has been a popular target for coronavirus inhibitors. It is very valuable for targeting PLpro to treat coronavirus infections, but no inhibitor has been approved by the FDA for marketing.
The screening results (Table 1 and supporting excel file PLpro.xlsx) showed that a series of anti-virus drugs (ribavirin, valganciclovir and thymidine), anti-bacterial drugs (chloramphenicol, cefamandole and tigecycline), muscle relaxant drug (chlorphenesin carbamate), anti-tussive drug (levodropropizine) may have high binding affinity to PLpro. The natural products (Table 2 and Supporting excel file PLpro_NP.xlsx), such as platycodin D from Platycodon grandiflorus, baicalin from Scutellaria baicalensis, sugetriol-3,9-diacetate from Cyperus rotundus, phaitanthrin D and 2,2-di (3-indolyl)-3-indolone from Isatis indigotica, catechin compounds ((–)-epigallocatechin gallate and 2-(3,4-dihydroxyphenyl)-2-[[2-(3,4-dihydroxyphenyl)-3,4-dihydro-5,7-dihydroxy-2H-1-benzopyran-3-yl]oxy]-3,4-dihydro-2H-1-benzopyran-3,4,5,7-tetrol) exhibited high binding affinity to PLpro protein, suggesting the potential utility of these compounds in the treatment of SARS-CoV-2.
Table 1.
Potential PLpro inhibitors from ZINC drug database.
| No. | Drug name | Structure | Pharmacological function |
|---|---|---|---|
| 1 | Ribavirin | 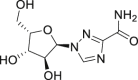 |
Anti-virus |
| 2 | Valganciclovir | 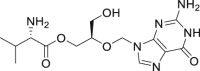 |
Anti-virus |
| 3 | β-Thymidine | 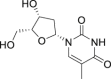 |
Anti-virus |
| 4 | Aspartame | 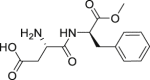 |
Non-carbohydrate sweetener |
| 5 | Oxprenolol |  |
Anti-hypertensive, anti-angina, anti-arrhythmic effects |
| 6 | Doxycycline | 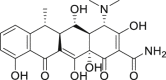 |
Anti-bacterial effect |
| 7 | Acetophenazine | 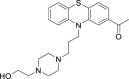 |
Anti-psychotic effect |
| 8 | Iopromide |  |
Low osmolar, non-ionic contrast agent |
| 9 | Riboflavin |  |
Treatment of vitamin B2 deficiency |
| 10 | Reproterol | 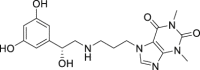 |
Treatment of bronchial asthma |
| 11 | 2,2′-Cyclocytidine | 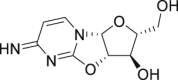 |
Anti-tumor |
| 12 | Chloramphenicol | 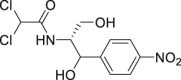 |
Anti-bacterial effect |
| 13 | Chlorphenesin carbamate |  |
Muscle relaxant effect |
| 14 | Levodropropizine |  |
Anti-tussive effect |
| 15 | Cefamandole | 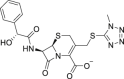 |
Anti-bacterial effect |
| 16 | Floxuridine |  |
Anti-tumor |
| 17 | Tigecycline | 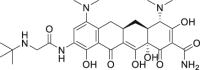 |
Anti-bacterial |
| 18 | Pemetrexed | 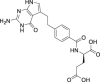 |
Anti-tumor |
| 19 | l (+)-Ascorbic acid | 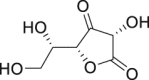 |
Anti-scorbutic |
| 20 | Glutathione |  |
Treatment of hepatic disease |
| 21 | Hesperetin | 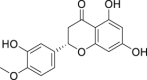 |
Anti-oxidation, anti-virus, anti-bacteria |
| 22 | Ademetionine | 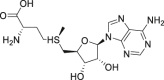 |
Cholagogue |
| 23 | Masoprocol | 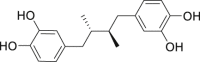 |
Treatment of actinic keratosis |
| 24 | Isotretinoin | 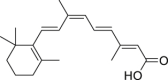 |
Anti-tumor |
| 25 | Dantrolene | 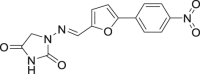 |
Muscle relaxant |
| 26 | Sulfasalazine | 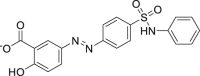 |
Anti-bacterial effect |
| 27 | Silybin | 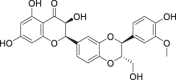 |
Hepatoprotective effect |
| 28 | Nicardipine |  |
Anti-hypertensive effect |
| 29 | Sildenafil |  |
Treatment of erectile dysfunction |
Table 2.
Potential PLpro inhibitors from in-house natural product database.
| No. | Compound name | Structure | Pharmacological function | Source |
|---|---|---|---|---|
| 1 | Platycodin D | 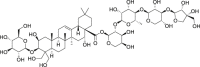 |
Anti-tumor, anti-inflammatory effect | Platycodon grandiflorus |
| 2 | Chrysin | 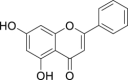 |
Anti-virus, anti-inflammatory effect | Scutellaria baicalensis |
| 3 | Neohesperidin | 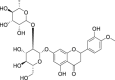 |
Anti-tumor, anti-allergic effect | Citrus aurantium L. |
| 4 | Baicalin | 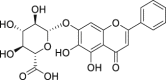 |
Anti-tumor, anti-inflammatory, anti-bacterial, anti-virus effect | Scutellaria baicalensis |
| 5 | Sugetriol-3,9-diacetate | 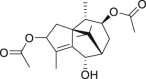 |
Anti-HBV, anti-HSV-1 |
Cyperus rotundus |
| 6 | (–)-Epigallocatechin gallate |  |
Anti-oxidation, anti-tumor, treatment of depression | Camellia sinensis |
| 7 | Phaitanthrin D |  |
Anti-virus | Isatis indigotica Fort. |
| 8 | 2-(3,4-Dihydroxyphenyl)-2-[[2-(3,4-dihydroxyphenyl)-3,4-dihydro-5,7-dihydroxy-2H-1-benzopyran-3-yl]oxy]-3,4-dihydro-2H-1-benzopyran-3,4,5,7-tetrol | 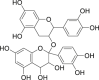 |
Anti-oxidant, anti-inflammatory, anti-tumor | Vitis vinifera |
| 9 | 2,2-Di (3-indolyl)-3-indolone | 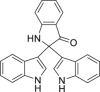 |
Anti-virus | Isatis indigotica Fort. |
| 10 | (S)-(1S,2R,4aS,5R,8aS)-1-Formamido-1,4a-dimethyl-6-methylene-5-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydronaphthalen-2-yl-2-amino-3-phenylpropanoate |  |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 11 | Piceatannol | 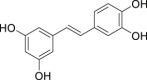 |
Anti-tumor, anti-virus effect | Vitis vinifera |
| 12 | Rosmarinic acid | 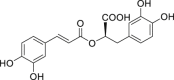 |
Anti-virus, anti-oxidant |
Salvia verticillata L. |
| 13 | Magnolol | 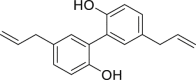 |
Anti-tumor, anti-microbial effect |
Magnolia officinalis |
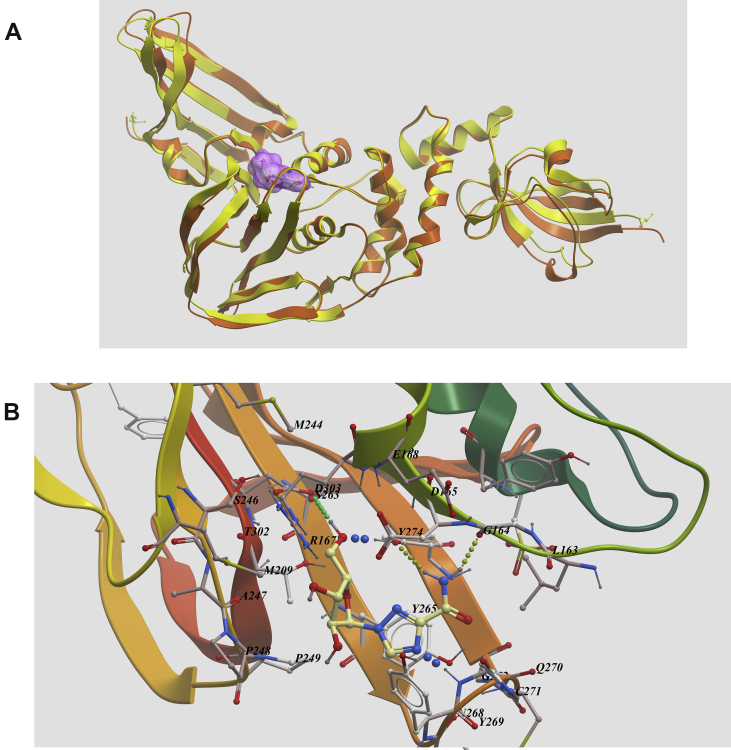
Anti-viral drug ribavirin was predicted to bind to PLpro with low binding energy (Scores = –38.58). From generated docking model, ribavirin was bound in the active site of the enzyme as reported SARS-PLpro inhibitors (PDB code 3e9s). Hydrogen bonds were predicted between Gly164, Gln270, Tyr274, Asp303 and the compound. Also, π–π stacking was found between Tyr265 and triazolering in the compound (Fig. 4A and B). The strong hydrogen bonding and hydrophobic interaction between ribavirin with the enzyme imply it may be a potent PLpro inhibitor.
Figure 4.
Low-energy binding conformations of ribavirin bound to PLpro generated by molecular docking. (A) Superimposing of 2019-nCoV PLpro (yellow) with the crystal structure of SARS PLpro (orange) (PDB code 3e9s). (B) Detailed view of rivavirin binding in the active site of the enzyme (Supporting PDB file SARS_CoV-2_PLpro_homo_Ribavirin.pdb).
3.2.1.2. 3C-like main protease (3CLpro)
The 3CLpro, also known as Nsp5, is first automatically cleaved from poly-proteins to produce mature enzymes, and then further cleaves downstream Nsps at 11 sites to release Nsp4–Nsp1631. 3CLpro directly mediates the maturation of Nsps, which is essential in the life cycle of the virus. The detailed investigation on the structure and catalytic mechanism of 3CLpro makes 3CLpro an attractive target for anti-coronavirus drug development. Inhibitors targeting at SARS-CoV 3CLpro mainly include peptide inhibitors and small-molecule inhibitors32.
3CLpro monomer has three domains, domain I (residues 8–101), domain II (residues 102–184) and domain III (residues 201–303), and a long loop (residues 185–200) connects domains II and III. The active site of 3CLpro is located in the gap between domains I and II, and has a CysHis catalytic dyad (Cys145 and His41)33. As shown in Table 3 and Supporting excel file 3CLpro.xlsx), anti-bacterial drugs (lymecycline, demeclocycline, doxycycline and oxytetracycline), anti-hypertensive drugs (nicardipine and telmisartan), and conivaptan treating hyponatremia show highest binding affinity to 3CLpro. Several natural compounds and derivatives with anti-virus and anti-inflammatory effects also exhibited high binding affinity to 3CLpro (Table 4 and Supporting excel file 3CLpro_NP.xlsx), including a series of andrographolide derivatives, chrysin-7-O-β-glucuronide from S. baicalensis, betulonal from Cassine xylocarpa, 2β-hydroxy-3,4-seco-friedelolactone-27-oic acid, isodecortinol and cerevisterol from Viola diffusa, hesperidin and neohesperidin from Citrus aurantium, kouitchenside I and deacetylcentapicrin from the plants of Swertia genus. The above results suggest that these small-molecule compounds might be the potential 3CLpro inhibitors and could probably be used for treating SARS-CoV-2.
Table 3.
Potential 3CLpro inhibitors from ZINC drug database.
| No. | Drug name | Structure | Pharmacological functions |
|---|---|---|---|
| 1 | Lymecycline | 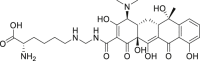 |
Anti-bacterial effect |
| 2 | Chlorhexidine | Anti-bacterial effect | |
| 3 | Alfuzosin | 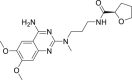 |
Anti-hypertensive agent, benign prostatic hyperplasia |
| 4 | Cilastatin | 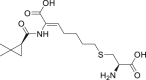 |
Renal peptidase inhibition |
| 5 | Famotidine | Anti-ulcerative activity | |
| 6 | Almitrine | 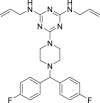 |
Treatment of cognitive and chronic sensory nerve impairment |
| 7 | Progabide | 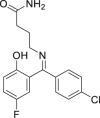 |
Anti-epileptic effect |
| 8 | Nepafenac | 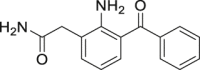 |
Treatment of pain and inflammation associated with cataract surgery |
| 9 | Carvedilol | 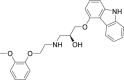 |
Vasodilator effect |
| 10 | Amprenavir |  |
HIV-1 protease inhibition |
| 11 | Tigecycline | 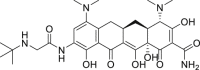 |
Anti-bacterial effect |
| 12 | Demeclocycline | 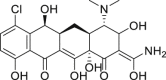 |
Anti-bacterial effect |
| 13 | Montelukast |  |
Anti-allergic, anti-asthmatic effects |
| 14 | Carminic acid | 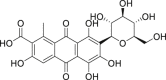 |
Food additive |
| 15 | Mimosine |  |
Depilatory effect |
| 16 | Flavin mononucleotide | 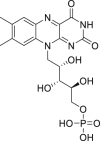 |
Electron transference in biological oxidation |
| 17 | Lutein | Vision protection, anti-oxidation |
|
| 18 | Cefpiramide |  |
Anti-bacterial effect |
| 19 | Phenethicillin | 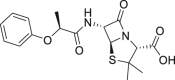 |
Anti-bacterial effect |
| 20 | Candoxatril |  |
Anti-hypertensive effect |
| 21 | Nicardipine | 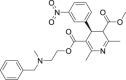 |
Anti-hypertensive effect |
| 22 | Estradiol valerate | 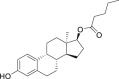 |
Treatment of estrogen deficiency |
| 23 | Pioglitazone | Anti-diabetic effect | |
| 24 | Conivaptan | 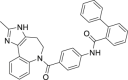 |
Treatment of hyponatremia |
| 25 | Telmisartan | 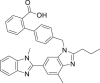 |
Anti-hypertensive effect |
| 26 | Doxycycline |  |
Anti-bacterial effect |
| 27 | Oxytetracycline |  |
Anti-bacterial effect |
Table 4.
Potential 3CLpro inhibitors from in-house natural product database.
| No. | Drug name | Structure | Pharmacological functions | Source |
|---|---|---|---|---|
| 1 | (1S,2R,4aS,5R,8aS)-1-Formamido-1,4a-dimethyl-6-methylene-5-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydronaphthalen-2-yl 5-((R)-1,2-dithiolan-3-yl) pentanoate | 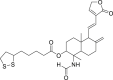 |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 2 | Betulonal |  |
Anti-HIV-1 | Cassine xylocarpa |
| 3 | Chrysin-7-O-β-glucuronide |  |
Anti-virus, anti-inflammatory effect | Scutellaria baicalensis |
| 4 | Andrographiside |  |
Anti-virus, anti-inflammatory effect | Andrographis paniculata |
| 5 | (1S,2R,4aS,5R,8aS)-1-Formamido-1,4a-dimethyl-6-methylene-5-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydronaphthalen-2-yl 2-nitrobenzoate |  |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 6 | 2β-Hydroxy-3,4-seco-friedelolactone-27-oic acid |  |
Anti-virus | Viola diffusa |
| 7 | (S)-(1S,2R,4aS,5R,8aS)-1-Formamido-1,4a-dimethyl-6-methylene-5-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydronaphthalen-2-yl-2-amino-3-phenylpropanoate | 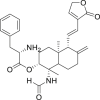 |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 8 | Isodecortinol | 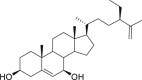 |
Anti-virus | Viola diffusa |
| 9 | Cerevisterol | 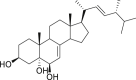 |
Anti-virus | Viola diffusa |
| 10 | Hesperidin | 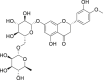 |
Anti-inflammatory, anti-oxidant effect | Citrus aurantium L. |
| 11 | Neohesperidin | 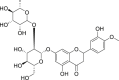 |
Anti-tumor, anti-allergic effect | Citrus aurantium L. |
| 12 | Andrograpanin | 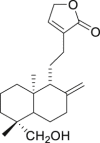 |
Anti-virus, anti-inflammatory effect | Andrographis paniculata |
| 13 | 2-((1R,5R,6R,8aS)-6-Hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethyl benzoate |  |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 14 | Cosmosiin |  |
Anti-inflammatory, anti-oxidant, anti-HIV effect |
Scutellaria baicalensis |
| 15 | Cleistocaltone A | 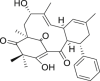 |
Anti-virus | Cleistocalyx operculatus |
| 16 | 2,2-Di(3-indolyl)-3-indolone | 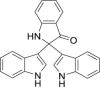 |
Anti-virus | Isatis indigotica Fort. |
| 17 | Biorobin |  |
Anti-virus | Ficus benjamina |
| 18 | Gnidicin | 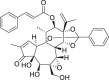 |
Anti-tumor | Gnidia lamprantha |
| 19 | Phyllaemblinol | 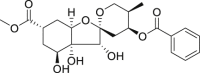 |
Anti-virus | Phyllanthus emblica |
| 20 | Theaflavin 3,3′-di-O-gallate | 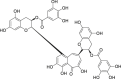 |
Anti-oxidant effect, anti-tumor, anti-virus |
Camellia sinensis |
| 21 | Rosmarinic acid |  |
Anti-viral, anti-oxidant |
Salvia verticillata L. |
| 22 | Kouitchenside I | 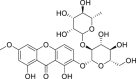 |
Anti-virus, anti-inflammatory effect | Swertia kouitchensis |
| 23 | Oleanolic acid | 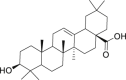 |
Anti-virus, anti-inflammatory effect | Swertia kouitchensis |
| 24 | Stigmast-5-en-3-ol | 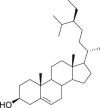 |
Anti-virus, anti-inflammatory effect | Swertia binchuanensis |
| 25 | Deacetylcentapicrin |  |
Anti-virus, anti-inflammatory effect | Swertia macrosperma |
| 26 | Berchemol | 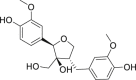 |
Anti-virus, anti-inflammatory effect | Swertiabi maculata |
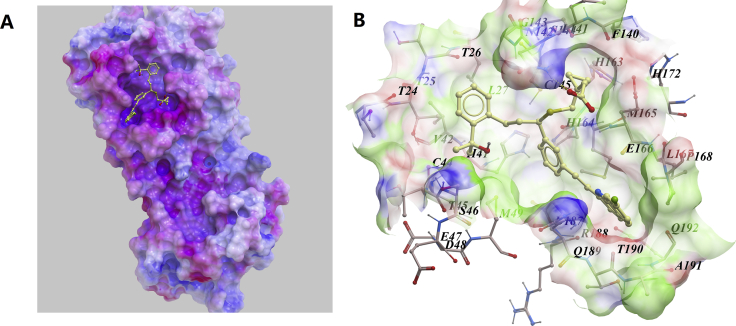
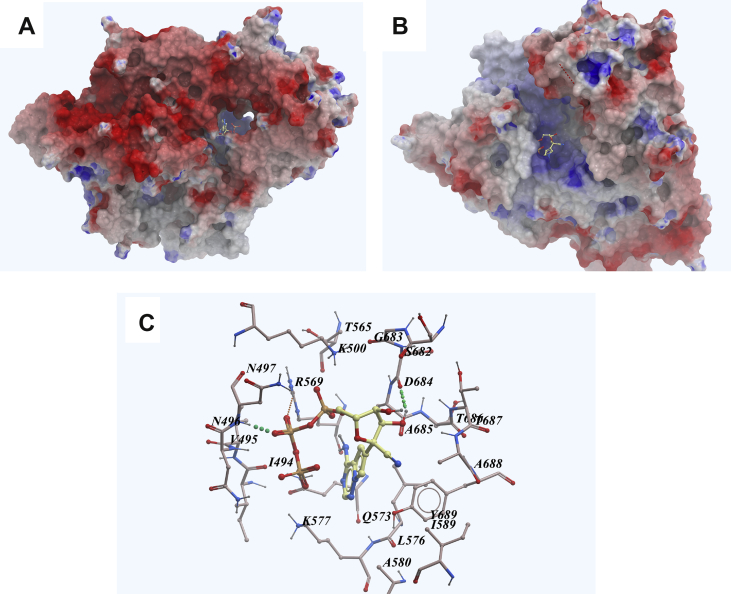
It's worth mentioning, anti-asthmatic drug montelukast also showed low binding energy to 3CLpro. As shown in Fig. 5A, montelukast was well fitted into the active pocket of 3CLpro, in which lots of hydrophobic amino acids, just like Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168 and His172 compose a relatively hydrophobic environment to contain the compound and stabilize its conformation. Hydrogen bonding was predicted between Asn142 and the carbonyl group of the compound (Fig. 5B).
Figure 5.
Low-energy binding conformations of Montelukast bound to 3CLpro generated by molecular docking. (A) Montelukast was fitted well in the active pocket of SARS-CoV-2 3CLpro, 3CLpro was shown as electrostatic surface model. (B) Detailed view of montelukastin binding in the active pocket of 3CLpro (Supporting PDB file SARS_CoV-2_3CLpro_homo_Montelukast.pdb).
3.2.1.3. RNA-dependent RNA polymerase (RdRp)
Nsp12, a conserved protein in coronavirus, is an RNA-dependent RNA polymerase (RdRp) and the vital enzyme of coronavirus replication/transcription complex. The RdRp domain of polymerase is located at the C-terminus and has a conserved Ser-Asp-Asp motif34. Nsp8 can de novo synthesize up to six nucleotides in length, which can be used as a primer for Nsp12-RdRp RNA synthesis. Further, the Nsp7_Nsp8 complex increases the binding of Nsp12 to RNA and enhances the RdRps enzyme activity of Nsp1235. In the research of SARS-CoV and MERS-CoV inhibitors, Nsp12-RdRp has been used as a very important drug target. In principle, targeted inhibition of Nsp12-RdRp could not cause significant toxicity and side effects on host cells, but no specific inhibitors have been found until now36.
Virtual screening results of RdRp demonstrated some drugs might be potential inhibitors with mfScores lower than −110, such as several anti-fungal drug itraconazole, anti-bacterial drug novobiocin, gallstone-dissolving drug chenodeoxycholic acid, anti-allergic drug cortisone, anti-tumor drug idarubicin, hepatoprotective drug silybin, muscle relaxant drug pancuronium bromide, and chronic enteritis, anti-coagulant drug dabigatran etexilate (Table 5 and Supporting excel file RdRp.xlsx). The natural products and derivatives with anti-virus, anti-inflammation and anti-tumor effects exhibited high binding affinity to RdRp, such as betulonal from C. xylocarpa, gnidicin and gniditrin from Gnidia lamprantha, 2β,30β-dihydroxy-3,4-seco-friedelolactone-27-lactone from V. diffusa, 14-deoxy-11,12-didehydroandrographolide from Andrographis paniculata, 1,7-dihydroxy-3- methoxyxanthone from Swerti apseudochinensis, theaflavin 3,3′-di-O-gallate from Camellia sinensis, and andrographolide derivative (R)-((1R,5aS,6R,9aS)-1,5a-dimethyl-7-methylene-3-oxo-6-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydro-1H-benzo[c]azepin-1-yl)methyl 2-amino-3-phenylpropanoate (Table 6 and Supporting excel file RdRp_NP.xlsx).
Table 5.
Potential RdRp inhibitors from ZINC drug database.
| No. | Drug name | Structure | Pharmacological function |
|---|---|---|---|
| 1 | Valganciclovir |  |
Anti-virus |
| 2 | Chlorhexidine | Anti-bacterial effect | |
| 3 | Ceftibuten | 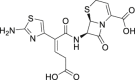 |
Anti-bacterial effect |
| 4 | Fenoterol | 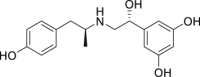 |
Treatment of bronchial asthma |
| 5 | Fludarabine | 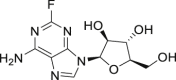 |
Anti-tumor |
| 6 | Itraconazole | 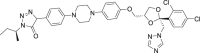 |
Anti-fungal effect |
| 7 | Cefuroxime | 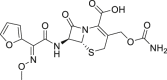 |
Anti-bacterial effect |
| 8 | Atovaquone |  |
Anti-malaria |
| 9 | Chenodeoxycholic acid | 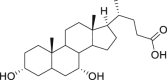 |
Dissolving gallstones |
| 10 | Cromolyn |  |
Treatment of allergic asthma |
| 11 | Pancuronium bromide | 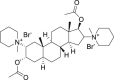 |
Muscle relaxant effect |
| 12 | Cortisone | 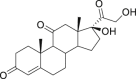 |
Anti-allergic effect |
| 13 | Tibolone | 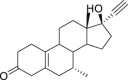 |
Contraceptive effect |
| 14 | Novobiocin |  |
Anti-bacterial effect |
| 15 | Silybin |  |
Hepatoprotective effect |
| 16 | Idarubicin | 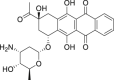 |
Anti-tumor |
| 17 | Bromocriptine | 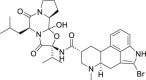 |
Treatment of amenorrhea |
| 18 | Diphenoxylate | 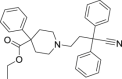 |
Treatment of diarrhea and chronic enteritis |
| 19 | Benzylpenicilloyl G | 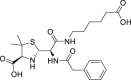 |
Skin-testing reagent to detect penicillin allergy |
| 20 | Dabigatran etexilate | 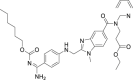 |
Anti-coagulant effect |
Table 6.
Potential RdRp inhibitors from in-house natural product database.
| No. | Drug name | Structure | Pharmacological function | Source |
|---|---|---|---|---|
| 1 | Betulonal | 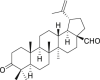 |
Anti-HIV-1 | Cassine xylocarpa |
| 2 | Gnidicin | 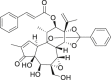 |
Anti-tumor | Gnidia lamprantha |
| 3 | 2β,30β-Dihydroxy-3,4-seco-friedelolactone-27-lactone | 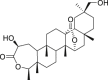 |
Anti-virus | Viola diffusa |
| 4 | 14-Deoxy-11,12-didehydroandrographolide | 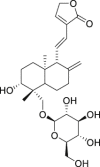 |
Anti-virus, anti-inflammatory effect | Andrographis paniculata |
| 5 | Gniditrin | 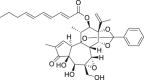 |
Anti-tumor | Gnidia lamprantha |
| 6 | Theaflavin 3,3′-di-O-gallate | 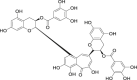 |
Anti-oxidant, anti-tumor, anti-virus |
Camellia sinensis |
| 7 | (R)-((1R,5aS,6R,9aS)-1,5a-Dimethyl-7-methylene-3-oxo-6-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydro-1H-benzo [c]azepin-1-yl)methyl 2-amino-3-phenylpropanoate | 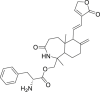 |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 8 | 2β-Hydroxy-3,4-seco-friedelolactone-27-oic acid |  |
Anti-virus | Viola diffusa |
| 9 | 2-(3,4-Dihydroxyphenyl)-2-[[2-(3,4-dihydroxyphenyl)-3,4-dihydro-5,7-dihydroxy-2H-1-benzopyran-3-yl]oxy]-3,4-dihydro-2H-1-benzopyran-3,4,5,7-tetrol |  |
Anti-oxidant, anti-inflammatory, anti-tumor | Vitis vinifera |
| 10 | Phyllaemblicin B |  |
Anti-virus | Phyllanthus emblica |
| 11 | 14-Hydroxycyperotundone |  |
Anti-HBV | Cyperus rotundus |
| 12 | Andrographiside |  |
Anti-virus, anti-inflammatory effect | Androgra phispaniculata |
| 13 | 2-((1R,5R,6R,8aS)-6-Hydroxy-5-(hydroxymethyl)-5,8a-dimethyl-2-methylenedecahydronaphthalen-1-yl)ethyl benzoate |  |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 14 | Sugetriol-3,9-diacetate |  |
Anti-HBV | Cyperus rotundus |
| 15 | Baicalin | 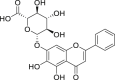 |
Anti-tumor, anti-inflammatory, anti-bacterial, anti-virus effect |
Scutellaria baicalensis |
| 16 | (1S,2R,4aS,5R,8aS)-1-Formamido-1,4a-dimethyl-6-methylene-5-((E)-2-(2-oxo-2,5-dihydrofuran-3-yl)ethenyl)decahydronaphthalen-2-yl 5-((R)-1,2-dithiolan-3-yl) pentanoate |  |
Anti-virus, anti-inflammatory effect | Andrographolide derivatives |
| 17 | 1,7-Dihydroxy-3-methoxyxanthone | 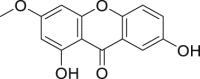 |
Anti-virus, anti-inflammatory effect | Swertia pseudochinensis |
| 18 | 1,2,6-Trimethoxy-8-[(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)oxy]-9H-xanthen-9-one | 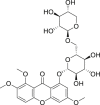 |
Anti-virus, anti-inflammatory effect | Swertia mussotii |
| 19 | 1,8-Dihydroxy-6-methoxy-2-[(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)oxy]-9H-xanthen-9-one | 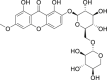 |
Anti-virus, anti-inflammatory effect | Swertia kouitchensis |
| 20 | 8-(β-d-Glucopyranosyloxy)-1,3,5-trihydroxy-9H-xanthen-9-one | 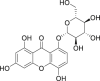 |
Anti-virus, anti-inflammatory effect | Swertia mussotii |
3.2.1.4. Helicase
Helicase (Nsp13), a multi-functional protein, include N-terminal metal binding domain (MBD) and helicase domain (Hel). N-terminal structure contains 26 cysteine residues to form a Zn2+ binding domain and helicase domain with a conserved motif at the C-terminus. Nsp13 can unravel double-stranded (ds) DNA and RNA along the 5′–3′ direction in an NTP-dependent manner37. Importantly, it has been reported that the SARS-Nsp13 sequence is conserved and indispensable, and is a necessary component for the replication of coronavirus. Therefore, it has been identified as a target for anti-viral drug discovery, but there are few reports about Nsp13 inhibitors38,39.
Based on structure modeling of helicase protein, anti-bacterial drugs (lymecycline, cefsulodine and rolitetracycline), anti-fungal drug itraconazole, anti-human immunodeficiency virus-1 (HIV-1) drug saquinavir, anti-coagulant drug dabigatran, and diuretic drug canrenoic acid were predicted to be helicase inhibitors with high mfScores through virtual ligand screening. The natural products, such as many flavanoids from different sources (α-glucosyl hesperidin, hesperidin, rutin, quercetagetin 6-O-β-d-glucopyranoside and homovitexin), xanthones such as 3,5-dimethoxy-1-[(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)oxy]-9H-xanthen-9-one, kouitchenside H, kouitchenside A, 8,2-dihydroxy-3,4,5-trimethoxy-1-[(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)oxy]-9H-xanthen-9-one, kouitchenside D, 1-hydroxy-2,6-dimethoxy-8-[(6-O-β-d-xylopyranosyl-β-d-glucopyranosyl)oxy]-9H-xanthen-9-one and triptexanthoside D from Swertia genus, phyllaemblicin B and phyllaemblinol from Phyllanthus emblica showed high binding affinity to this target.
Besides the above targets, some non-structural proteins, including Nsp3b, Nsp3e, Nsp7_Nsp8 complex, Nsp9, Nsp10, Nsp14, Nsp15, and Nsp16, also play an important role in the virus RNA synthesis and replication, suggesting these proteins may be useful targets for the anti-viral drug discovery. The virtual screening results showed many anti-bacterial, anti-viral, or anti-inflammatory drugs from ZINC drug database and our in-house natural products/derivatives database displayed potential good affinity to these targets, and the detailed information of virtual screening results is shown in Supporting excel files (for ZDD screening results, file names as target.xlsx; for natural products screening results, file names as target_NP.xlsx).
3.2.2. Targets inhibiting virus structural proteins
Spike is the main structural protein of coronavirus and assembles into a special corolla structure on the surface of the virus as a trimer. Spike is a main protein that interacts with the host by binding to host cell receptors to mediate virus invasion and determine viral tissue or host tropism40. Spike is cleaved into S1 and S2 by the host cell protease like TMPRSS2, etc. The main function of S1 is to bind with host cell surface receptors, and the S2 subunit mediates virus–cell and cell–cell membrane fusion. Spike structural integrity and cleavage activation play a key role in virus invasion and virulence41. Therapeutic strategies to block coronavirus from entering host cells by targeting Spike proteins or specific receptors on the host surface are valuable for the development of anti-viral drugs.
On the basis of virtual screening results of small-molecule compounds against Spike protein, some drugs showed high binding affinity (mfScore<–150 or score<–35), such as anti-hypertensive drugs (rescinnamine, iloprost and prazosin), anti-fungal drugs (posaconazole and itraconazole), anti-bacterial drug (sulfasalazine, azlocillin, penicillin and cefsulodin) and anti-coagulant drug dabigatran etexilate. Some natural flavanoids, licoflavonol from Glycyrrhiza uralensis, cosmosiin from S. baicalensis, neohesperidin from C. aurantium, mangostin from Garcinia mangostana, kouitchenside D from Swertia kouitchensis, excoecariatoxin from Excoecaria agallocha, phyllaemblicin G7 from P. emblica, and piceatannol from Vitis vinifera, exhibited high binding affinity. The detailed virtual screening results are shown in Supporting excel files.
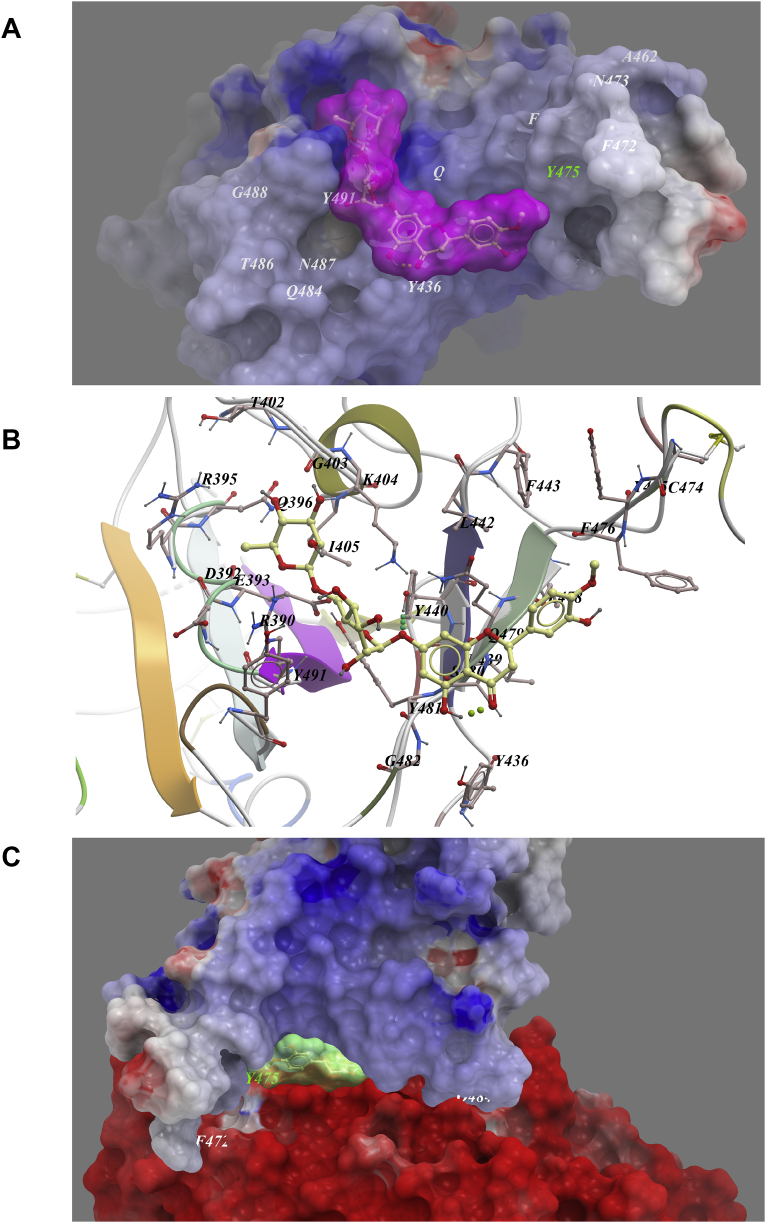
However, most of above compounds were not predicted to bind with the binding interface of the Spike–ACE2 complex. The only compound that could target the binding interface between Spike and ACE2 was hesperidin, as shown in Fig. 6A. Hesperidin was predicted to lie on the middle shallow pit of the surface of RBD of Spike, where the dihydroflavone part of the compound went parallel with the β-6 sheet of RBD. And the sugar part was inserted into the shallow pit in the direction away from ACE2, where a few hydrophobic amino acids, including Tyr436, Try440, Leu442, Phe443, Phe476, Try475, Try481 and Tyr49 form a relatively hydrophobic shallow pocket to contain the compound (Fig. 6B). Hydrogen bonding was predicted between Tyr440 and the compound. By superimposing the ACE2–RBD complex to the hesperidin–RBD complex, a distinct overlap of hesperidin with the interface of ACE2 could be observed (Fig. 6C), suggesting hesperidin may disrupt the interaction of ACE2 with RBD.
Figure 6.
Low-energy binding conformation of hesperidin bound to Spike RBD generated by molecular docking. (A) Hesperidinwas fitted into the shallow pocket in the surface of SARS-CoV-2 Spike RBD. (B) Detailed view of hesperidinbinding in the pocket of Spike RBD. (C) Hesperidin blocks the interface of ACE2 and Spike RBD binding (Supporting PDB file SARS_CoV-2 _Spike_RBD_homo_Hesperidin.pdb).
Except for Spike protein, E protein (E-channel) possesses important biological functions for the structural integrity of coronavirus and host virulence. NRBD and CRBD of coronavirus N protein are needed for N proteins in host cells to bind with coronavirus RNA efficiently. Therefore, E protein or N protein (NRBD and CRBD domains) can be used as targets for the discovery of anti-viral drugs. Through virtual screening, many anti-bacterial, anti-viral, anti-tumor, anti-asthmatic, and anti-inflammatory drugs, etc. from ZINC database and our in-house natural products/derivatives database were found to display relatively good affinity to these targets. And the detailed results of virtual screening are given in Supplementary excel files.
3.2.3. Targets inhibiting virulence factor
There are three coronavirus virulence factors Nsp1, Nsp3c and ORF7a related to interfering host's innate immunity and assisting coronavirus immune escape. Nsp1 interacts with host 40S ribosomal subunit that induces specifically host mRNA degradation42 and also inhibits type-I interferon production43. Nsp3c has ability to bind with host's ADP-ribose to help coronavirus resist host innate immunity44. Bone marrow matrix antigen 2 (BST-2) can inhibit the release of newly-assembled coronavirus from host cells. SARS-CoV ORF7a directly binds to BST-2 and inhibits its activity by blocking the glycosylation of BST-245. These evidences suggest that Nsp1, Nsp3c and ORF7a may be potential targets for anti-viral drug discovery.
The detailed screening results of Nsp1, Nsp3c, and ORF7a showed that a series of clinical drugs and natural products with anti-bacterial and anti-inflammatory effects exhibited relatively high binding affinity to these three target proteins, such as piperacillin, cefpiramide, streptomycin, lymecycline, tetracycline, platycodin D from Platycodon grandifloras, wogonoside from S. baicalensis, vitexin from Vitex negundo, andrographolide derivatives, and xanthones from Swertia genus. The detailed results of virtual screening are shown in Supporting excel files.
3.2.4. Targets blocking hose specific receptor or enzymes
The host ACE2 has been proved by many studies to be the specific receptor for the Spike RBD of SARS-CoV12. The latest research shows that the host receptor of SARS-CoV-2 is consistent with SARS-CoV, exhibiting that the Spike RBD sequence of SARS-CoV-2 is similar to SARS-CoV RBD and there are important interactions between several key amino acid residues of RBD receptor-binding motif and ACE224. Based on the current research progress, ACE2 is considered as a host target for the treatment of coronavirus infection to block SARS-CoV-2 from entering host cells.
Based on the virtual screening results of ACE2 protein, anti-diabetes drug troglitazone, anti-hypertensive drug losartan, analgesia drug ergotamine, anti-bacterial drug cefmenoxime, and hepatoprotective drug silybin, etc., were predicted to bind with ACE2 with low energy. The natural products, such as phyllaemblicin G7 from P. emblica, xanthones from the plants of Swertiagenus, neohesperidin and hesperidin from C. aurantium, exhibited potentially high binding affinity to ACE2 protein. However, none of above ACE2 binding compounds was predicted to target the ACE2–RBD interface.
In addition, TMPRSS2 was known to cut the Spike to trigger the infection of SARS-CoV and MERS-CoV. Studies have shown that inhibiting the enzyme activity of TMPRSS2 can prevent some coronaviruses from entering host cells46. As a possible target for anti-viral drug discovery, the virtual screen results (shown in Supporting excel files) predicted many anti-bacterial drugs (pivampicillin, hetacillin, cefoperazone and clindamycin) and anti-virus natural compounds (phyllaemblicin G7, neoandrographolide, kouitchenside I), etc. to be potential TMPRSS2 inhibitors.
3.3. Virtual screening and target identification of common anti-viral drugs
In order to further verify the screening results of ZINC drug database and utilize the current resource of anti-viral drugs immediately, we constructed a database of 78 anti-viral drugs for deep calculation, including compounds already on the market and currently undergoing clinical trials to treat SARS-CoV-2 infections. The compounds were docked with 19 constructed targets of SARS-CoV-2 and two human targets to predict their possible targets, and also to search possible binding partners of certain important targets. Special attentions were paid to the drugs currently in clinical trials. All results of calculated docking scores were listed in the Supporting excel file (anti-virals_vs_targets_scores), significant scores (score<–32, mfScore<–110) were highlighted in brighten yellow.
Remdesivir (GS-5734), a nucleoside analogue, is an RdRp inhibitor. It can inhibit virus by inhibiting synthesis of viral nucleic acid and has not yet been approved for marketing in any country. On January 31 of this year, the New England Journal of Medicine reported the diagnosis and treatment of the first SARS-CoV-2 patient in the United States47. Remdesivir showed some potential in the treatment of the first patient with novel coronavirus infection. On Vero E6 cells, the EC50 of remdesivir for SARS-CoV-2 is 0.77 μmol/L, and the selection index SI is greater than 12948. Remdesivir is currently undergoing a randomized, double-blind, controlled phase III clinical study in China. Our molecular docking results show that the potential targets of remdesivir is Nsp3b (score = –36.5), RdRp (mfScores = –112.8), E-channel (mfScore = –125.1), and TMPRSS2 (score = –36.23, mfScores = –109.4).
For RdRp, from generated docking model, remdesivir could bind to the RNA-binding channel of the SARS-CoV-2 RdRp, and the binding mode and site were highly similar to that of coxsackievirus B3 (CVB3) RdRp inhibitor GPC-N11449.The binding pocket of remdesivir was in the bottom of the RNA template channel, which position was for the acceptor template nucleotide (Fig. 7A and B). The compound was well fitted with the shape of the pocket, where it formed three hydrogen bonds with Asn497, Arg569 and Asp684. In addition, hydrophobic interactions with Leu576, Ala685 and Tyr687 may further direct the favorite conformation of remdesivir (Fig. 7C).
Figure 7.
Low-energy binding conformation of remdesivir bound to SARS-CoV-2 RdRp generated by molecular docking. (A) Remdesivir was fitted in the bottom of the RNA template channel (top view). (B) Remdesivir was fitted in the bottom of the RNA template channel (bottom view). (C) Detailed view of remdesivir binding with RdRp (Supporting PDB file SARS_CoV-2_RdRp_homo_Remdesivir.pdb).
Interestingly, remdesivir was predicted to bind with the target TMPRSS2 with low binding energy for both score and mfScore. As shown in Fig. 8A, remdesivir was bound in a relatively positively-charged allosteric pocket which is far away from the enzyme active center. Asn84 and Arg405 formed two hydrogen bonds with the phosphate groups of the compound. Weak hydrophobic interaction between the pyrrolotriazine ring of remdesivir with Tyr131 and Try401, and side chains of some polar amino acids may further stabilize the compound conformation (Fig. 8B).
Figure 8.
Low-energy binding conformation of remdesivir bound to human TMPRSS2 generated by molecular docking. (A) Remdesivir was fitted in an allosteric pocket far away from the enzyme active center (bottom). (B) Detailed view of remdesivir binding with TMPRSS2 (Supporting PDB file HS_TMPRSS2_homo_Remdesivir.pdb).
Lopinavir and ritonavir have been marketed in China and are mainly used to treat HIV-1 infection in adults and children over 2 years of age. In vitro studies have shown that lopinavir and ritonavir can inhibit the replication of MERS-CoV and SARS-CoV to exert anti-viral effects. The drug is listed as a recommended drug in the Anti-viral Drugs section of the New Coronavirus Infected Pneumonia Diagnosis and Treatment Program. The molecular docking results showed that ritonavir's possible target is Nsp3c or E-channel with the mfScores of −152.100 and −277.769, respectively. Lopinavir's possible target is Nsp3b, Nsp3c, helicase, NRBD or E-channel with the mfScores of −158.050, −189.140, −114.018, −171.127, and −221.785, respectively.
Arbidol is a broad-spectrum anti-viral drug, mainly for the treatment of upper respiratory tract infections caused by influenza A and B viruses, etc. In recent years, many studies have proven its effectiveness against both SARS-CoV and MERS-CoV. Arbidol hydrochloride can block virus replication by inhibiting the fusion of the lipid membrane of the virus with the host cells. Compared with the untreated control group, arbidol can effectively inhibit coronavirus up to 60 times at a concentration of 10–30 μmol/L, and significantly inhibit the virus's pathological effects on cells. The docking results of arbidol with the possible drug targets of the new coronavirus showed that it may interact with Nsp7_Nsp8 complex, Nsp14, Nsp15, E-channel, or Spike with the mfScores of −136.087, −118.253, −118.253, −117.879, and −145.125, respectively.
Darunavir is an HIV-1 protease inhibitor that selectively inhibits the cleavage of HIV-encoded Gag-Pol polyprotein in virally infected cells, thereby preventing the formation of mature infectious virus particles. At a concentration of 300 μmol/L, darunavir can significantly inhibit virus replication, and the inhibition efficiency is 280 times compared with the untreated group. Our docking results showed that the possible targets of darunavir are Nsp3c, PLpro, E-channel or Spike proteins with the mfScores of −126.149, −110.759, −157.184, and −111.865, respectively.
Favipiravir, a broad-spectrum anti-viral drug, is used to treat flu. The Shenzhen Health Commission has now initiated clinical studies on the use of favipiravir to treat SARS-CoV-2 infections. While, the scores of favipiravir docking with the targets in our virtual screening are relatively low.
Chloroquine phosphate has been used in the treatment of malaria since the 1940s and later in rheumatoid arthritis. Chloroquine has been reported in some earlier studies to have direct anti-viral effects, such as inhibiting flavivirus, retrovirus (such as HIV), and many coronaviruses. Recent study showed that with EC50 of 1.13 μmol/L and SI greater than 88, chloroquine can effectively inhibit SARS-CoV-2 in the cell level. Its efficiency in the human body for SARS-CoV-2 infection has not yet been clinically proven. Chloroquine phosphate was turned into chloroquine in the body to play therapeutic effect. Our docking results showed that the possible target of chloroquine is Nsp3b or E-channel with the docking mfScores of −130.355 and −107.889, respectively.
We also performed the longitudinal analysis on the drugs against 21 targets, and the results showed that only tenofovir, disoproxil, fumarate, and beclabuvir may bind to Nsp1. Fewer compounds were predicted to act on some targets, such as Nsp1, Nsp3e, Nsp9, Nsp10, Nsp16, NRBD, CRBD, ORF7a, and TMPRSS2, showing that these selected anti-viral drugs are unlikely to acting on the above targets of the new coronavirus, which provides a meaningful reference for our future research.
For other targets, such as Nsp3b, Nsp3c, Nsp7_Nsp8 complex, Nsp14, Nsp15, PLpro, 3CLpro, RdRp, helicase, E-channel, Spike and ACE2, more anti-viral drugs were predicted to bind with them, especially E-channel, RdRp, 3CLpro and PLpro, indicating that these targets are more likely to be useful for the discovery of SARS-CoV-2 therapeutic drugs from known anti-virals, and should be the focus of our subsequent research.
4. Discussion
The ongoing SARS-CoV-2 epidemic makes us painfully realize that our current options for treating life-threatening zoonotic coronavirus infections are very limited. Although the outbreaks of SARS in 2003 and MERS-CoV in 2012 triggered extensive research efforts, there are currently no drugs that can treat any zoonotic coronavirus. The transient nature of this epidemic is one of the major reasons why no prototype coronavirus inhibitor has progressed to the (early) preclinical stage to date. As with the SARS virus 17 years ago and the current SARS-CoV-2, emerging coronaviruses in the future may continue to pose a threat to global public health. Therefore, finding broad-spectrum inhibitors that may reduce the effects of human coronavirus infection remains a challenging research focus. Given the time-consuming nature of anti-viral drug development and registration, existing treatments for other diseases may be the only fastest treatment option for emerging infectious diseases. For most of these drugs that have been prepared, the medication has sufficient experience and dosage, and their safety and ADME situation are well known.
In this study, based on the results of bioinformatics analysis, 20 homology structures of 18 SARS-CoV-2 proteins and one human protein were built, plus human ACE2 structure, totally 21 targets were setup for high throughput virtual ligand screening. The special double scoring system ICM-Pro soft allowed us to evaluate the docking results more accurately. The score of ICM software is calculated by the overall empirical function of predicted physical interaction, whiles the mfScore (mean force Score) is computed by the knowledge-based potential functions derived from statistics of ligand–receptor complex in PDB50,51.
From protein–protein docking results, we can deduce that SARS-CoV-2 Spike protein have strong binding affinity to human ACE2, although weaker than that of SARS-CoV Spike protein. Unexpectedly, Bat-CoV RaTG13 Spike protein only has slightly weaker binding affinity with ACE2 compared to SARS-CoV-2. It is speculated that the virus may be capable of infecting humans directly, or it is close. If SARS-CoV-2 evolved from Bat-CoV RaTG13, there may be no more than one or two host evolutions involved. But there are no published articles that directly related to intermediate hosts yet. According to the most recent research paper on the clinical characteristics of SARS-CoV-2 infection52, SARS-CoV-2 can spread rapidly by human-to-human transmission, this was consistent with observed protein–protein docking results of SARS-CoV-2 Spike RBD and human ACE2, for both of ours and previous prediction25. However, the fast growing incidents of SARS-CoV-2 pneumonia and related deaths implied the weaker binding of SARS-CoV-2 Spike with human ACE2 compared to SARS Spike may not fully explained the current situation of this epidemic. There must have some other receptors of SARS-CoV-2 or infection facilitating mechanisms in the human host, or SARS-CoV-2 Spike-RBD may take other lower energy binding conformation different from current SARS-CoV Spike-RBD conformation found in the crystal structure, probably by induce fitting. These need to be further investigated.
To search potential coronavirus therapeutic drugs as soon as possible, we first screened potential compounds from a ZINC drug database (2924 compounds) and a small in-house database of natural products (about 1066 compounds). A series of clinical drugs and natural products with anti-viral, anti-bacterial and anti-inflammatory effects exhibited high binding affinity to different target proteins, indicating their potential utility for treating SARS-CoV-2.
We have identified a number of compounds that might have anti-viral activity from the approved drugs library, such as anti-virus drugs (ribavirin, valganciclovir and thymidine), anti-bacterial drugs (cefpiramide, sulfasalazine, phenethicillin, lymecycline, demeclocycline, doxycycline, oxytetracycline and tigecycline), anti-asthmatic drugs (montelukast, fenoterol and reproterol), and hepatoprotective drug silybin. The original pharmacological actions of these drugs could be helpful for the therapy of viral infection pneumonia. The natural products, such as flavanoids like neohesperidin, hesperidin, baicalin, kaempferol 3-O-rutinoside and rutin from different sources, andrographolide, neoandrographolide and 14-deoxy-11,12-didehydroandrographolide from A. paniculata, and a series of xanthones from the plants of Swertia genus, with anti-virus, anti-bacteria and anti-inflammation activity could effectively interact with these targets of SARS-CoV-2. Therefore, the herbal medicines containing these compounds as major components might be meaningful for the treatment of SARS-CoV-2 infections.
For ACE2 target, although several compounds could bind with ACE2 through virtual screening in our studies, no compound was found to bind with the contact surface of ACE2–Spike complex, suggesting that these compounds are only the inhibitors of ACE2 enzyme activities, rather than inhibitors of ACE2 driven virus infections. Just like what described in recently published research53, most of selected compounds are also unable to bind with the contact surface of ACE2–Spike complex. Actually, these potential ACE2 inhibitors may not be suitable to use as drugs for treating SARS-CoV-2 infection because the poor prognosis would be induced by the inhibition of ACE2 enzyme activities, for ACE2 was considered as a protective factor of lung injury54.
For those targets which are difficult to find direct inhibitors, or non-druggable targets, just like Nsp1, Nsp3b, Nsp3c, and E-channel, etc., currently popular PROTAC technology may be a good strategy to degrade these proteins and then inhibit the virus. The potential binding compounds found in this study for these targets might be a good start point.
For Spike protein, we found only one compound, natural hesperidin was targeting the binding between Spike RBD and human ACE2. However, not like the ACE2 binding compounds, non-interface binding compound may still meaningful applications, considering that the fusion of CoVs membrane with host cell membrane need the big conformational change of remained Spike part after RBD removal55. Any small molecule bound to Spike at this time may interfere the re-folding of Spike therefore inhibits the viral infection process. Furthermore, small molecule that can target any part of Spike protein may be a good start point to design PROTAC based therapy.
Also, we dock existing anti-viral drugs with our targets, analyze the possible targets of each anti-viral drug horizontally, and analyze the drugs that may interact with 21 targets vertically. We analyzed 21 targets based on the docking results and found that Nsp3b, Nsp3c, Nsp7_Nsp8 complex, Nsp14, Nsp15, PLpro, 3CLpro, RdRp, helicase, E-channel, Spike and ACE2 are more likely to be therapeutic targets of anti-viral drugs. The three targets Nsp3b, Nsp3c, and E-channel are screened more anti-viral drugs. This may be due to the model problem because of flexible small protein (Nsp3b and Nsp3c) or partial model (E-channel). Whether the screened anti-viral drugs really work on these targets needs further verification. We also do not recommend the application of new coronavirus pneumonia to compounds for which no target has been predicted.
The triphosphate nucleotide product of remdesivir, remdesivir-TP, competes with RdRp for substrate ATP, so it can interfere with viral RNA synthesis. Our docking results show that remdesivir-TP binds to SARS-CoV-2 RdRp, with a score of −112.8, and the docking results are consistent with its original anti-viral mechanism, so we think remdesivir may be good in treating SARS-CoV-2 pneumonia. In addition, remdesivir also predicted to bind with the human TMPRSS2, a protein facilitating the virus infection, this is a new discovery and provides ideas for subsequent research.
Chloroquine phosphate has shown better anti-SARS-CoV-2 effects in recent studies, but this drug has no clear target of action. In our docking results, chloroquine phosphate is predicted to possibly combine with Nsp3b and E-channel. But we need to do further experiments to verify this conclusion.
In response to the recently reported anti-AIDS drugs lopinavir and ritonavir tablets, which have a poor effect on the treatment of novel coronavirus pneumonia and have toxic side effects, we analyzed it in conjunction with the docking results. The molecular docking results show that ritonavir's possible target is Nsp3c or E-channel. Lopinavir's possible target is Nsp3b, Nsp3c, helicase, NRBD or E-channel. Some of these targets (such as Nsp3b, Nsp3c, E-channel) may be false positives due to the model inaccuracy for small flexible protein or partial model. For both lopinavir and ritonavir, we did not observe possible binding to major targets like 3CLpro, PLpro, RdRp, and so on. This docking result implies lopinavir and ritonavir tablets may not be suitable for treatment of SARS-CoV-2 infections.
The results of the entire article are based on computer virtual screening. We have not conduct further in vivo and in vitro anti-viral experiments yet, because we want to share our results with scientists in anti-SARS-CoV-2 research as soon as possible. Our subsequent research will try to solve the three-dimensional structures of all 24 proteins of SARS-CoV-2 and their drug complexes, providing more target information for drug intervention and long-term drug design, perform in vivo and in vitro evaluations for candidate drugs obtained in this study, and prepare for clinical trial applications.
Acknowledgments
We acknowledge support from National Mega-project for Innovative Drugs (grant number 2019ZX09721001-004-007, China), National Natural Science Foundation of China (NSFC, grant numbers U1803122, 81773637, 81773594, and U1703111), and the Fundamental Research Fund for the Central Universities (HUST COVID-19 Rapid Response Call, No. 2020kfyXGYJ037, China). This article is dedicated to the heroic medical workers who are fighting in the front line of anti epidemic and the people of Wuhan who made great sacrifices.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.02.008.
Contributor Information
Xingzhou Li, Email: xingzhouli@aliyun.com.
Mengzhu Zheng, Email: mengzhu_zheng@hust.edu.cn.
Lixia Chen, Email: syzyclx@163.com.
Hua Li, Email: li_hua@hust.edu.cn.
Author contributions
Hua Li, Lixia Chen, Mengzhu Zheng and Xingzhou Li designed the research, performed virtual screening and revised the manuscript. Canrong Wu, Yang Liu, and Yueying Yang performed the bioinformatics analysis, analyzed the virtual screening experiment data and drafted the manuscript. Peng Zhang, Wu Zhong, Yali Wang, Qiqi Wang, Yang Xu, and Mingxue Li checked the structures and made the Excel forms. All authors have read and approved the final manuscript.
Conflicts of interest
The authors claim that the researchers in this study have no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.-W., Wong B.H.L. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusken C.B., Haagmans B.L., Müller M.A., Gutierrez C., Godeke G.-J., Meyer B. El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch B-J, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MPG. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. China Novel Coronavirus I, Research T. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omrani A.S., Saad M.M., Baig K., Bahloul A., Abdul-Matin M., Alaidaroos A.Y. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis. 2014;14:1090–1095. doi: 10.1016/S1473-3099(14)70920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agrawal A.S., Garron T., Tao X., Peng B.H., Wakamiya M., Chan T.S. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol. 2015;89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan J.F., Chan K.H., Kao R.Y., To K.K., Zheng B.J., Li C.P.Y. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East Respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;14:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abagyan R., Totrov M., Kuznetsov D. ICM—A new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- 24.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 27.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING–TRAF3–TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan L., Chen Z., Song S., Wang S., Tian C., Xing G. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J Biol Chem. 2015;290:3172–3182. doi: 10.1074/jbc.M114.619890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S.W., Wang C.Y., Jou Y.J., Huang S.H., Hsiao L.H., Wan L. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.-P. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu C.K., Gadthula S., Chen X., Choo H., Olgen S., Barnard D.L. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-CoV) Antivir Chem Chemother. 2006;17:285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- 37.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shum K.T., Tanner J.A. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008;9:3037–3045. doi: 10.1002/cbic.200800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem Biophys Res Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. 2006;103:12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan K., Huang C., Lokugamage K., Kamitani W., Ikegami T., Tseng C.T.K. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forni D., Cagliani R., Mozzi A., Pozzoli U., Al-Daghri N., Clerici M., Sironi M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J Virol. 2015;89:11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holshue M.L., De Bolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H. Washington State 2019-nCoV Case Investigation Team. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Linden L., Vives-Adrián L., Selisko B., Ferrer-Orta C., Liu X., Lanke K. The RNA template channel of the RNA dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PLoS Pathog. 2015;11:e1004733. doi: 10.1371/journal.ppat.1004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muegge I., Martin Y.C. A general and fast scoring function for protein ligand interactions: a simplified potential approach. J Med Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 51.Neves M.A., Totrov M., Abagyan R. Docking and scoring with ICM: thebenchmarking results and strategies for improvement. J Comput Aided Mol Des. 2012;26:675–686. doi: 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H., Du Q. Potential natural compounds for preventing 2019-nCoV infection. Preprints. 2020 [Google Scholar]
- 54.Ye R., Liu Z. ACE2 exhibits protective effects against LPS-induced acute lung injury in mice by inhibiting the LPS-TLR4 pathway. Exp Mol Pathol. 2019;113:104350. doi: 10.1016/j.yexmp.2019.104350. [DOI] [PubMed] [Google Scholar]
- 55.Heald-Sargent T., Gallagher I. Ready. Set, Fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.