Abstract
Helicobacter pylori colonizes human stomach mucosa and its infection causes gastrointestinal diseases with variable severity. Bacterial infection stimulates autophagy, which is a part of innate immunity used to eliminate intracellular pathogens. Several intracellular bacteria have evolved multipronged strategies to circumvent this conserved system and thereby enhance their chance of intracellular survival. Nonetheless, studies on H. pylori have produced inconsistent results, showing either elevated or reduced clearance efficiency of intracellular bacteria through autophagy. In this review, we summarize recent studies on the mechanisms involved in autophagy induced by H. pylori and the fate of intracellular bacteria.
Keywords: Helicobacter pylori, Autophagy, Virulence factor, Intracellular survival
1. Introduction
The immune system of healthy individuals comprises potent mechanisms to protect the body against microbial infections. Autophagy is an evolutionarily conserved process to eliminate pathogens [1]. This catabolic process sequesters cytosolic components including misfolded proteins, injured organelles, and intracellular pathogens through a lysosome-dependent pathway [2]. Accumulated evidence shows that successful pathogens can hijack autophagy for their own replication [3]. This review focuses on a chronic gastric pathogen, Helicobacter pylori, which often inhabits mucosal layers and causes dyspeptic symptoms of varying severity. Findings of previous studies indicate that autophagy plays an important role in host defense, lessening H. pylori burden [[4], [5], [6], [7]]. However, H. pylori is a facultative intracellular pathogen and is capable of invading and surviving not only in non-phagocytic cells but also in professional phagocytes [[8], [9], [10], [11]]. In addition, H. pylori is detected in double-layered autophagosomes, indicating that autophagosomes serve as a peculiar niche for H. pylori multiplication to sustain intracellular infections [8,12,13]. However, the underlying mechanisms through which H. pylori coopts the autophagy machinery to affect bacterial clearance efficiency in host cells remain to be elucidated.
2. H. pylori virulence factors
2.1. Adhesion molecules
Microorganisms use bacterial adhesins to bind to host cells, protecting themselves from mechanical attack, such as acidic pH and mucosal liquids [14]. Genomic analysis reveals that H pylori contains more than 30 outer membrane protein (omp) genes including the Helicobacter OMPs (Hop) and the hop-related group (hor) [15]. The two most important adhesins in H. pylori are Lewis b (Leb) blood group antigen-binding adhesin (BabA) and sialyl Lewis X antigen-binding adhesin (SabA), and both these adhesins are Hop proteins [[16], [17], [18]]. Thus, the host and pathogen interact partly through the binding of adhesins to specific carbohydrate moieties of the gastric epithelium, promoting infection and inflammatory processes in the gastrointestinal tract.
2.2. Vacuolating cytotoxin A (VacA)
VacA, which causes massive vacuolation in cultured cells, was first observed in culture supernatants of toxic H. pylori isolates [19]. Purification and subsequent studies on this unique toxin showed that VacA is synthesized as a 140-kDa polypeptide precursor, which undergoes trimming to yield a mature 95-kDa toxin and is secreted via an auto-transporter encoded in its C-terminal domain [[20], [21], [22]]. It can assemble into water-soluble oligomeric forms including a single layer structure consisting of hexamers/heptamers and a double-layered assembly [23]. It is interesting that VacA disassembles at acidic pH, interacts with the lipid membrane and reassembles into a pore-forming hexameric structure in membranes, enabling the transport of chloride ions [22,24]. This unique channel-formation activity contributes to vacuolation in late endosomal and lysosomal compartments as well as increases in mitochondrial membrane permeability which leads to cytochrome c release [22,23].
Several surface molecules have been reported as VacA receptors. Receptor-like protein tyrosine phosphatase β (RPTPβ) binds to VacA through its terminal sialic acid domain in gastric epithelial cells [25,26]. RPTPα also serves as a functional VacA receptor in human kidney tumor cell G401 that lacks RPTPβ [27]. The interaction of VacA and RPTPα activates Src phosphorylation, resulting in CagA phosphorylation at Tyr972 in AZ-521 cells [28]. Notably, VacA is able to target the membrane of multiple cell types including gastric epithelial cells, T cells, and parietal cells. Therefore, VacA with high pore-forming activity is associated with increased disease severity [29]. It has been described that removing the membrane-associated region of VacA blocks its vacuolating activity and prevents VacA from integrating into the inner-mitochondrial membrane [30]. Recent evidence also shows that VacA binds to low-density lipoprotein receptor-related protein 1 (LRP1) and is involved in triggering autophagy as discussed below [31].
2.3. CagA and type IV secretion system (T4SS)
CagA is the gene product encoded within the 40-kb cag-pathogenicity island (cag-PAI) which also includes 31 additional genes that encode the T4SS [32]. The mechanisms that pathogens have exploited in order to introduce the virulence factors into the host cells. In another study conducted, H. pylori cag-T4SS interacts with the β1 integrin of the host cells in an arginine-glycine-aspartate (RGD)-independent manner, which initiates effector protein translocation [33]. When H. pylori contacts with the host membrane, CagA interacts with the externalized phosphatidylserine to facilitate its entry into target cells, which is important for the pathophysiological activity of CagA [34]. In addition to β1 integrin, a recent study by Königer et. al. describes that HopQ specifically interacts with human carcinoembryonic antigen-related cell adhesion molecule family (CEACAMs), facilitating CagA translocation into host cells [35]. These studies suggest that H. pylori exploits multiple ways to exert CagA translocation into the host cells. The translocated CagA can be phosphorylated and interacted with several host signaling proteins, thus interfering with host cell signaling [36]. It is known that CagA translocation and phosphorylation are mediated through cholesterol-rich microdomains present on the cytoplasm membrane (also referred as lipid rafts) [34,[37], [38], [39]]. Lipid rafts are important for efficient T4SS-mediated CagA translocation, inducing an elevated level of the cell hummingbird phenotype and increased IL-8 production [37]. The host membrane lipid phosphatidylserine plays a crucial role in CagA delivery and localization [34,40]. Moreover, simvastatin, a HMG-CoA reductase inhibitor, which reduces the level of cellular cholesterol and decreases translocation and phosphorylation of CagA, indicating that cholesterol is crucial for CagA-mediated actions [39].
2.4. Cholesterol-α-glucosyltransferase (CGT)
The O-Glycans of the human gastric mucosa diminish the levels of H. pylori infection by inhibiting the bacterial cholesterol-modifying enzyme, cholesterol-α-glucosyltransferase (CGT) encoded by the type 1 capsular polysaccharide biosynthesis protein J (capJ) [41]. CGT catalyzes the conversion of host cholesterol into cholesteryl-α-glucoside (αCG) [41]. Further modification of αCG yields cholesteryl-6′-O-acyl-α-glucoside, cholesteryl-6′-O-phosphatidyl-α-glucoside, and cholesteryl-6′-O-lysophosphatidyl-α-glucoside, which constitute a large proportion of the glycolipids in the cell wall [41]. Cholesterol glucosylation of H. pylori by CGT, interestingly, promotes H. pylori evasion of immune defenses [42]. We have previously shown that knockout of capJ (ΔCapJ), which abolishes the production of cholesteryl glucosides (CGs), greatly decreases the degree of T4SS-induced activities, including CagA translocation/phosphorylation, subsequent signaling events, the scattering hummingbird morphology in AGS cells, and IL-8 secretion [43]. This is mainly because CGs of the wild-type H. pylori-infected AGS cells are essential for reorganizing the lipid-raft membranes for efficient T4SS function [43]. Furthermore, cholesterol-glucosylated H. pylori is largely internalized via a lipid-raft-dependent pathway, which delays the maturation of phagosomes and facilitates the evasion of macrophages [44], suggesting a role in interfering with autophagy, which is discussed in a later section [45]. H. pylori harboring CGT, inhibits interferon gamma-induced signaling, which avoids host inflammatory responses [46]. Noticeably, CGs also contribute to cell wall integrity, morphology, and antibiotic resistance [47]. Collectively, these findings highlight the multiple strategies employed by H. pylori for persistent colonization.
2.5. Other virulence factors
Other virulence factors are also engaged in H. pylori-induced pathogenesis, such as urease, flagella, and heat-shock proteins (Hsp). Infection of H. pylori upholds the periplasmic pH to adapt to the acidic environment of stomach by means of urease [48]. The increased levels of ammonia from hydrolysis of urea by urease as well as the metabolites from recruited neutrophils disrupts the gastric epithelium, which synergistically facilitates the development of gastric malignancies [49,50]. In addition, H. pylori possesses four to eight unipolar flagella that are important in bacterial colonization and induce inflammatory response [51]. The flagellar sheath of H. pylori provides the protection of acid-labile flagellar structure from the attack of the acid environment [52]. Additionally, H. pylori harbors two main heat-shock proteins, GroES-like HspA (Hsp10), and GroEL-like HspB (Hsp60). The regulation of heat-shock proteins expression by H. pylori is crucial for bacteria to adapt and survive in the hostile environment of the stomach [53].
3. Autophagy in bacterial infection and pathogenesis
3.1. Composition, structure, and functions of autophagy
Autophagy is an intracellular degradation process by which cytoplasmic constituents are degraded by the lysosome [54]. The three major forms of autophagy include microautophagy, macroautophagy, and chaperone-mediated autophagy [55,56]. Microautophagy and chaperone-mediated autophagy are directly recruited to a lysosome, which engulfs small portions of cytosol and receives chaperone-associated cargo [57]. Macroautophagy can sequester cytosolic molecules, including damaged organelles or pathogens, in the autophagosome [57]. In mammalian cells, macroautophagy occurs in the cytoplasm; the organelles are surrounded by a phagophore or cytoplasmic double-membrane-bound structure, which start growing at both ends to create an autophagosome [58,59]. The autophagosome subsequently fuses with a lysosome, forming a single, large, and membrane-surrounded vesicle (autophagolysosome); both the membrane and its contents are then degraded by lytic enzymes [60,61]. Autophagy has been demonstrated to play crucial roles in biological processes, such as cell protection, response to starvation, recycling nutrients from digested organelles and macromolecules by removing damaged organelles and aberrantly folded proteins, diseases, and host defenses [62].
3.2. Bacterial infection and the autophagy pathway
The large-scale degradation of invading pathogens by autophagy, called xenophagy, has been commonly used to describe the host defense response to effectively eliminate the invading pathogens including intracellular bacteria [63]. Various studies have been conducted on different pathogens and how their virulence factors manipulate the autophagy pathways of their hosts to promote their intracellular survival and multiplication. Examples of such pathogens include Mycobacterium tuberculosis, Streptococcus pyogenes, Shigella flexneri, Salmonella typhimurium, Listeria monocytogenes, and Legionella pneumophila, all of which have been extensively studied to elucidate the strategies they use to alter the host autophagy pathway (Table 1 ). The details of the underlying mechanism will be covered later.
Table 1.
Bacterial utilization of host autophagy for their intracellular survival and pathogenesis.
| Bacteria growth restricted by autophagy components | Bacteria evades being targeted by host’s autophagy | Bacteria exploit autophagy components for its replication and pathogenesis |
|---|---|---|
| Mycobacterium tuberculosis [97] | Shigella flexneri [119] | Brucella abortus [120] |
| Mycobacterium marinum [121] | Listeria monocytogenes [122] | Coxiella burnetiid [123] |
| Group A Streptococcus [124] | Burkholderia pseudomallei [125] | Legionella pneumophila [126] |
| Salmonella enterica serovar Typhimurium [127] | Staphylococcus aureus [128] | |
| Francisella tularensis [129] |
3.3. Adhesion and invasion of cells
Pathogenic microbes exhibit numerous approaches for evading immune defenses and developing a protected niche within the host. Therefore, the interplay between the initial entry of these pathogenic microbes into the host cells and the response of the host cells plays a critical role in determining the fate of the microbes. Wang and Hajishengallis previously reported that Porphyromonas gingivalis relies on the lipid rafts of the macrophages to enhance its intracellular bacterial load and survival [64]. There is mounting evidence that shows parasites [[65], [66], [67]], viruses [68,69], and prions [70,71] target the lipid rafts of the host cells, which benefits infections. Lipids cluster together to form microdomains in cell membranes, i.e., lipid rafts. Lipid rafts composed of high concentrations of cholesterol and glycosphingolipids [72], that modulate a wide array of cellular processes such as signal transduction [73], membrane trafficking [74,75], and cytoskeletal organization [76,77].
Various studies have been shown that lipid rafts are common platforms for bacteria to adhere and invade to host cells [78]. A study conducted by Shin et al. demonstrated that lipid rafts serve as important docking points for bacteria that express FimH [79]. Furthermore, these bacteria reside in vacuoles containing the lipid rafts, which is important for promoting their intracellular survival. H. pylori also exploits lipid rafts for initial adhesion and invasion. A study conducted by Wunder et al. demonstrated that H. pylori is auxotrophic for cholesterol and is able to extract cholesterol from the lipid rafts of the host, resulting in immune evasion [42]. This study also corroborates our previous findings that lipid rafts of the epithelial cells play a key role in H. pylori internalization [37]. In addition to bacteria, viruses also rely on lipid rafts for attachment and invasion. The coronavirus, infectious bronchitis virus is reliant on the host’s lipid rafts for attachment and thereby promoting viral infection [80].
3.4. Cytoplasmic trafficking
Upon invasion of host cells by the pathogens, they can modulate the host cell trafficking machinery to promote evasion from the host innate immune response through Rab GTPases which are proteins responsible for vesicular trafficking, at least in part [81]. Rab proteins, when activated, interact with a large number of Rab effector proteins to regulate different cellular functions such as membrane trafficking and intracellular signaling [82]. Rab proteins reside in intracellular membrane compartments of the plasma membrane, mitochondria, and the nucleus and are involved in a large number of cellular functions [83]. Above all, Rab proteins are known for their roles in the endocytic/exocytic pathways, the retrograde/anterograde secretory pathways, and membrane recycling [84].
As xenophagy involves the trafficking of pathogens in the host cells to the lysosomes for degradation, various studies have demonstrated that different pathogens target different Rab proteins to promote their intracellular survival. For instance, M. tuberculosis modifies Rab22, which is associated with the maturation of phagosome [85], and promotes the accumulation of Rab14 in M. tuberculosis containing phagosomes, preventing fusion of the phagosome with lysosome [86].
While Rab proteins serve as one of the key targets for pathogens, other molecules are also triggered by pathogens to manipulate the host’s cytoplasmic trafficking. Salmonella secretes secretor effector protein (SifA) via its type 3 secretion system (T3SS) from the Salmonella-containing vacuoles to the cytoplasm [87]. SifA then binds to kinesin-interacting protein (SKIP), a host protein, which downregulates the recruitment of kinesin to the bacterial vacuoles, thereby promoting the membrane dynamics of vacuoles [88]. Upon invading the host cells, a few pathogens escape from the vacuoles and replicate in the host cytoplasm. These pathogens express factors that initiate the process of actin polymerization, facilitating their propagation in the cells. For example, L. monocytogenes secretes the virulence factor listeriolysin O (Llo), a pore-forming exotoxin which degrades the membrane of internalized vacuoles in the infected host cells [89], eventually resulting in vacuolar dissolution, and thereby promoting intracellular survival [90]. Furthermore, the pathogen is able to translocate across the infected cell wall to spread and invade adjacent cells through actin-based motility [91]. This phenomenon is attributable to the presence of the protein actin assembly-inducing protein (ActA) on the bacterial surface. ActA acts as a nucleator, which induces actin polymerization allowing the bacteria to move forward in the host cells [92].
3.5. Bacterial survival or elimination in host cells
Upon entering host cells, several pathogens evade phagosome fusion with the degradative lysosome through different mechanisms thereby increasing their survival and bacterial load in the host cells. One of the mechanisms by which pathogens promote their intracellular survival is by arresting phagosome maturation. To achieve this, pathogens are capable of obstructing phagosomal lipid metabolism. One example is M. tuberculosis, which produces a phosphatase identified as secreted acid phosphatase (SapM). SapM is capable of converting PI(3)P to PI, inhibiting late phagosome endosome fusion which in turn inhibits phagosomal maturation [93]. Another example is Salmonella enterica, which injects effector sigma D factor (SigD) into host cells, leading to the modulation of phosphatidylinositol 3-kinase (VPs34) and increasing vacuolar PI(3)P, thereby averting phagosome maturation and preventing fusion with a lysosome [94].
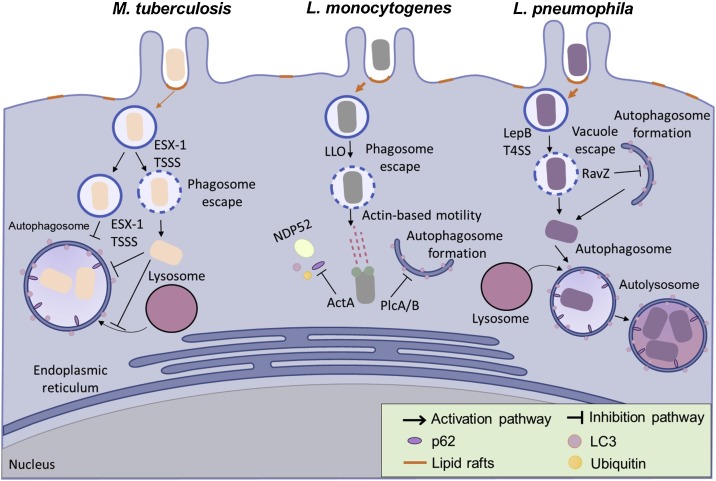
A second method used by pathogens to evade host cell degradation is escaping from the host cell phagosome. Studies on Mycobacteria species have shown that they activate host cytosolic phospholipase A2 to assist the microbe in escaping the phagosome [95]. This clearly demonstrates the highly flexible adaptation strategy of Mycobacteria for surviving in host cells (Fig. 1 ). Recently, another survival strategy employed by the pathogen has been discovered. While employing this strategy, the pathogen directly interferes with the maturation of the autolysosome. In a very recent study conducted by Zhang et al., H. pylori was able to inhibit lysosomal function and suppress the process of autolysosomal maturation, thereby circumventing autophagic degradation [96].
Fig. 1.
Bacteria hijack lipid rafts to induce autophagy as part of their infectious strategy. Mycobacterium tuberculosis is capable of surviving in the phagosome after being internalized into the host cell by preventing phagosomal maturation and inhibiting autophagosome fusion with lysosome. M. tuberculosis is able to survive and replicate in the phagosome. However, cytosolic translocation of M. tuberculosis can occur when it expresses the early secretory antigenic target of M. tuberculosis (ESAT-6) secretion system (ESX-1) and type VII secretion system (TSSS). Listeria monocytogenes employs various virulence factors in its escape from autophagy. Upon escaping from the phagosome using its pore forming toxin (Listeriolysin O), L. monocytogenes expresses another virulence factor ActA to inhibit the recruitment of p62 and NDP52 to its surface. Another group of virulence factors, two phospholipases C (PlcA/B) then prevent the formation of the autophagosomal membrane. In Legionella infections, L. pneumophila escapes from the vacuole and enters the host’s cytosol via LepB using a type IV secretion system. The cytosolic L. pneumophila is subsequently recognized by the autophagic machinery but delays phagocytosis by using its effector protein RavZ. In the autophagosome L. pneumophila also delays the fusion of the autophagosome with lysosome to allow it time to develop into an acid-resistant form. This acid-resistant form of L. pneumophila can then replicate in the acidic autophagolysosome.
In contrast to the abovementioned examples where pathogens subvert autophagy to promote their intracellular survival, some pathogens that are eliminated by activating autophagy. Gutierrez et al. demonstrated that activation of autophagy by starvation or treatment with rapamycin triggers an effective clearance of M. tuberculosis in macrophages [97]. Similarly, using rapamycin to activate autophagy led to an enhancement of targeting Burkholderia cepacia to lysosomal compartments, resulting in better prognosis in patients with cystic fibrosis [98]. Notably, a new LC3-associated phagocytosis (LAP) autophagic machinery has been recently identified [99]. The key function of LAP is to enhance fusion of phagosomes and lysosomes, which promotes pathogen killing [100]. However, the detailed mechanism by which LAP enhances phago-lysosome fusion remains to be studied.
4. H. pylori modulates autophagy
Autophagy as an intracellular defense to eliminate pathogens can be induced by H. pylori in both gastric epithelial cells [13,96,101,102] and professional phagocytes [103,104]. Recently accumulating evidence has shown that H. pylori exploits host autophagy to survive within cells and even multiply [10,105,106]. However, the mechanisms vary depending on the host cells or bacterial strains. In this section, we summarize recent studies conducted to elucidate the different mechanisms of autophagy of H. pylori in gastric epithelial cells and professional phagocytes.
4.1. H. pylori infects gastric epithelial cells
Several virulence factors and host genes have been shown to affect H. pylori-induced autophagy (Table 2 ). Of these, VacA was first reported by Terebiznik et al. to induce autophagy in the AGS cell line [101]. H. pylori infection of AGS cells increases LC3 puncta and promotes the conversion of LC3-I to LC3-II. Moreover, formation of LC3 puncta was decreased in atg5 −/− mouse embryonic fibroblasts (MEFs) challenged with H. pylori, indicating that H. pylori-induced autophagy in AGS cells follows the canonical pathway [101]. Treatment of AGS cells with purified toxin or conditioned culture media supernatants revealed that VacA but not CagA, CagE, urease, or the T4SS is essential and sufficient to induce autophagy [101]. In contrast, another study using AZ-521 cells found that treatment with 3-methyladenine (3-MA), a PtdIns3K inhibitor, or knockdown of BECN1 did not inhibit VacA-induced autophagy, suggesting that it does not use the canonical pathway [31]. This finding contradicts the results in AGS cells reported by Tang et al. [102], possibly due to the different cell lines used. Interestingly, autophagy in turns modulates the activity of intracellular VacA, indicating that autophagy protects host cells against H. pylori infection. However, the mechanism by which these virulence factors induce autophagy remains to be investigated. One clue is that VacA damages mitochondrial function and causes nutrient depletion which may be linked to inhibition of mTOR signaling and induction of autophagy [107].
Table 2.
H. pylori manipulates autophagy in phagocytic and non-phagocytic cells.
| Bacterial virulence factors (V) or host determinants (H) | Factor | Cell line used | Role in intracellular survival and/or autophagic processes | Reference |
|---|---|---|---|---|
| Epithelial cells | ||||
| V | CagA | AGS | Does not involve in H. pylori-induced autophagy | [101] |
| Promote H. pylori intracellular survival | [13] | |||
| CagE | AGS | Does not involve in H. pylori-induced autophagy | [101] | |
| VacA | AGS | VacA channel-forming activity is essentitial for the intudtion of autophagy | [101] | |
| AZ521 | Induce autophagy by binding to LRP1 | [31] | ||
| AGS | Prolong expossure may cause autophagy disruption due to lack of cathepsin D in lysosome | [107] | ||
| AGS | Inhibit H. pylori internalization and invasion | [13] | ||
| BabA | AGS | Promote H. pylori intracellular survival | [13] | |
| Urease | AGS | Does not involve in H. pylori-induced autophagy | [101] | |
| H | ATG5 | AGS | Essential for H. pylori-induced autophagy | [101] |
| AGS | Decrease the intracellular survival of VacA+ H. pylori | [107] | ||
| ATG12 | AGS | Essential for H. pylori-induced autophagy | [101,102,107] | |
| ATG16L1 | AGS | Single nucleotide polymorphism ATG16L1300A may increase the intracellular survival of H. pylori | [107] | |
| BECN1 | AGS | Essential for H. pylori-induced autophagy | [102] | |
| AZ521 | Does not involve in VacA-induced autophagy | [31] | ||
| HFE145 | Promote H. pylori intracellular survival by inhibiting autophagosome formation | [96] | ||
| LRP1 | AZ521 | Induce autophagy by binding to VacA | [31] | |
| MIR30B | AGS | Inhibit H. pylori-induced autophagy by downregulating ATG12 and BECN1 | [102] | |
| MIR30D | AGS, GES1 | Inhibit H. pylori-induced autophagy by downregulating genes involved in autophagy pathway | [112] | |
| PtdIns3K | AZ521 | Does not involve in VacA-induced autophagy | [31] | |
| Professional phagocytes | ||||
| V | CagA | THP-1 | Promote H. pylori internalization and intracellular survival | [103] |
| BMDCs | Promote H. pylori intracellular survival | [104] | ||
| VacA | THP-1 | Promote H. pylori internalization and intracellular survival | [103] | |
| BMDCs | Promote H. pylori intracellular survival | [104] | ||
| CapJ | J774A.1 | Delay the fusion of autophagosome and lysosome | [45] | |
| J774A.1 | Delay H. pylori internalization | [44] | ||
| H | ATG16L1 | PMBCs | Single nucleotide polymorphism ATG16L1300A may increase the intracellular survival of H. pylori | [107] |
| PtdIns3K | THP-1 | Inhibit the bacterial multiplication | [103] | |
| RAW264.7 | Does not inhibit the bacterial multiplication | [103] | ||
Many studies have reported that H. pylori can prevent its degradation, allowing it to reside and multiply in autophagosomes. Raju et. al. showed that prolonged exposure (24 h) to a toxin disrupts subsequent autophagosome-lysosome fusion and causes the accumulation of autophagsomes in the cells [108]. This observation suggests that the underlying mechanisms of autophagy are not the same in acute and chronic exposure (Fig. 2 ). The final step of autophagy is formation of autolysosomes, which are the fusion of autophagosomes and lysosomes. For instance, after prolonged exposure to conditioned culture media, AGS cells show a reduced level of the lysosomal protease cathepsin D, due to disruption of lysosome acidification [108]. The defective autophagosomes in AGS cells caused by VacA may lead to the accumulation of SQSTM1, a major cargo ubiquitin-binding receptor. This phenomenon is also observed in patient biopsies [108]. Other reports also showed the accumulation of different proteins due to VacA-disrupted autophagy, leading to changes in cellular ROS levels [109] and gene mutations [110]. The accumulation of p62 activates nuclear factor erythroid 2-related factor (NRF2) which controls the expression of ROS detoxification genes in AGS cells [108,110]. A contradictory result showed that CagA and HspB, but not VacA, regulate the antioxidant response [111]. Therefore, a detailed investigation of the effect of p62 and NRF2 on H. pylori virulence factors that regulate autophagy is warranted.
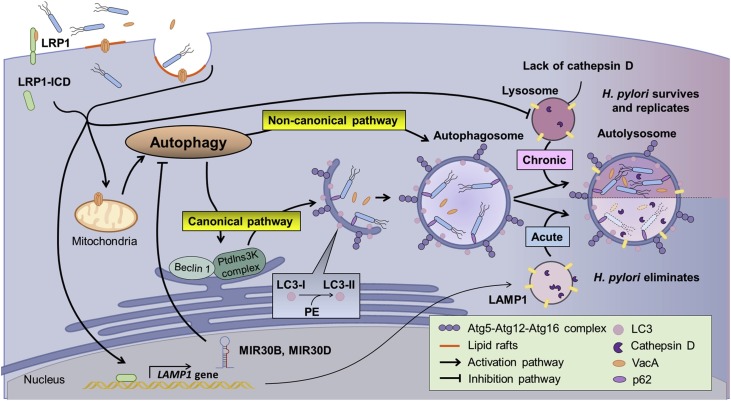
Fig. 2.
H. pylori modulates autophagy in epithelial cells. VacA is an essential virulence factor for modulating H. pylori induced autophagy via various mechanisms. The initiation of infection starts with the internalization of H. pylori and VacA, the formation of VacA pores or the binding of VacA to LRP1. The internalization of H. pylori and VacA may induce autophagy. VacA forms pores in the mitochondria, causing nutrient depletion which induces autophagy, mediated through the inhibition of mammalian target of rapamycin complex 1 (mTORC1). Autophagy may proceed through either the canonical or non-canonical pathway depending on the cell lines used. In addition to inducing autophagy, H. pylori can hijack the autophagy process. H. pylori infection may induce the expression of MIR30B and MIR30D, which subsequently inhibit autophagy by targeting different autophagy-related genes. Also, the acidification of lysosome is disrupted by chronic exposure to VacA, causing the accumulation of pro-cathepsin D. These mechanisms allow H. pylori to survive and replicate in the autophagosome.
Consistent with the previous findings, a recent study has also shown reduced activity of lysosomal proteases including cathepsin D, N-acetyl-β-d-glucosaminidase (β-NAG), and acid phosphatase in HFE-145 cells infected by H. pylori [96]. Disruption of lysosome acidification causes the repression of the retrograde trafficking of mannose-6-phosphate receptors which play an important role in carrying lysosomal hydrolytic enzymes in the trans-Golgi network [96]. However, the mechanism underlying the disruption of lysosome acidification in H. pylori infected cells is still unclear.
The internalization of VacA or H. pylori plays a critical role at the beginning of autophagy. There is evidence that VacA induces autophagy by binding to LRP1 [31]. Interestingly, s1m1 VacA, but not s1m2 or s2m2 interacts with LRP1 and stimulates autophagy [31]. Moreover, knockdown of LRP1 significantly reduces autophagy, indicating that VacA internalization is crucial for the induction of autophagy. Internalized VacA may further alter the membrane permeability of mitochondria, causing the release of cytochrome c and leading to the induction of mitophagy and apoptosis [29,111]. In addition, the internalization of H. pylori is unaffected by VacA and CagA, but is correlated with H. pylori invasin NudA in AGS cells [112]. Notably, the internalization of H. pylori also requires the activation of PI3-kinase and PKC in AGS cells [105].
In addition, although VacA induces autophagy, H. pylori may evade autophagy process by altering the expression of various microRNAs, including MIR30B and MIR30D [102,112]. H. pylori infection may increase the expression of MIR30B in different gastric cell lines (i.e. MIR30D in AGS and GES1 cells). Both MIR30B and MIR30D target several genes involved in autophagy, such as ATG12 and BECN1. Interestingly, a very recent report by Tsugawa et al. showed that overexpression of capping actin protein of muscle Z-line alpha subunit 1 (CAPZA1) blocks the CagA-degraded autophagy by downregulating the expression of lysosomal associated membrane protein 1 (LAMP1) in H. pylori-infected cells [113]. Moreover, Tsugawa et al. showed that CagA accumulation in CAPZA1-overexpressing cells is essential for the upregulation of CD44v9 which is a cancer stem-cell marker, suggesting that overexpression of CAPZA1 could be a key risk factor in gastric cancer development [114]. This notion is supported by an earlier study that the abundance of CD44v9 is a superior prognostic predictor than the presence of open-type gastric mucosal atrophy for the recurrence of gastric cancer in patients after the resection of early gastric cancer [115]. These lines of evidence together suggest that H. pylori-infected gastric mucosa exhibits negative regulation of autophagy through an orchestrated array of actions from multiple virulence factors. As such, H. pylori-mediated defective autophagy links to an increased gastric cancer risk, thereby promoting the development of various gastrointestinal diseases.
4.2. H. pylori manipulates autophagy in professional phagocytes
Compared to gastric epithelial cells, the effects of H. pylori-induced autophagy by professional phagocytes are much more complicated due to differences between the cell lines and bacterial strains used. The primary role of phagocytes is to ensure the efficient clearance of harmful foreign particles, bacteria, and dead or dying cells. In the case of H. pylori infection, various studies have demonstrated its capability to delay phagocytosis by phagocytes.
Wang et. al. first reported that H. pylori induces activation of autophagy in macrophages [103]. In another previous study, H. pylori infection was found to increase the number of LC3-puncta in both RAW264.7 and THP-1 cells [12]. In addition to activation of autophagy, Wang et. al. further demonstrated that H. pylori multiplies in human monocytic THP-1 or U937 cells, but not in murine macrophage RAW264.7 cells. Another report showed that H. pylori could replicate in the autophagosomes of mouse bone marrow-derived dendritic cells (BMDCs) [104]. Moreover, H. pylori infection of BMDCs may impair MHC class II molecule surface expression which is negatively regulated by Toll-like receptor (TLR)-2 and 4. The correlation of TLR4 polymorphism and gastric cancer may deserve further attention [116]. Interestingly, both studies show that CagA and VacA are important for the survival and multiplication of H. pylori in autophagosomes [103,104]. These results contradicted the observations in gastric epithelial cells, which may be due to the different types of cell lines and H. pylori strains used.
This observation corroborates with another study where pre-loading H. pylori with cholesterol enhances the rate of H. pylori internalization [117]. We recently have shown that CGT, a main factor in H. pylori cholesterol glucosylation, promotes autophagosome formation, and simultaneously inhibits lysosome formation in murine macrophages to improve its intracellular survival [45]. This study also demonstrated that ATG12 is involved in the autophagy pathway, which is essential for H. pylori intracellular survival [45]. In addition, treatment of macrophages with simvastatin promoted autophagy and thereby reduced intracellular bacterial burden [7] (Fig. 3 ).
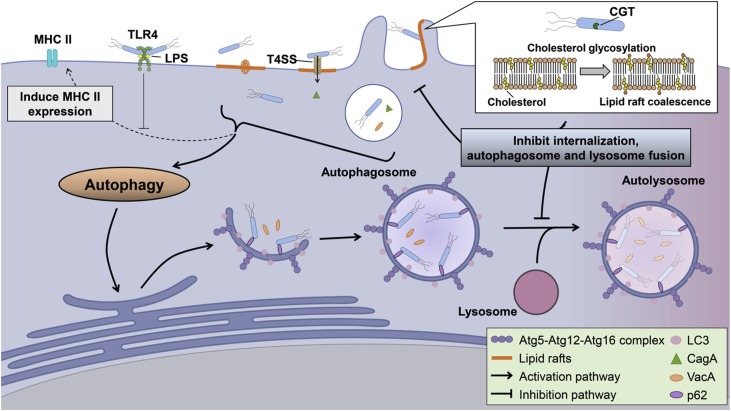
Fig. 3.
H. pylori regulates autophagy in professional phagocytes. Unlike epithelial cells, both VacA, CagA and T4SS play an important role in H. pylori-induced autophagy. Recent studies have shown that HP0421, a cholesterol-α-glucosyltransferase (CGT), modulates autophagy and promotes bacterial survival in macrophages by clustering lipid rafts. The disrupted lipid rafts result in a lower internalization of H. pylori and interfere with autophagosome fusion with lysosomes. Another study has shown that H. pylori infection may increase the expression of major histocompatibility complex II (MHC II). However, TLR4 plays a negative role on BMDC maturation by inhibiting this process.
H. pylori induced autophagy in phagocytes is affected by the host genotype. For instance, several gene polymorphisms are associated with Crohn disease, an inflammatory bowel disorder, which affects H. pylori induced autophagy. ATG16L1, which is involved in autophagosome formation with the T300A mutation makes host more susceptible to H. pylori infection and reduces H. pylori-induced autophagy [108]. However, the detailed underlying mechanism remains to be explored. Another gene involved in this autophagy is the nucleotide-binding oligomerization domain-containing protein 2 (NOD2), a pattern recognition receptor. During H. pylori infection, NOD2 activates nuclear factor-kappa B (NF-κB) signaling which induces inflammation. However, the R702W mutant cannot activate NF-κB signaling [118]. Therefore, the mutation is associated with gastric MALT lymphoma, which is usually associated with H. pylori infection [118]. In addition to NOD2, TLR4, a pattern recognition receptor, also plays an important role in stimulating macrophage autophagy by binding to lipopolysaccharides on the bacteria [7,45]. TLR4 polymorphisms have been shown to correlate with gastric cancer [116]. However, further experiments are needed to reveal the underlying mechanisms.
5. Conclusions and perspectives
Autophagy is increasingly recognized as a vital route for host defense. Thus, it is not surprising that successful intracellular pathogens are evolved to dysregulate autophagy, contributing to disease pathogenesis. H. pylori, a small pathogen, shows remarkable capabilities to enhance its intracellular survival and propagation by means of a limited number of virulence factors, exemplified by multiple strategies that have evolved to negatively regulate autophagy. This, in turn, increases the risk of developing H. pylori-associated diseases. However, there are discrepancies in the results obtained to date, mainly due to the different types of cell models and/or bacterial strains used. Furthermore, the connection between virulence factors, dysregulated intracellular trafficking, disease etiology and its clinical relevance remains largely unanswered. For instance, does CagA, VacA, or CGT mediate membrane remodeling and interfere with intracellular trafficking, and how does this relate to a defective autophagy? Although VacA-induced autophagy is dependent on LRP1, the detailed mechanisms and signaling underlying the autophagic trafficking remains to be clarified. Negative regulation of autophagy in H. pylori-infected gastric biopsies harboring abundance of CAPZA1, accumulation of CagA, and expression of CD44v9 is considered to increase gastric cancer risk. It will be of interest to characterize CagA accumulation in CD44v9-negative gastric epithelial cells. It is important to examine these data using cell models, in vivo studies, and in clinical settings. Further investigations are required to provide a mechanistic basis for better understanding the host-pathogen interplay in the context of autophagy and disease progression. This may facilitate the development of new strategies for combating diseases affected by infection and autophagy.
Declaration of Competing Interest
The authors did not have conflicts of interest to declare for this work.
Acknowledgments
The authors would like to thank the editors and reviewers for the editorial assistance and their valuable comments. This study is supported by Taiwan Ministry of Science and Technology (MOST108-2811-B-007-507, 108-2321-B007-004, 107-2627-M-007-004, and 106-2320-B-182-012-MY3), Chang Gung Memorial Hospital (CMRPD1I0061-3, CMRPD1J0021-3, and BMRPE90), and Tomorrow Medical Foundation.
Contributor Information
Chih-Ho Lai, Email: chlai@mail.cgu.edu.tw.
Wen-Ching Wang, Email: wcwang@life.nthu.edu.tw.
References
- 1.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siqueira Mda S., Ribeiro Rde M., Travassos L.H. Autophagy and its interaction with intracellular bacterial pathogens. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V., Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5(6):527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Z., Fux B., Goodwin M., Dunay I.R., Strong D., Miller B.C. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J.C., Chien C.T. A new approach for the prevention and treatment of Helicobacter pylori infection via upregulation of autophagy and downregulation of apoptosis. Autophagy. 2009;5(3):413–414. doi: 10.4161/auto.5.3.7826. [DOI] [PubMed] [Google Scholar]
- 7.Liao W.C., Huang M.Z., Wang M.L., Lin C.J., Lu T.L., Lo H.R. Statin decreases Helicobacter pylori burden in macrophages by promoting autophagy. Front. Cell. Infect. Microbiol. 2017;6:203. doi: 10.3389/fcimb.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y.H., Wu J.J., Lei H.Y. When Helicobacter pylori invades and replicates in the cells. Autophagy. 2009;5(4):540–542. doi: 10.4161/auto.5.4.8167. [DOI] [PubMed] [Google Scholar]
- 9.Petersen A.M., Krogfelt K.A. Helicobacter pylori: an invading microorganism? A review. FEMS Immunol. Med. Microbiol. 2003;36(3):117–126. doi: 10.1016/S0928-8244(03)00020-8. [DOI] [PubMed] [Google Scholar]
- 10.Dubois A., Boren T. Helicobacter pylori is invasive and it may be a facultative intracellular organism. Cell. Microbiol. 2007;9(5):1108–1116. doi: 10.1111/j.1462-5822.2007.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai C.H., Kuo C.H., Chen P.Y., Poon S.K., Chang C.S., Wang W.C. Association of antibiotic resistance and higher internalization activity in resistant Helicobacter pylori isolates. J. Antimicrob. Chemother. 2006;57(3):466–471. doi: 10.1093/jac/dki479. [DOI] [PubMed] [Google Scholar]
- 12.Deen N.S., Gong L., Naderer T., Devenish R.J., Kwok T. Analysis of the relative contribution of phagocytosis, LC3-Associated phagocytosis, and canonical autophagy during Helicobacter pylori infection of macrophages. Helicobacter. 2015;20(6):449–459. doi: 10.1111/hel.12223. [DOI] [PubMed] [Google Scholar]
- 13.Chu Y.T., Wang Y.H., Wu J.J., Lei H.Y. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect. Immun. 2010 doi: 10.1128/IAI.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smolka A.J., Backert S. How Helicobacter pylori infection controls gastric acid secretion. J. Gastroenterol. 2012;47(6):609–618. doi: 10.1007/s00535-012-0592-1. [DOI] [PubMed] [Google Scholar]
- 15.Odenbreit S., Swoboda K., Barwig I., Ruhl S., Boren T., Koletzko S. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect. Immun. 2009;77(9):3782–3790. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilver D., Arnqvist A., Ogren J., Frick I.M., Kersulyte D., Incecik E.T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279(5349):373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 17.Mahdavi J., Sonden B., Hurtig M., Olfat F.O., Forsberg L., Roche N. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297(5581):573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peck B., Ortkamp M., Diehl K.D., Hundt E., Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27(16):3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leunk R.D., Johnson P.T., David B.C., Kraft W.G., Morgan D.R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 1988;26(2):93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 20.Cover T.L., Blaser M.J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 1992;267(15):10570–10575. [PubMed] [Google Scholar]
- 21.Wang X., Wattiez R., Paggliacia C., Telford J.L., Ruysschaert J., Cabiaux V. Membrane topology of VacA cytotoxin from H. Pylori. FEBS Lett. 2000;481(2):96–100. doi: 10.1016/s0014-5793(00)01978-5. [DOI] [PubMed] [Google Scholar]
- 22.Cover T.L., Blanke S.R. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 2005;3(4):320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 23.Foegeding N.J., Caston R.R., Mcclain M.S., Ohi M.D., Cover T.L. An overview of Helicobacter pylori VacA toxin biology. Toxins (Basel) 2016;8(6) doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czajkowsky D.M., Iwamoto H., Cover T.L., Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc. Natl. Acad. Sci. U. S. A. 1999;96(5):2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yahiro K., Niidome T., Kimura M., Hatakeyama T., Aoyagi H., Kurazono H. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J. Biol. Chem. 1999;274(51):36693–36699. doi: 10.1074/jbc.274.51.36693. [DOI] [PubMed] [Google Scholar]
- 26.Yahiro K., Wada A., Yamasaki E., Nakayama M., Nishi Y., Hisatsune J. Essential domain of receptor tyrosine phosphatase beta (RPTPbeta) for interaction with Helicobacter pylori vacuolating cytotoxin. J. Biol. Chem. 2004;279(49):51013–51021. doi: 10.1074/jbc.M406473200. [DOI] [PubMed] [Google Scholar]
- 27.Yahiro K., Wada A., Nakayama M., Kimura T., Ogushi K., Niidome T. Protein-tyrosine phosphatase alpha, RPTP alpha, is a Helicobacter pylori VacA receptor. J. Biol. Chem. 2003;278(21):19183–19189. doi: 10.1074/jbc.M300117200. [DOI] [PubMed] [Google Scholar]
- 28.Nakano M., Yahiro K., Yamasaki E., Kurazono H., Akada J., Yamaoka Y. Helicobacter pylori VacA, acting through receptor protein tyrosine phosphatase alpha, is crucial for CagA phosphorylation in human duodenum carcinoma cell line AZ-521. Dis. Model. Mech. 2016;9(12):1473–1481. doi: 10.1242/dmm.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palframan S.L., Kwok T., Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front. Cell. Infect. Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foo J.H., Culvenor J.G., Ferrero R.L., Kwok T., Lithgow T., Gabriel K. Both the p33 and p55 subunits of the Helicobacter pylori VacA toxin are targeted to mammalian mitochondria. J. Mol. Biol. 2010;401(5):792–798. doi: 10.1016/j.jmb.2010.06.065. [DOI] [PubMed] [Google Scholar]
- 31.Yahiro K., Satoh M., Nakano M., Hisatsune J., Isomoto H., Sap J. Low-density lipoprotein receptor-related protein-1 (LRP1) mediates autophagy and apoptosis caused by Helicobacter pylori VacA. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.387498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viala J., Chaput C., Boneca I.G., Cardona A., Girardin S.E., Moran A.P. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004;5(11):1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez-Soto L.F., Kutter S., Sewald X., Ertl C., Weiss E., Kapp U. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5(12) doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murata-Kamiya N., Kikuchi K., Hayashi T., Higashi H., Hatakeyama M. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe. 2010;7(5):399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Koniger V., Holsten L., Harrison U., Busch B., Loell E., Zhao Q. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat. Microbiol. 2016;2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]
- 36.Backert S., Ziska E., Brinkmann V., Zimny-Arndt U., Fauconnier A., Jungblut P.R. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2000;2(2):155–164. doi: 10.1046/j.1462-5822.2000.00043.x. [DOI] [PubMed] [Google Scholar]
- 37.Lai C.H., Chang Y.C., Du S.Y., Wang H.J., Kuo C.H., Fang S.H. Cholesterol depletion reduces Helicobacter pylori CagA translocation and CagA-induced responses in AGS cells. Infect. Immun. 2008;76(7):3293–3303. doi: 10.1128/IAI.00365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai C.H., Wang H.J., Chang Y.C., Hsieh W.C., Lin H.J., Tang C.H. Helicobacter pylori CagA-mediated IL-8 induction in gastric epithelial cells is cholesterol-dependent and requires the C-terminal tyrosine phosphorylation-containing domain. FEMS Microbiol. Lett. 2011;323(2):155–163. doi: 10.1111/j.1574-6968.2011.02372.x. [DOI] [PubMed] [Google Scholar]
- 39.Lin C.J., Liao W.C., Lin H.J., Hsu Y.M., Lin C.L., Chen Y.A. Statins attenuate Helicobacter pylori CagA translocation and reduce incidence of gastric Cancer: In vitro and population-based case-control studies. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tohidpour A., Gorrell R.J., Roujeinikova A., Kwok T. The middle fragment of Helicobacter pylori CagA induces actin rearrangement and triggers its own uptake into gastric epithelial cells. Toxins (Basel) 2017;9(8) doi: 10.3390/toxins9080237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebrun A.H., Wunder C., Hildebrand J., Churin Y., Zahringer U., Lindner B. Cloning of a cholesterol-alpha-glucosyltransferase from Helicobacter pylori. J. Biol. Chem. 2006;281(38):27765–27772. doi: 10.1074/jbc.M603345200. [DOI] [PubMed] [Google Scholar]
- 42.Wunder C., Churin Y., Winau F., Warnecke D., Vieth M., Lindner B. Cholesterol glucosylation promotes immune evasion by Helicobacter pylori. Nat. Med. 2006;12(9):1030–1038. doi: 10.1038/nm1480. [DOI] [PubMed] [Google Scholar]
- 43.Wang H.J., Cheng W.C., Cheng H.H., Lai C.H., Wang W.C. Helicobacter pylori cholesteryl glucosides interfere with host membrane phase and affect type IV secretion system function during infection in AGS cells. Mol. Microbiol. 2012;83(1):67–84. doi: 10.1111/j.1365-2958.2011.07910.x. [DOI] [PubMed] [Google Scholar]
- 44.Du S.Y., Wang H.J., Cheng H.H., Chen S.D., Wang L.H.C., Wang W.C. Cholesterol glucosylation by Helicobacter pylori delays internalization and arrests phagosome maturation in macrophages. J. Microbiol. Immunol. Infect. 2016 doi: 10.1016/j.jmii.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Lai C.H., Huang J.C., Cheng H.H., Wu M.C., Huang M.Z., Hsu H.Y. Helicobacter pylori cholesterol glucosylation modulates autophagy for increasing intracellular survival in macrophages. Cell. Microbiol. 2018 doi: 10.1111/cmi.12947. [DOI] [PubMed] [Google Scholar]
- 46.Morey P., Pfannkuch L., Pang E., Boccellato F., Sigal M., Imai-Matsushima A. Helicobacter pylori depletes cholesterol in gastric glands to prevent interferon gamma signaling and escape the inflammatory response. Gastroenterology. 2018;154(5) doi: 10.1053/j.gastro.2017.12.008. 1391-404.e9. [DOI] [PubMed] [Google Scholar]
- 47.Qaria M.A., Kumar N., Hussain A., Qumar S., Doddam S.N., Sepe L.P. Roles of cholesteryl-alpha-Glucoside transferase and cholesteryl glucosides in maintenance of Helicobacter pylori morphology, cell wall integrity, and resistance to antibiotics. MBio. 2018;9(6) doi: 10.1128/mBio.01523-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Athmann C., Zeng N., Kang T., Marcus E.A., Scott D.R., Rektorschek M. Local pH elevation mediated by the intrabacterial urease of Helicobacter pylori cocultured with gastric cells. J. Clin. Invest. 2000;106(3):339–347. doi: 10.1172/JCI9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Megraud F., Neman-Simha V., Brugmann D. Further evidence of the toxic effect of ammonia produced by Helicobacter pylori urease on human epithelial cells. Infect. Immun. 1992;60(5):1858–1863. doi: 10.1128/iai.60.5.1858-1863.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki M., Miura S., Suematsu M., Fukumura D., Kurose I., Suzuki H. Helicobacter pylori-associated ammonia production enhances neutrophil-dependent gastric mucosal cell injury. Am. J. Physiol. 1992;263(5 Pt 1):G719–25. doi: 10.1152/ajpgi.1992.263.5.G719. [DOI] [PubMed] [Google Scholar]
- 51.Gu H. Role of flagella in the pathogenesis of Helicobacter pylori. Curr. Microbiol. 2017;74(7):863–869. doi: 10.1007/s00284-017-1256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geis G., Suerbaum S., Forsthoff B., Leying H., Opferkuch W. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J. Med. Microbiol. 1993;38(5):371–377. doi: 10.1099/00222615-38-5-371. [DOI] [PubMed] [Google Scholar]
- 53.Roncarati D., Scarlato V. The interplay between two transcriptional repressors and chaperones orchestrates Helicobacter pylori heat-shock response. Int. J. Mol. Sci. 2018;19(6) doi: 10.3390/ijms19061702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eskelinen E.L., Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta. 2009;1793(4):664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Klionsky D.J. The molecular machinery of autophagy: unanswered questions. J. Cell. Sci. 2005;118(Pt 1):7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massey A.C., Zhang C., Cuervo A.M. Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 57.Bejarano E., Cuervo A.M. Chaperone-mediated autophagy. Proc. Am. Thorac. Soc. 2010;7(1):29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z., Klionsky D.J. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anding A.L., Baehrecke E.H. Cleaning house: selective autophagy of organelles. Dev. Cell. 2017;41(1):10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cecconi F., Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 2008;15(3):344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dall’armi C., Devereaux K.A., Di Paolo G. The role of lipids in the control of autophagy. Curr. Biol. 2013;23(1):R33–45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao K., Klionsky D.J. Xenophagy: a battlefield between host and microbe, and a possible avenue for cancer treatment. Autophagy. 2016 doi: 10.1080/15548627.2016.1267075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang M., Hajishengallis G. Lipid raft‐dependent uptake, signalling and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cell. Microbiol. 2008;10(10):2029–2042. doi: 10.1111/j.1462-5822.2008.01185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders P.R., Gilson P.R., Cantin G.T., Greenbaum D.C., Nebl T., Carucci D.J. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 2005;280(48):40169–40176. doi: 10.1074/jbc.M509631200. [DOI] [PubMed] [Google Scholar]
- 66.Yoneyama K.A., Tanaka A.K., Silveira T.G., Takahashi H.K., Straus A.H. Characterization of Leishmania (Viannia) braziliensis membrane microdomains, and their role in macrophage infectivity. J. Lipid Res. 2006;47(10):2171–2178. doi: 10.1194/jlr.M600285-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes M.C., Cortez M., Geraldo Yoneyama K.A., Straus A.H., Yoshida N., Mortara R.A. Novel strategy in Trypanosoma cruzi cell invasion: implication of cholesterol and host cell microdomains. Int. J. Parasitol. 2007;37(13):1431–1441. doi: 10.1016/j.ijpara.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 68.Dykstra M.L., Longnecker R., Pierce S.K. Epstein-Barr virus coopts lipid rafts to block the signaling and antigen transport functions of the BCR. Immunity. 2001;14(1):57–67. doi: 10.1016/s1074-7613(01)00089-9. [DOI] [PubMed] [Google Scholar]
- 69.Graham D.R., Chertova E., Hilburn J.M., Arthur L.O., Hildreth J.E. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J. Virol. 2003;77(15):8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor D.R., Whitehouse I.J., Hooper N.M. Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PLoS Pathog. 2009;5(11) doi: 10.1371/journal.ppat.1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wadia J.S., Schaller M., Williamson R.A., Dowdy S.F. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS One. 2008;3(10):e3314. doi: 10.1371/journal.pone.0003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pike L.J. Lipid rafts: bringing order to chaos. J. Lipid Res. 2003;44(4):655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 74.Alonso M.A., Millan J. The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J. Cell. Sci. 2001;114(Pt 22):3957–3965. doi: 10.1242/jcs.114.22.3957. [DOI] [PubMed] [Google Scholar]
- 75.Hanzal-Bayer M.F., Hancock J.F. Lipid rafts and membrane traffic. FEBS Lett. 2007;581(11):2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 76.Chichili G.R., Rodgers W. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. 2009;66(14):2319–2328. doi: 10.1007/s00018-009-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Head B.P., Patel H.H., Insel P.A. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta. 2014;1838(2):532–545. doi: 10.1016/j.bbamem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenberger C.M., Brumell J.H., Finlay B.B. Microbial pathogenesis: lipid rafts as pathogen portals. Curr. Biol. 2000;10(22):R823–5. doi: 10.1016/s0960-9822(00)00788-0. [DOI] [PubMed] [Google Scholar]
- 79.Shin J.S., Gao Z., Abraham S.N. Involvement of cellular caveolae in bacterial entry into mast cells. Science. 2000;289(5480):785–788. doi: 10.1126/science.289.5480.785. [DOI] [PubMed] [Google Scholar]
- 80.Guo H., Huang M., Yuan Q., Wei Y., Gao Y., Mao L. The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS One. 2017 doi: 10.1371/journal.pone.0170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spano S., Galan J.E. Taking control: hijacking of Rab GTPases by intracellular bacterial pathogens. Small GTPases. 2018;9(1–2):182–191. doi: 10.1080/21541248.2017.1336192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zerial M., Mcbride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 83.Schwartz S.L., Cao C., Pylypenko O., Rak A., Wandinger-Ness A. RabGT Pases at a glance. J. Cell. Sci. 2007;120(Pt 22):3905–3910. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 84.Galvez T., Gilleron J., Zerial M., O’sullivan G.A. SnapShot: mammalian Rab proteins in endocytic trafficking. Cell. 2012;151(1) doi: 10.1016/j.cell.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 85.Roberts E.A., Chua J., Kyei G.B., Deretic V. Higher order Rab programming in phagolysosome biogenesis. J. Cell Biol. 2006;174(7):923–929. doi: 10.1083/jcb.200603026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kyei G.B., Vergne I., Chua J., Roberts E., Harris J., Junutula J.R. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J. 2006;25(22):5250–5259. doi: 10.1038/sj.emboj.7601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao W., Moest T., Zhao Y., Guilhon A.A., Buffat C., Gorvel J.P. The Salmonella effector protein SifA plays a dual role in virulence. Sci. Rep. 2015;5:12979. doi: 10.1038/srep12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boucrot E., Henry T., Borg J.P., Gorvel J.P., Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308(5725):1174–1178. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 89.Gedde M.M., Higgins D.E., Tilney L.G., Portnoy D.A. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 2000;68(2):999–1003. doi: 10.1128/iai.68.2.999-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burrack L.S., Harper J.W., Higgins D.E. Perturbation of vacuolar maturation promotes listeriolysin O-independent vacuolar escape during Listeria monocytogenes infection of human cells. Cell. Microbiol. 2009;11(9):1382–1398. doi: 10.1111/j.1462-5822.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lambrechts A., Gevaert K., Cossart P., Vandekerckhove J., Van Troys M. Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 2008;18(5):220–227. doi: 10.1016/j.tcb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 92.Welch M.D., Way M. Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe. 2013;14(3):242–255. doi: 10.1016/j.chom.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vergne I., Chua J., Lee H.H., Lucas M., Belisle J., Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2005;102(11):4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steinberg B.E., Grinstein S. Pathogen destruction versus intracellular survival: the role of lipids as phagosomal fate determinants. J. Clin. Invest. 2008:2002–2011. doi: 10.1172/JCI35433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jamwal S.V., Mehrotra P., Singh A., Siddiqui Z., Basu A., Rao K.V. Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Sci. Rep. 2016 doi: 10.1038/srep23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang L., Hu W., Cho C.H., Chan F.K., Yu J., Fitzgerald J.R. Reduced lysosomal clearance of autophagosomes promotes survival and colonization of Helicobacter pylori. J. Pathol. 2018;244(4):432–444. doi: 10.1002/path.5033. [DOI] [PubMed] [Google Scholar]
- 97.Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 98.Abdulrahman B.A., Khweek A.A., Akhter A., Caution K., Kotrange S., Abdelaziz D.H. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy. 2011;7(11):1359–1370. doi: 10.4161/auto.7.11.17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez J., Malireddi R.K., Lu Q., Cunha L.D., Pelletier S., Gingras S. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015;17(7):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Herb M., Gluschko A., Schramm M. LC3-associated phagocytosis - the highway to hell for phagocytosed microbes. Semin. Cell Dev. Biol. 2019 doi: 10.1016/j.semcdb.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 101.Terebiznik M.R., Raju D., Vázquez C.L., Torbricki K., Kulkarni R., Blanke S.R. Effect of Helicobacter pylori's vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009 doi: 10.4161/auto.5.3.7663. [DOI] [PubMed] [Google Scholar]
- 102.Tang B., Li N., Gu J., Zhuang Y., Li Q., Wang H.G. Compromised autophagy by MIR30B benefits the intracellular survival of Helicobacter pylori. Autophagy. 2012 doi: 10.4161/auto.20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y.-H., Wu J.-J., Lei H.-Y. The autophagic induction in Helicobacter pylori-Infected macrophage. Exp. Biol. Med. 2009 doi: 10.3181/0808-RM-252. [DOI] [PubMed] [Google Scholar]
- 104.Wang Y.H., Gorvel J.P., Chu Y.T., Wu J.J., Lei H.Y. Helicobacter pylori lmpairs murine dendritic cell responses to infection. PLoS One. 2010 doi: 10.1371/journal.pone.0010844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kwok T., Backert S., Schwarz H., Berger J., Meyer T.F. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 2002 doi: 10.1128/IAI.70.4.2108-2120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Amieva M.R., Salama N.R., Tompkins L.S., Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 2002 doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 107.Kim I.J., Lee J., Oh S.J., Yoon M.S., Jang S.S., Holland R.L. Helicobacter pylori infection modulates host cell metabolism through VacA-Dependent inhibition of mTORC1. Cell Host Microbe. 2018 doi: 10.1016/j.chom.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raju D., Hussey S., Ang M., Terebiznik M.R., Sibony M., Galindo-Mata E. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsugawa H., Suzuki H., Saya H., Hatakeyama M., Hirayama T., Hirata K. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe. 2012 doi: 10.1016/j.chom.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 110.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012 doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buommino E., Donnarumma G., Manente L., De Filippis A., Silvestri F., Iaquinto S. The HelicobacterPylori Protein HspB Interferes With Nrf2/Keap1 Pathway Altering The Antioxidant Response of Ags Cells. Helicobacter. 2012 doi: 10.1111/j.1523-5378.2012.00973.x. [DOI] [PubMed] [Google Scholar]
- 112.Liu H., Semino-Mora C., Dubois A. Mechanism of H. Pylori intracellular entry: an in vitro study. Front. Cell. Infect. Microbiol. 2012 doi: 10.3389/fcimb.2012.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsugawa H., Mori H., Matsuzaki J., Sato A., Saito Y., Imoto M. CAPZA1 determines the risk of gastric carcinogenesis by inhibiting Helicobacter pylori CagA-degraded autophagy. Autophagy. 2019;15(2):242–258. doi: 10.1080/15548627.2018.1515530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsugawa H., Kato C., Mori H., Matsuzaki J., Kameyama K., Saya H. Cancer stem-cell marker CD44v9-Positive cells arise from Helicobacter pylori-Infected CAPZA1-Overexpressing cells. Cell. Mol. Gastroenterol. Hepatol. 2019;8(3):319–334. doi: 10.1016/j.jcmgh.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hirata K., Suzuki H., Imaeda H., Matsuzaki J., Tsugawa H., Nagano O. CD44 variant 9 expression in primary early gastric cancer as a predictive marker for recurrence. Br. J. Cancer. 2013;109(2):379–386. doi: 10.1038/bjc.2013.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fukata M., Abreu M.T. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008 doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wunder C. Cholesterol glycosylation glucosylation promotes H pylori ulcer infection. Nat. Med. 2006 doi: 10.1038/nm1480. [DOI] [PubMed] [Google Scholar]
- 118.Rosenstiel P., Hellmig S., Hampe J., Ott S., Till A., Fischbach W. Influence of polymorphisms in the NOD1/CARD4 and NOD2/CARD15 genes on the clinical outcome of Helicobacter pylori infection. Cell. Microbiol. 2006 doi: 10.1111/j.1462-5822.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- 119.Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307(5710):727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 120.Starr T., Child R., Wehrly T.D., Hansen B., Hwang S., Lopez-Otin C. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Collins C.A., De Maziere A., Van Dijk S., Carlsson F., Klumperman J., Brown E.J. Atg5-independent sequestration of ubiquitinated mycobacteria. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Birmingham C.L., Canadien V., Gouin E., Troy E.B., Yoshimori T., Cossart P. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3(5):442–451. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 123.Vazquez C.L., Colombo M.I. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ. 2010;17(3):421–438. doi: 10.1038/cdd.2009.129. [DOI] [PubMed] [Google Scholar]
- 124.Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306(5698):1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 125.Gong L., Cullinane M., Treerat P., Ramm G., Prescott M., Adler B. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Amer A.O., Swanson M.S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell. Microbiol. 2005;7(6):765–778. doi: 10.1111/j.1462-5822.2005.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Birmingham C.L., Smith A.C., Bakowski M.A., Yoshimori T., Brumell J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281(16):11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 128.Schnaith A., Kashkar H., Leggio S.A., Addicks K., Kronke M., Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 2007;282(4):2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 129.Checroun C., Wehrly T.D., Fischer E.R., Hayes S.F., Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 2006;103(39):14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]