Abstract
Objectives
Understanding the novel coronavirus (COVID-19) mode of host cell recognition may help to fight the disease and save lives. The spike protein of coronaviruses is the main driving force for host cell recognition.
Methods
In this study, the COVID-19 spike binding site to the cell-surface receptor (Glucose Regulated Protein 78 (GRP78)) is predicted using combined molecular modeling docking and structural bioinformatics. The COVID-19 spike protein is modeled using its counterpart, the SARS spike.
Results
Sequence and structural alignments show that four regions, in addition to its cyclic nature have sequence and physicochemical similarities to the cyclic Pep42. Protein-protein docking was performed to test the four regions of the spike that fit tightly in the GRP78 Substrate Binding Domain β (SBDβ). The docking pose revealed the involvement of the SBDβ of GRP78 and the receptor-binding domain of the coronavirus spike protein in recognition of the host cell receptor.
Conclusions
We reveal that the binding is more favorable between regions III (C391-C525) and IV (C480-C488) of the spike protein model and GRP78. Region IV is the main driving force for GRP78 binding with the predicted binding affinity of -9.8 kcal/mol. These nine residues can be used to develop therapeutics specific against COVID-19.
Keywords: GRP78, BiP, COVID-19 spike, Protein-protein docking, Structural bioinformatics, Pep42
Introduction
In late December 2019, it was noticed that several people in Wuhan city of Hubei Province, China, were suffering from SARS-like pneumonia, which the World Health Organization (WHO) would later name COVID-19.1 , 2 According to the WHO surveillance draft, in January 2020, any resident or citizen in transit through Wuhan city 14 days before the onset of the symptoms is suspected to be infected by COVID-19.2 , 3 Additionally, WHO distributed interim guidance for laboratories that carry out the testing for the newly emerged outbreak and infection prevention and control guidance.4 , 5 The COVID-19 virus is suspected of having emerged in an unknown animal (perhaps a bat) and to have subsequently been transmitted to humans in the seafood and wild animal market.1 All over the world, there are surveillance borders to prevent the spread of the new unknown coronavirus, while some countries stopped flights to and from China.6 By the first week of the year 2020, 41 cases were confirmed to be COVID-19 positive, leaving one person dead and seven in critical care.7 This number is continuously increasing on a daily basis. The number of confirmed cases at the time of writing this manuscript exceeded 77,000, and there were confirmed more than 2200 deaths, mainly in mainland China. On January 20, 2020, the National Health Commission of China confirmed the human-to-human transmission of the new coronavirus outbreak.7 Ten days later, WHO declared COVID-19 as a Public Health Emergency of International Concern (PHEIC). COVID-19 symptoms include fever, malaise, dry cough, shortness of breath, and respiratory distress.1
COVID-19 is a member of Betacoronaviruses, like the former human coronaviruses SARS and MERS.8 , 9 With this novel human coronavirus, there are now seven different strains of Human coronaviruses (HCoVs), namely, 229E and NL63 strains of HCoVs (Alphacoronaviruses), OC43, HKU1, SARS, MERS, and COVID-19 HCoVs (Betacoronaviruses).1 , 8 , 10 SARS and MERS HCoV are the widely known strains of coronaviruses, and each has caused about 800 deaths. According to WHO, the mortality rates for SARS and MERS HCoV are 10% and 36%, respectively.8 , 10, 11, 12, 13 COVID-19 has a 2% mortality rate, but only in a few months will we know how fast the new virus spreads.
HCoVs are positive-sense, long (30,000 bp) single-stranded RNA viruses. Two groups of proteins characterize HCoVs; structural proteins such as Spike (S) that characterize all coronaviruses, Nucleocapsid (N), Matrix (M), and Envelope (E), in addition to the non-structural proteins, such as proteases (nsp3 and nsp5) and RdRp (nsp12).8 , 14 The Spike protein is a crucial recognition factor for virus attachment and entry to the host cells. It is present on the virion's outer surface in a homo-trimeric state.14 , 15
The Glucose Regulating Protein 78 (GRP78) or Binding immunoglobulin protein (BiP) is the master chaperone protein of the unfolded protein response (when unfolded or misfolded proteins accumulate).16, 17, 18, 19 Under reasonable conditions, GRP78 is found in the lumen of the Endoplasmic Reticulum (ER) bound to and inactivating three enzymes responsible for cell death or differentiation. These enzymes are Activating Transcription Factor 6 (ATF6), Protein kinase RNA-like Endoplasmic Reticulum Kinase (PERK), and Inositol-requiring Enzyme 1 (IRE1).14 Above a threshold of accumulated unfolded proteins, GRP78 releases ATF6, PERK, and IRE1, leading to their activation. Inhibition of protein synthesis and enhancement of the refolding is the end result of the enzymes’ activation.14 , 20 Overexpression of GRP78 is also initiated upon cell stress, which increases the chance for GRP78 to escape ER retention and translocate to the cell membrane. Once translocated to the cell membrane, GRP78 is susceptible to virus recognition by its substrate-binding domain (SBD), and it can mediate the virus entry in the cell.14 Pep42 is a cyclic peptide that has been reported to bind the GRP78 overexpressed and expressed at the surface of cancer cells.21
In this study, the spike protein of COVID-19 was modeled using solved structures in the protein data bank.22 After model validation, molecular docking was performed to test its binding affinity against GRP78. We hypothesized that GRP78 binds to COVID-19, as it happens in the case of the MERS-CoV coronavirus,23 and we tried to predict the binding site using the similarity between Pep42 and the COVID-19 Spike protein.14 Four regions of the spike were predicted to be the binding site to GRP78 based on sequence and structural similarity. The results are promising and suggest the possible recognition of the COVID-19 spike by the cell-surface GRP78 upon cell stress.
Materials and methods
All human coronaviruses (HCoV) spike proteins were downloaded from the National Center for Biotechnology Information (NCBI).24 Multiple Sequence Alignment (MSA) of all of the sequences was performed using the Clustal Omega web server of the European Bioinformatics Institute (EMBL-EBI).25 ESpript 3 software was used to represent the MSA.26 The pairwise sequence identity between COVID-19 spike protein and each of the other HCoV spike proteins was calculated using the same method.
A model was built for the COVID-19 spike using the SWISS-MODEL web server.27 The SARS HCoV spike (PDB ID: 6ACD, chain C), as the closest spike protein to COVID-19 among all of the human coronaviruses, was used as a template for building the COVID-19 spike. The structural superposition of the SARS spike (6ACD, chain C) and COVID-19 spike model was performed using PyMOL software.28
Four regions are selected from the COVID-19 spike protein receptor-binding domain (RBD). These regions are cyclic (starting and ending with Cysteine residues connected by a disulfide bond) and were hypothesized to be a possible binding site to GRP78 based on the alignment with the Pep42 cyclic peptide. Pairwise sequence alignment was performed for the Pep42 peptide against the four predicted regions of the COVID-19 spike utilizing the Clustal Omega web server. ProtScale web server of the ExPASy bioinformatic resource portal was used to compare the Pep42 sequence to the four regions of the COVID-19 spike that we hypothesized to be the binding site to GRP78.29 The Kyte & Doolittle hydrophobicity index was calculated for each residue of the peptides, while the grand average hydrophobicity (GRAVY) was calculated for each region of the spike and Pep42.
Molecular docking was utilized to test the binding affinity of the four regions of the COVID-19 spike against the solved structure of GRP78 Substrate Binding Domain (SBD) β. The only solved structure of wild-type, full-length GRP78 bound to ATP in the open configuration found in the PDB database is 5E84.30 , 31 The coordinates of 5E84 were downloaded and prepared for the docking experiment by removing water ions and the ligand. The solvated docking software, HADDOCK, was used in this study to dock the four regions of the spike model for COVID-19 against the solved structure of GRP78.32 The easy interface was utilized since no restraints are defined.33 GRP78 active residues (I426, T428, V429, V432, T434, F451, S452, V457, and I459) were retrieved from the literature.30 The active residues from the COVID-19 spike protein were chosen for each region of the spike to be the hydrophobic residues. In both proteins, the residues surrounding the active residues were selected as passive in HADDOCK. Active residues are the amino acid residues from the two interacting proteins’ binding sites that take part in direct interaction with the other protein partner while passive residues are the residues that can interact indirectly in HADDOCK.33, 34, 35, 36
Furthermore, Pep42 is tested against the GRP78 solved structure to compare its binding affinity to that of the four regions of the spike protein. The Pep42 3D structure was generated using the I-Tasser web server.37 Pep42 was treated as cyclic during the docking experiment (a distance restraint is added to HADDOCK) since the cyclic form of Pep42 is the selectivity determinant against GRP78 recognition.38 , 39
After docking, the docking complexes were analyzed by the aid of the Protein-Ligand Interaction Profiler (PLIP) web server of Technical University, Dresden.40 Two main types of interactions are established upon docking: H-bonding and hydrophobic interactions. PRODIGY software was used to predict the binding affinity for each region of the spike to GRP78. The average docking scores and the residues that take part in the interactions are discussed.
Results and discussion
Sequence and structural alignment
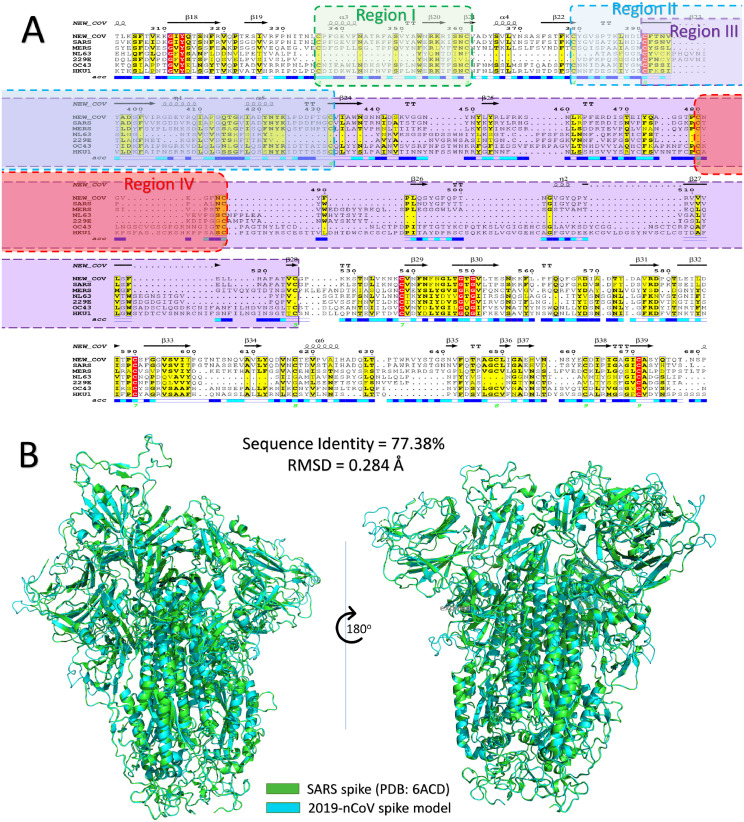
Fig. 1 A shows part of the Multiple Sequence Alignment (MSA) between the seven human coronaviruses (229E, NL63, OC43, HKU1, SARS, MERS, and COVID-19) spike proteins performed by Clustal Omega web server and visualized by ESpript software. The secondary structure for the COVID-19 spike model is displayed at the top of the MSA, and residual surface accessibility is present at the bottom. Alpha helices are shown by helix while arrows show beta-sheets on the top of the MSA. The residues that are surface accessible are in blue, while buried residues are in white at the bottom of the MSA. Identical residues are highlighted in red, while similar residues are highlighted in yellow. The positions of the disulfide bonds are marked by the green numbers below the accessibility rows in the MSA. 13 disulfide bonds are found in the spike protein from which we predict four regions to be the binding site with cell surface GRP78. These four regions of the spike protein, identified with the disulfides numbers 3, 4, 5, and 6 are marked in the MSA with green, blue, magenta, and red dashed lines, respectively. A complete MSA for the spike protein (1273 residues) is found in supplementary figure S1.
Fig. 1.
(A) Part of the multiple sequence alignment for the spike protein of all of the currently reported human coronaviruses strains (COVID-19, SARS, MERS, NL63, 229E, OC43, and HKU1). The alignment is made using the Clustal Omega web server and is displayed by ESpript 3 software. The red highlighted residues are identical, while yellow highlighted residues are conserved among the seven HCoVs. Secondary structures are represented at the top of the MSA for the COVID-19 spike, while the surface accessibility is shown at the bottom (blue, surface accessible, cyan, partially accessible, and white for buried residues). The four regions of the spike protein are shaded with green, blue, magenta, and red for regions I, II, III, and IV, respectively. (B) Structural superposition of SARS spike structure (green cartoon) and COVID-19 spike model (cyan cartoon). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The SARS spike protein sequence is the closest to the COVID-19 spike, with 77.38% identity. In contrast, OC43, MERS, HKU1, 229E, and NL63 share only 32.81%, 32.79%, 31.86%, 30.35%, and 28.28%, respectively, with COVID-19 spike. Fig. 1B shows the superposition of the homo-trimeric COVID-19 spike model (cyan cartoon) and SARS spike structure (PDB ID: 6ACD) (green cartoon). Two views are shown with a vertical axis rotation of 180°. The Root Mean Square Deviation (RMSD) between the two structures is only 0.284 Å, while the sequence identity is 77.38%.
Pep42 versus spike regions
Pep42 is reported to specifically target the cell-surface GRP78 in cancer cells.14 Its selectivity against GRP78 has been reported for its cyclic form but not for the extended form. This may be due to the rigidity of the cyclic structure of the peptide, which causes the stabilization of the hydrophobic patch formed by C1, V3, A4, L5, V10, V12, and C13. These residues become closer to each other by the aid of the disulfide bond, making the cyclic peptide the perfect docking platform for GRP78 SBDβ.
We found 13 disulfide bonds in the COVID-19 spike protein model that form 13 different cyclic regions that may resemble the cyclic Pep42. Four of these disulfides are found in the outer surface of the spike receptor-binding domain that faces the outside part of the virion, a region that has been targeted with neutralizing antibodies against the SARS and MERS spikes. These four regions, namely, the region I C336: C361 (26 residues), region II C379: C432 (54 residues), region III C391: C525 (135 residues), and region IV C480: C488 (9 residues), are marked in Fig. 1A.
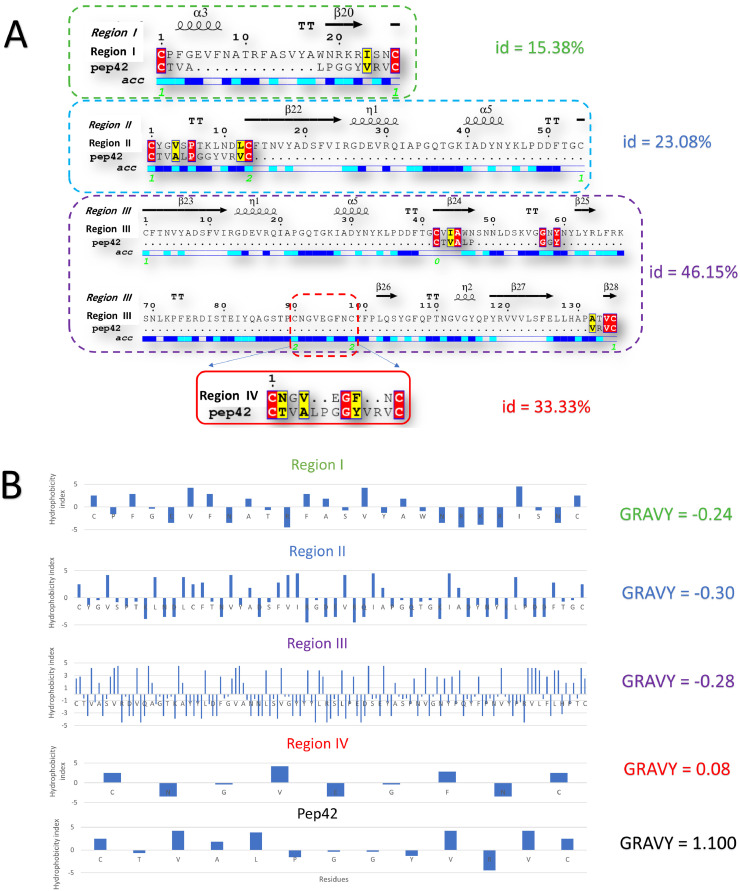
Fig. 2 A shows the pairwise sequence alignments between each spike region and Pep42. The percentage of pairwise sequence identity is listed on the right side of each alignment. The percent identity for region III is the most significant (46.15%) compared to other regions (15.38%, 23.08%, and 33.33% for the regions I, II, and IV, respectively). As shown in Fig. 2A, region IV is part of region III. Moreover, regions II and III share some residues. Again, identical residues are highlighted in red, while the conserved residues are highlighted in yellow. The secondary structure is shown at the top of the alignment and the surface accessibility at the bottom. Region IV has all of its residues exposed at the surface (either blue, meaning surface accessible, or cyan, for partially accessible residues). For other regions, some residues are surface exposed (blue or cyan), while others are buried (in white).
Fig. 2.
(A) Pairwise sequence alignment between Pep42 and four regions of the COVID-19 spike protein model. Secondary structures and surface accessibility are shown at the top and the bottom of the alignment, respectively. The percentage of identity arrows shows beta-sheets on the right side of each alignment. Identical and similar residues are highlighted in red and yellow, respectively. (B) The hydrophobicity index (Kyte & Doolittle) for the four regions of the spike protein model for COVID-19 and the Pep42 peptide. Grand average hydrophobicity (GRAVY) is shown on the right side for each peptide. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2B shows the hydrophobicity index (Kyte & Doolittle) for each suggested region and the Pep42. The grand average hydrophobicity index for each region (GRAVY) is listed in front of each peptide. Regions I, II, and III have negative values of GRAVY (−0.24, −0.30, and −0.28, respectively). In contrast, region IV has a positive value (0.08), which means that it has a slightly more hydrophobic character compared to other regions. Pep42 has a highly hydrophobic character (GRAVY value of 1.1) that enables it to be recognized by the cell-surface GRP78.14 , 38 , 39
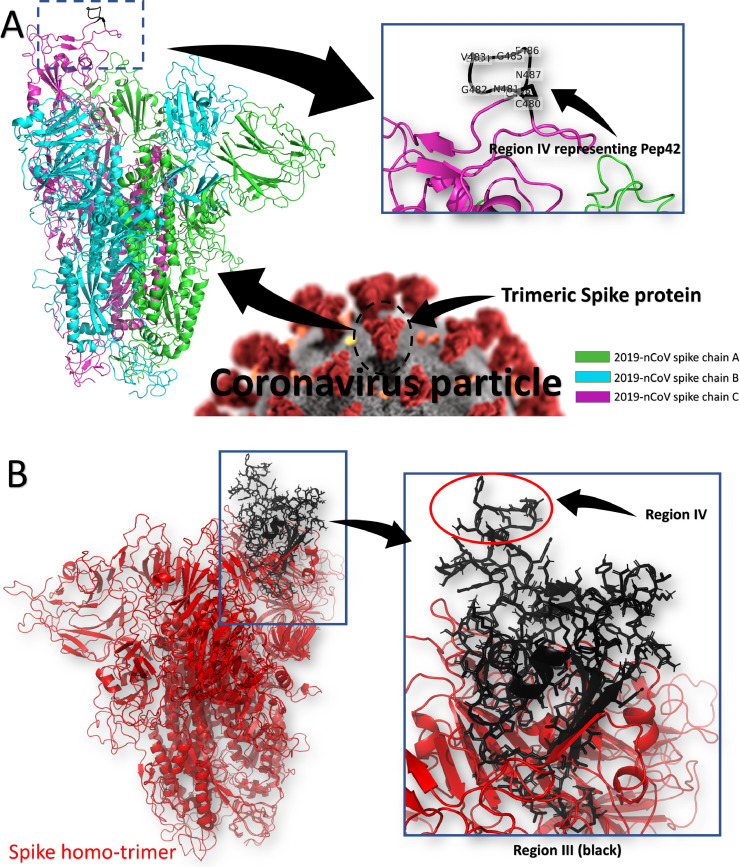
Fig. 3 A shows the structure model of the COVID-19 spike protein model (homo-trimeric) in a colored cartoon representation. Region IV of the spike (C480: C488) is not only cyclic but also surface-accessible and protrudes to the outer side of the spike, i.e., facing the target cell. It has a slightly hydrophobic character, hence resembling the Pep42 cyclic peptide, and it seems suitable to be the binding site to the cell-surface GRP78. Fig. 3B shows the region III of the spike (black cartoon). As shown in the enlarged panel, region IV is part of region III and it is the most surface-exposed part of the spike receptor-binding domain. The contribution of region IV (C480: C488) in binding region III to GRP78 is high (−9.8 out of −14.0 kcal/mol)
Fig. 3.
(A) The structure of the spike protein model of COVID-19 in its homotrimer state (colored cartoon). Two chains, A (green) and B (cyan) are in the closed conformation, while chain C (magenta) is the open configuration that makes it able to recognize the host cell receptor. Region IV of the spike (C480-C488), which we suggest is the recognition site for cell-surface GRP78, is shown in the black cartoon in the enlarged panel. (B) The structure of the region III (black carton). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Binding mode of spike-GRP78
GRP78-COVID-19 spike protein docking was performed using the HADDOCK software in four different ways. Each region of the spike (predicted to be the binding site to GRP78) was used as the binding site to GRP78, using its active residues selected to be that have hydrophobic character. The active residues for region I are: C336, F338, V341, F342, A344, R347, F348, S350, A352, I358, and C361. For region II, the active residues are: C379, V382, L387, I390, C391, F392, V395, A397, F400, V401, I402, V407, I410, A411, I418, A419, L425, F429, and C432. For region III, the active residues are: C391, F392, V395, A397, F400, V401, I402, V407, I410, A411, I418, A419, L425, F429, C432, V433, I434, A435, L441, V445, L452, L455, F456, L461, F464, I468, I472, A475, C480, V483, F486, C488, F490, L492, F497, V503, V510, V511, V512, L513, F515, L517, L518, A520, A522, V524, and C52. Finally, for region IV, the active residues are C480, V483, F486, and C488. For GRP78, the active site residues are retrieved from previous work to be I426, T428, V429, V432, T434, F451, S452, V457, and I459.
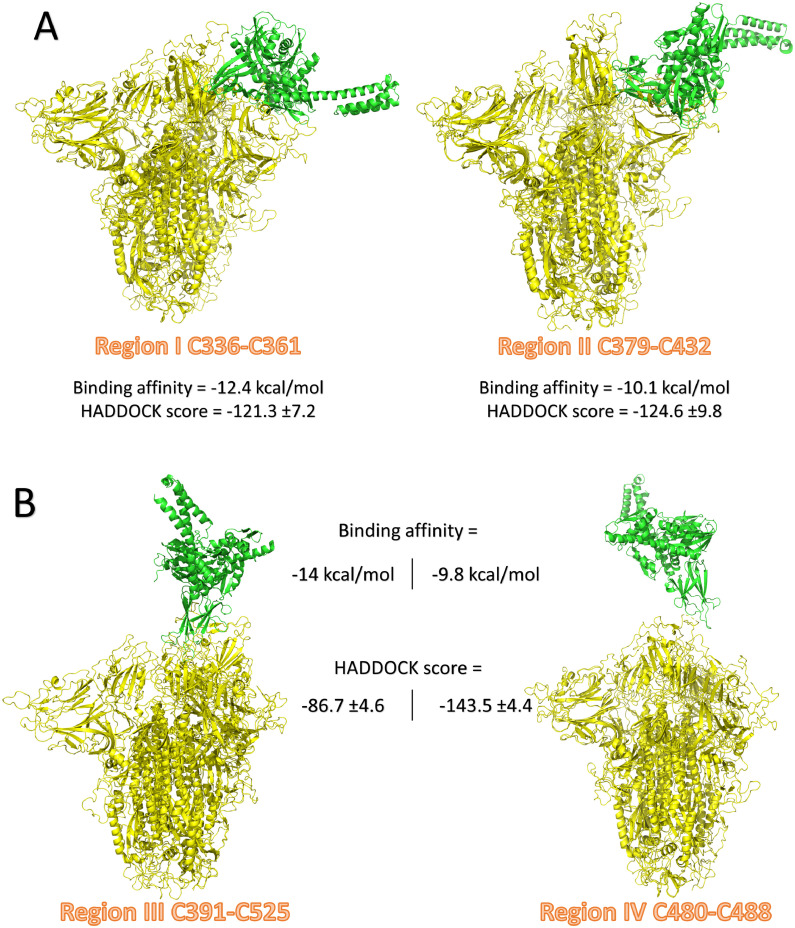
Figs. 4 A and 4B show the binding mode for each of the docking trials (the best-formed complexes from each docking experiment) with green cartoon representing GRP78 and yellow cartoon representing the homo-trimeric COVID-19 spike. All the docking trials proved the possibility of fitting the GRP78 SBDβ to the spike with binding affinities (predicted by PRODIGY) ranging from −9.8 up to −14 kcal/mol. In terms of the orientation of the two interacting proteins, regions III and IV are accepted to be the docking platform. In these two trials, the GRP78 and spike can interact in a head-to-head fashion.
Fig. 4.
The structure of the docking complexes of GRP78 (green cartoon) and COVID-19 spike (yellow cartoon) regions I and II (A) and regions III and IV (B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1 summarizes the docking trials of the four regions of the COVID-19 spike protein against GRP78. The docking scores are listed, while the interaction pattern is analyzed by PLIP software and listed in the table. As shown from the docking scores, region IV of the spike is the best docking platform to GRP78, with a score of −143.5 ± 4.4. This score is lower (better) than other regions by 18.3%, 15.2%, and 65.5% for the region I, region II, and region III, respectively. The PRODIGY binding affinities are also listed in Table 1. The PLIP analysis partially explains the binding affinity. Region IV of the spike interacts with the substrate-binding domain β of GRP78 with five H-bonds (through P479, N481, E484, and N487) and four hydrophobic interactions (through T478, E484, and F486). The average H-bond length for the docking trial of region IV is 2.26 ± 0.54, while the average hydrophobic contact length is 3.66 ± 0.18. These values are less than other docking trials using other regions.
Table 1.
The interactions formed between the spike protein of COVID-19 and cell-surface GRP78 SBDβ upon docking with HADDOCK.
| Pep42 conformation at | HADDOCK score | PRODIGY binding affinity (kcal/mol) | H-bonding |
Hydrophobic interaction |
||||
|---|---|---|---|---|---|---|---|---|
| number | Residues from the spike | Residues from GRP78 | number | Residues from the spike | Residues from GRP78 | |||
| Region I | −121.3 ±7.2 |
−12.4 | 11 | R466 N343 N354 N354 D467 E340 G339 I468 I468 K356 T345 |
T434 G489 S452 V453 T434 V490 G489 T434 K447 G454 G489 |
7 | E465 I468 W353 W353 A344 R346 R466 |
V432 L436 T428 V429 V490 Q492 T428 |
| Region II | −124.6 ±9.8 |
−10.1 | 12 | Y396 Y396 R357 R357 R466 R466 R466 F464 E516 E516 K356 N354 |
S452 S452 Q449 I450 G454 T456 T456 G454 S452 Q492 T428 G430 |
10 | E340 E516 K356 K356 F429 W353 N354 R355 R355 R357 |
V432 V490 T428 V429 V490 V453 V429 F451 V453 F451 |
| Region III | −86.7 ±4.6 |
−14.0 | 9 | N460 N487 D420 L455 K417 Y421 Y421 Y473 Y505 |
Q492 K447 S452 Q449 E427 I450 I450 S448 G430 |
3 | E406 K417 F486 |
V429 V429 T441 |
| Region IV | −143.5 ±4.4 |
−9.8 | 5 | E484 E484 N481 P479 N487 |
T428 V429 S452 S452 T458 |
4 | E484 E484 F486 T478 |
T428 V429 V457 V490 |
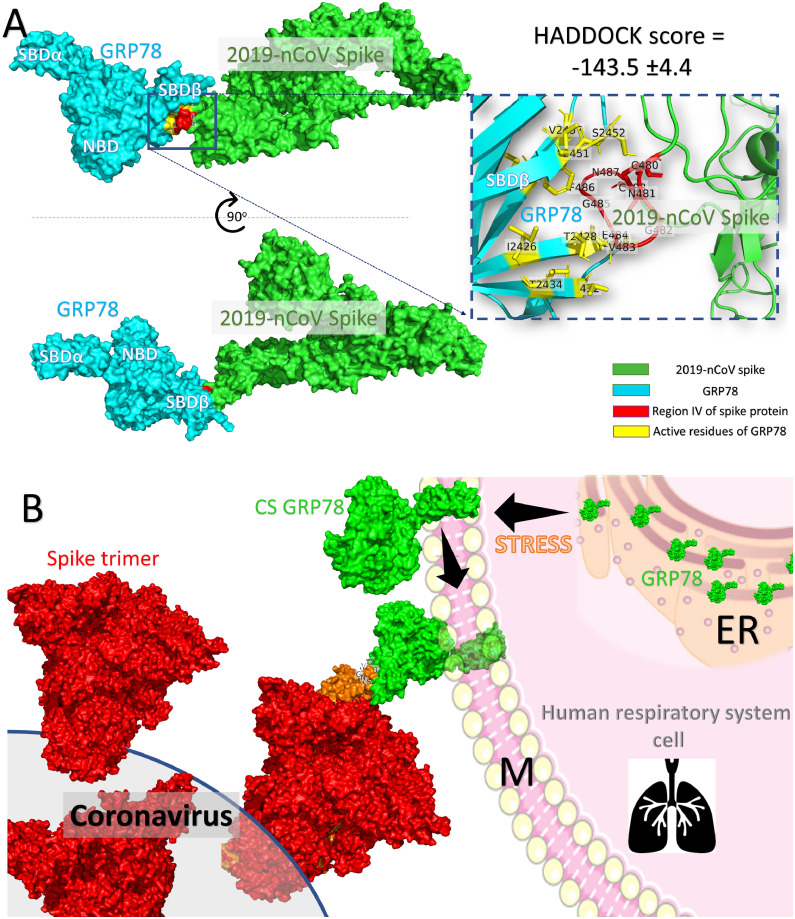
Fig. 5 A shows the predicted binding mode of the GRP78 (cyan surface) to the spike of the newly emerged coronavirus (green surface) using region IV of the spike (red surface) as the docking platform. The interacting residues of GRP78 and the spike proteins are shown in yellow and red, respectively. The enlarged panel shows in more detail the interacting residues of GRP78 (yellow sticks) and the region IV of the spike (red cartoon) labeled with its one-letter codes. This binding mode is acceptable since the two proteins are interacting, as when the virus is approaching the target cell (respiratory system cells) expressing its cell-surface receptor, GRP78. Fig. 5B shows a hypothetical binding model showing the homotrimer spike (red surface) protein of the COVID-19 bound to a respiratory system cell exposing the GRP78 protein (green surface). This scenario could occur in stressed cells when GRP78 is overexpressed and translocated from the Endoplasmic Reticulum (ER) to the cell membrane (M).
Fig. 5.
(A) The proposed binding mode of the host cell GRP78 (cyan surface) and the COVID-19 spike model (green surface) through region IV (C480-C488) (red surface). The amino acids from the GRP78 SBDβ that interact with the spike protein region IV (red cartoon) are labeled and represented in yellow sticks in the enlarged panel. (B) The proposed recognition mode of the COVID-19 spike (red surface) and cell-surface GRP78 (green surface) through the spike protein region C480-C488. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The predicted binding site of the Spike protein to GRP7 found in this study is in good agreement with studies that identified the spike receptor-binding domain using antibodies.23 , 41 Knowledge of this binding site could open the door for further experimental and simulation studies on the mode of envelope protein recognition by the highly dynamical GRP78 substrate-binding domain.
Conclusion
Spike protein is an essential viral element that helps in the attachment and virus internalization to the host cell. A vast amount of host cell receptors are targets for viruses, including the cell-surface GRP78. Inhibiting the interaction that occurs between the COVID-19 spike protein and the host cell receptor GRP78 would probably decrease the rate of viral infection. Furthermore, a vaccine against the COVID-19 spike protein would likely prevent viral infection. The present in silico perspective suggests the existence of a COVID-19 spike protein-GRP78 binding site, thus paving the route for drug designers to develop suitable inhibitors to prevent the binding and hence the infection. Future work involving the dynamics of GRP78 and the experimental validation is required to suggest potent peptidomimetic inhibitors.
Declaration of Competing Interests
All of the authors declare that there is no competing interest in this work.
Acknowledgments
Acknowledgments
We want to acknowledge Mrs. Alaa Ismail for helpful discussions and suggestions. We thank LetPub for its linguistic assistance during the preparation of this manuscript.
Data Availability
The docking structures are available upon request from the corresponding author.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.02.026.
Appendix. Supplementary materials
References
- 1.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med. 2020 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Organization WH. World Health Organization; 2020. Surveillance case definitions for human infection with novel coronavirus ( nCoV): interim guidance v1, January 2020. [Google Scholar]
- 4.Organization WH . World Health Organization; 2020. Laboratory testing of human suspected cases of novel coronavirus ( nCoV) infection: interim guidance, 10 January 2020. [Google Scholar]
- 5.Organization WH . World Health Organization; 2020. Infection prevention and control during health care when novel coronavirus ( nCoV) infection is suspected: interim guidance, January 2020. [Google Scholar]
- 6.Parr J. British Medical Journal Publishing Group; 2020. Pneumonia in China: lack of information raises concerns among Hong Kong health workers. [DOI] [PubMed] [Google Scholar]
- 7.Yang L. 2020. China confirms human-to-human transmission of coronavirus. [Google Scholar]
- 8.Elfiky A.A., Mahdy S.M., Elshemey W.M. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J Med Virol. 2017;89(6):1040–1047. doi: 10.1002/jmv.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. Middle East respiratory syndrome coronavirus (MERS-CoV). 23 june 2016 2016 (accessed 8 October 2016 2016).
- 11.Hemida M.G., Alnaeem A. Some one health based control strategies for the middle east respiratory syndrome coronavirus. One Health. 2019;8 doi: 10.1016/j.onehlt.2019.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the middle east respiratory syndrome coronavirus. J Virol. 2014;88(21):12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Organization W.H. World Health Organization; 2019. Clinical management of severe acute respiratory infection when Middle East respiratory syndrome coronavirus ( MERS-CoV) infection is suspected: interim guidance. [Google Scholar]
- 14.Ibrahim I.M., Abdelmalek D.H., Elfiky A.A. GRP78: a cell's response to stress. Life Sci. 2019;226:156–163. doi: 10.1016/j.lfs.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee A.S. The ER chaperone and signaling regulator GRP78/BIP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Lee A.S. Stress induction of GRP78/BIP and its role in cancer. Curr Mol Med. 2006;6(1):45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 18.Rao R.V., Peel A., Logvinova A. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514(2–3):122–128. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinones Q.J., Ridder G.G.D., Pizzo S.V. GRP78, a chaperone with diverse roles beyond the endoplasmic reticulum. Histol Histopathol. 2008 doi: 10.14670/HH-23.1409. [DOI] [PubMed] [Google Scholar]
- 20.Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BIP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y., Lillo A.M., Steiniger S.C. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45(31):9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 22.Berman H., Henrick K., Nakamura H. Announcing the worldwide protein data bank. Nat Struct Mol Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 23.Chu H., Chan C.-.M., Zhang X. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J Biol Chem. 2018;293(30):11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.NCBI. National Center of Biotechnology Informatics (NCBI) database websitehttp://www.ncbi.nlm.nih.gov/. 2020. http://www.ncbi.nlm.nih.gov/2020).
- 25.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 26.Gouet P., Courcelle E., Stuart D.I., M√©toz F. ESPript: analysis of multiple sequence alignments in postscript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 27.Biasini M., Bienert S., Waterhouse A. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(W1):W252–W2W8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1.7.6V. The PyMOL Molecular Graphics System, Version 1.7.6 Schrödinger, LLC.
- 29.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The proteomics protocols handbook. Springer; 2005. Protein identification and analysis tools on the Expasy server; pp. 571–607. [Google Scholar]
- 30.Yang J., Nune M., Zong Y., Zhou L., Liu Q. Close and allosteric opening of the polypeptide-binding site in a human HSP70 chaperone bip. Structure. 2015;23(12):2191–2203. doi: 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Zong Y., Su J. Conformation transitions of the polypeptide-binding pocket support an active substrate release from HSP70S. Nat Commun. 2017;8(1):1201. doi: 10.1038/s41467-017-01310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dijk A.D., Bonvin A.M. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22(19):2340–2347. doi: 10.1093/bioinformatics/btl395. [DOI] [PubMed] [Google Scholar]
- 33.de Vries S.J., van Dijk M., Bonvin A.M. The Haddock web server for data-driven biomolecular docking. Nat Protoc. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 34.de Vries S.J., van Dijk A.D.J., Krzeminski M. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the Capri targets. Proteins. 2007;69(4):726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 35.van Dijk A.D., de Vries S.J., Dominguez C., Chen H., Zhou H.X., Bonvin A.M. Data-driven docking: HADDOCK's adventures in Capri. Proteins. 2005;60(2):232–238. doi: 10.1002/prot.20563. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoneda Y., Steiniger S.C., Capkova K. A cell-penetrating peptidic GRP78 ligand for tumor cell-specific prodrug therapy. Bioorg Med Chem Lett. 2008;18(5):1632–1636. doi: 10.1016/j.bmcl.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y., Lillo A.M., Steiniger S.C. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45(31):9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 40.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W4W7. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pujhari S., Macias V.M., Nissly R.H., Nomura M., Kuchipudi S.V., Rasgon J.L. Heat shock protein 70 (Hsp70) is involved in the Zika virus cellular infection process. bioRxiv. 2017 doi: 10.1080/22221751.2018.1557988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The docking structures are available upon request from the corresponding author.