Abstract
Introduction
Sources describing the global burden of emerging diseases accurately are still limited. We reviewed coronavirus infections reported by ProMED and assessed the reliability of the data retrieved compared to published reports. We evaluated the effectiveness of ProMED as a source of epidemiological data on coronavirus.
Methods
Using the keyword “coronavirus” in the ProMED search engine, we reviewed all the information from the reports and collected data using a structured form, including year, country, gender, occupation, the number of infected individuals, and the number of fatal cases.
Results
We identified 109 entries reported between February 29, 2000 and January 22, 2020. A total of 966 cases were reported, with death reported in 188 cases, suggesting an overall case fatality rate (CFR) of 19.5%. Of 70 cases for which the gender was reported, 47 (67.1%) were male. Most of the cases were reported from China, the United Arab Emirates, and Saudi Arabia, with reports from other countries, including imported cases in Europe and North America.
Conclusions
Internet-based reporting systems such as ProMED are useful to gather information and synthesize knowledge on emerging infections. Although certain areas need to be improved, ProMED provided useful information about coronaviruses especially during outbreaks.
Keywords: Coronavirus, Outbreak, ProMED, Epidemiology
1. Introduction
Coronaviruses are enveloped non-segmented positive-sense RNA viruses belonging to the family Coronaviridae (subfamily Orthocoronavirinae) and the order Nidovirales and broadly distributed in humans and other mammals [1,2], including four genuses, Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus. Although most human coronavirus infections are mild, the previous epidemics of two betacoronaviruses, the severe acute respiratory syndrome coronavirus (SARS-CoV) [[3], [4], [5], [6], [7]] and Middle East respiratory syndrome coronavirus (MERS-CoV) [[8], [9], [10], [11]], have caused more than 10,000 cumulative cases in the past two decades, with mortality rates of 10% for SARS-CoV and 37% for MERS-CoV. The coronaviruses already identified might only be the tip of the iceberg, with potentially more novel and severe zoonotic events to be revealed [1]. Now, the 2019 novel CoV (2019-nCoV) have led to more than 17,400 cases and 362 death till February 2, 2020.
The global burden of coronavirus needs to be determined. In 2018, the World Health Organization (WHO) held its annual review of the Blueprint List of Priority Diseases, where Coronaviruses were considered and included. These diseases, given their potential to cause a public health emergency and the absence of efficacious drug and vaccine, are considered to need accelerated research and development [12]. Given the current situation of the 2019nCoV in China and multiple countries in different continents affected, the WHO declared this outbreak as a Public Health Emergency of International Concern (PHEIC) on Thursday, January 30, 2020 [13,14].
ProMED (The Program for Monitoring Emerging Diseases) is an internet-based reporting system for emerging infectious or toxin-mediated diseases [[15], [16], [17], [18], [19], [20]]. It was founded in 1994 and is currently a program of the International Society for Infectious Diseases [[15], [16], [17],21]. Its mission is to serve global health by the immediate dissemination of information, which may lead to new preventive measures, prevent the spread of outbreaks, and provide more accurate disease control. Its sources of information include media reports, official reports, online summaries, reports from local observers and others [21]. The reports are reviewed and edited by expert moderators in the field of infectious diseases and are then submitted to the mail server and published on the website and disseminated to subscribers by email. Being a non-profit, non-governmental system, it is free from governmental constraints [[15], [16], [17],21]. In 2014 had 70,000 subscribers in over 185 countries [21], and now (2020) is more 80,000 subscribers, representing almost every country in the world, with ProMED staff, moderators, and team, located across 32 countries and are constantly scanning for, reviewing and posting information related to global health security [22].
We reviewed the coronaviruses incidents reported by ProMED and assessed the reliability of the data retrieved compared to published reports, as has been previously performed for Crimean Congo Hemorrhagic Fever [15] and melioidosis [23]. We thus evaluated the effectiveness of ProMED as an epidemiological data source by focusing on coronaviruses.
2. Methods
The keyword “coronaviruses” was used in the ProMED search engine, looking at ProMED-mail (English). We reviewed all the information in the individual reports and collected data using a structured form, including year, country, gender, occupation, the number of infected individuals, and the number of fatal cases.
We also looked into reports of animal disease made during the study period and recorded in ProMED.
3. Results
One hundred and nine entries reported between February 29, 2000 and January 22, 2020 were identified (Table 1 ), 80.9% of them about MERS-CoV. A total of 966 cases were reported, with 188 cases recorded as being fatal, giving a reported CFR of 19.5% (Table 1). From the total number, 61.6% of cases corresponded to 2019-nCoV, 35.4% to MERS-CoV, 0.8% to SARS-CoV, and 2.2% for the rest of coronaviruses (Table 1). Gender was only recorded in 70 cases, of whom 47 (67.1%) were male. Of 54 cases for which the age was reported, the median was 60 years (interquartile range 49.0–70.8). Twenty cases were reported as imported, in the United Kingdom, Thailand, France, and the USA, among others (Table 1).
Table 1.
The number of infected and fatal coronavirus cases reported to ProMED between 2000 and 2020.
| Country | Infected patients, n | Country of origin, imported cases | Reported deaths, n | CFR% | % SARS-CoV | % MERS-CoV | % 2019-nCoV | % Others |
|---|---|---|---|---|---|---|---|---|
| China | 589 | 34 | 5.8 | 0.2 | 0 | 99.8 | 0 | |
| United Arab Emirates | 183 | 80 | 43.7 | 0 | 100 | 0 | 0 | |
| Saudi Arabia | 158 | 71 | 44.9 | 1.3 | 95.6 | 0 | 3.8 | |
| Jordan | 11 | 2 | 18.2 | 0 | 0 | 0 | 100.0 | |
| United Kingdoma | 9 | 4 from Qatar, 2 from Saudi Arabia (1 died) | 1 | 11.1 | 44.4 | 11.11 | 0 | 44.4 |
| Thailanda | 5 | China | 0 | 0.0 | 0 | 0 | 100.0 | 0 |
| Francea | 2 | 1 from Saudi Arabia and 1 from United Arab Emirates | 0 | 0.0 | 0 | 100.0 | 0 | 0 |
| Qatar | 1 | 0 | 0.0 | 100.0 | 0 | 0 | 0 | |
| Japana | 1 | China | 0 | 0.0 | 0 | 100.0 | 0 | 0 |
| Nepala | 1 | China | 0 | 0.0 | 0 | 100.0 | 0 | 0 |
| South Koreaa | 1 | China | 0 | 0.0 | 0 | 100.0 | 0 | 0 |
| USAa | 1 | China | 0 | 0.0 | 0 | 100.0 | 0 | 0 |
CFR%, case fatality rate.
imported cases.
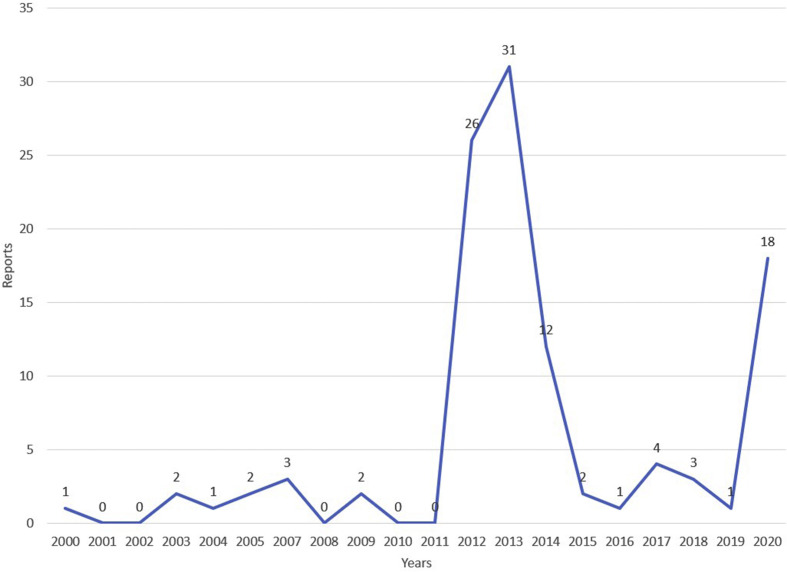
Most of the cases were reported from China, the United Arab Emirates, and Saudi Arabia (Table 1). The lowest fatality rates occurred among the imported cases (<15%). The highest number of cases were reported from Saudi Arabia (44.9%) and the United Arab Emirates (43.7%). The number of reports was higher in 2012–2013 and 2020 (Fig. 1 ).
Fig. 1.
Number of reports on “coronavirus” per year, ProMED, 2000–2020.
Regarding animal disease, thirty-five entries were reported, with 17 reports from USA (48.5%), four from China and four from Brazil, among others (Table 2 ). Among those CoV, characterized, most reports corresponded with equine CoV (20.0%), 11.4% with porcine deltacoronavirus (PDCoV), 8.6% with swine delta coronavirus (SDCV), and 5.4% with the porcine epidemic diarrhea virus (PEDV), among others (Table 2). From the reports, 36.3% informed on pigs infections, 24.2% on bats and 18.18% on horses, among others.
Table 2.
Coronavirus cases reported to ProMED in animals between 2000 and 2020.
| Country | Reports | % | % |
n, animals | |||||
|---|---|---|---|---|---|---|---|---|---|
| ECoV | SDCV | PDCoV | PEDV | MERS-CoV | Others CoV | ||||
| USA | 17 | 48.57 | 41.2 | 17.6 | 11.8 | 11.8 | 0.0 | 17.6 | 10154 |
| China | 4 | 11.43 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 294 |
| Brazil | 4 | 11.42 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 8 |
| Saudi Arabia | 3 | 8.57 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 1 |
| Canada | 2 | 5.71 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | N/A |
| Ecuador | 1 | 2.86 | 0.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1341 |
| Ireland | 1 | 2.86 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5 |
| Myanmar | 1 | 2.86 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | N/A |
| Qatar | 1 | 2.86 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 1 |
| UAE | 1 | 2.86 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 0.0 | 14 |
PDCoV, porcine deltacoronavirus; SDCV, swine delta coronavirus; PEDV, porcine epidemic diarrhea virus; MERS, Middle East Respiratory Syndrome.
4. Discussion
Track infectious diseases trends is a paramount work, especially globally speaking, depending on multiple sources, counting on accurately information when possible, timely and appropriate disseminated to people and healthcare workers that will use it, and this is also a complex case for CoV infections, where systems such as ProMED provide an opportunity to alert the international community to the occurrence of cases around the world, including the identification of new foci of outbreaks [24]. Coronaviruses cause respiratory and intestinal infections in animals and humans. They were not considered to be highly pathogenic to humans until the outbreak of severe acute respiratory syndrome (SARS) in 2002 and 2003 in Guangdong province, China, as the coronaviruses that circulated before that time in humans mostly caused mild infections in immunocompetent people [25]. After that, with MERS-CoV, and now in 2020 with the outbreak of the 2019nCoV, there is an increasing concern on highly pathogenic coronaviruses, such as these, but also other human and animal coronaviruses, should be highlighted. As observed in the results of this study, other human coronaviruses, such as HCoV-229E, -HKU1, -NL63 and -OC43 [25], are poorly reported in posts of ProMED, but also would consequence of weak interest on them, as has been recently evidenced in a bibliometric analysis [26]. The three highly pathogenic viruses (SARS-CoV, MERS-CoV and 2019-nCoV), cause severe respiratory syndrome in humans, and the other four human coronaviruses (HCoV- NL63, HCoV-229E, HCoV- OC43 and HCoV-HKU1) induce only mild upper respiratory diseases in immunocompetent hosts, although some of them can cause severe infections in infants, young children and elderly individuals [25], and in immunocompromised patients.
When compared with the information provided by reports in the subject literature, the most notable finding was that the CFR identifiable from ProMED was low in comparison with the published literature 10% for SARS-CoV and 37% for MERS-CoV. The coronaviruses already identified might only be the tip of the iceberg, with potentially more novel and severe zoonotic events to be revealed [1], but under-reporting of fatal cases by ProMED is not surprising, as the purpose is to act as an early alert system, and outcomes may not be known at the time of posting [23]. Besides, the data were incomplete with regards to characteristics such as gender, age and occupation (last, no analyzed).
In the present study, the highest numbers of reported cases corresponded with countries well known for having outbreaks of SARS-CoV, MERS-CoV, and the recent 2019-nCoV, autochthonous or imported, which is consistent with the known epidemiology of the disease, then becoming ProMED as a relevant source of epidemiological alerts. Although that ProMED cannot always be regarded as an accurate source of information about the patient outcome, which is not surprising as that is not its purpose.
The current 2019nCoV outbreaks are affecting 27 countries, 16 in Asia (including China), 8 in Europe, 2 in the Americas, and Australia in Oceania. There is still much global concern, considering the PHEIC declared by WHO, but also for the uncertainty of many questions about the clinical features and especially how to deal individually and collectively with the managing of them, including preventive and therapeutically [1,5,[27], [28], [29], [30]].
Although ProMED proved to be a useful source of information about coronavirus, it was evident that it gave a misleading impression of the distribution and mortality in some areas, as has been previously reported for other infections [15]. We think it would be useful to consider the adoption of standardized templates for reporting to ProMED in order to enhance the standardization of data and provide clear, accurate and reliable information that could be used in conducting epidemiological analyses to establish the status of emerging diseases and assist in their recognition and control, as has been previously suggested [23]. Besides, we think it would be useful for reports in one language to be mirrored to ProMED in other languages, especially when it is the language of the country from which the report originated.
Also, ProMED would be partially useful as a source of information for travel medicine practice [3,31]. Clinicians in this setting need to consider coronavirus in the evaluation and treatment of patients with febrile illnesses returning from endemic countries, particularly from China, Southeast Asia and Middle East countries, as has been seen in this study. A more accurate knowledge of this disease among physicians in non-endemic areas is needed for better management of imported cases and professional exposures especially with emerging viruses, such as the one causing the 2019 novel CoV [1,27], and ProMED would also be a source of epidemiological information when giving pre-travel advice and post-travel consultation.
Also, to all of this, another critical aspect, in the context of reports on CoV at ProMED, is to have them on the occurrence of infections in animals. Currently, there is significant concern about the animal origin of the 2019nCoV, probably on bats, but also suggested on other mammals, birds and even snakes [25,[32], [33], [34]]. It is imperative that animal reservoirs that transmitted the 2019nCoV to humans be identified as quickly as possible to aid in the control of this disease. It is thus urgent that the results of animal testing from the seafood market in Wuhan, where the virus was initially isolated, be released as soon as possible [34].
In different regions, strategies for surveillance and recognition of coronavirus infections need to be strengthened. Working groups of national infectious disease societies could help to increase awareness among general and infectious disease practitioners, but might also influence public health authorities by suggesting that CoV infections become notifiable diseases in each country.
It should be highlighted that the reliability of ProMED information when issuing from the general press, could be questioned. However, also uses as sources the WHO, the Pan American Health Organization/WHO (PAHO/WHO), other WHO regional offices, the U.S. Centers for Disease Control and Prevention (CDC), among other official agencies. Sometimes, also papers from scientific literature, are highlighted and posted in ProMED.
Rapid dissemination as trying continuously ProMED since 1994, in emerging infectious diseases, such as is the case of SARS-CoV, MERS-CoV and now 2019-nCoV, is remarkably. Even also, now scientific and especially medical journals are open to make a rapid dissemination of information, expeditely peer-reviewed, in order to place online relevant information during the epidemics [35]. This has been the case of the Lancet, the New England Journal of Medicine, F1000 Research, Viruses, and Travel Medicine and Infectious Diseases, among other journals [34,36,37].
In conclusion, information systems such as ProMED provide insight through daily reports of the occurrence of both individual cases and clusters of infectious diseases. This helps the rapid dissemination of knowledge of the current global situation of emerging infectious diseases and their outcomes in terms of morbidity, disability, and death. However, the data on ProMED is often incomplete and it is essential to continue improving and supplementing these systems to allow a more precise knowledge of the worldwide epidemiology of emerging diseases such as coronavirus infections.
Funding
Universidad Tecnologica de Pereira, Pereira, Risaralda, Colombia.
CRediT authorship contribution statement
D. Katterine Bonilla-Aldana: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Yeimer Holguin-Rivera: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. Isabella Cortes-Bonilla: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. María C. Cardona-Trujillo: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. Alejandra García-Barco: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. Hugo A. Bedoya-Arias: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. Ali A. Rabaan: Formal analysis, Writing - review & editing. Ranjit Sah: Formal analysis, Writing - review & editing. Alfonso J. Rodriguez-Morales: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of competing interest
There is no conflict of interest.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P. Coronavirus infections and immune responses. J Med Virol. 2020 doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tawfiq J.A., Zumla A., Memish Z.A. Travel implications of emerging coronaviruses: SARS and MERS-CoV. Trav Med Infect Dis. 2014;12:422–428. doi: 10.1016/j.tmaid.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilder-Smith A. The severe acute respiratory syndrome: impact on travel and tourism. Trav Med Infect Dis. 2006;4:53–60. doi: 10.1016/j.tmaid.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Al-Tawfiq J.A., Gautret P. Asymptomatic Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection: extent and implications for infection control: a systematic review. Trav Med Infect Dis. 2019;27:27–32. doi: 10.1016/j.tmaid.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baharoon S., Memish Z.A. MERS-CoV as an emerging respiratory illness: a review of prevention methods. Trav Med Infect Dis. 2019:101520. doi: 10.1016/j.tmaid.2019.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization . WHO; 2018.. List of Blueprint priority diseases.https://www.who.int/docs/default-source/blue-print/2018-annual-review-of-diseases-prioritized-under-the-research-and-development-blueprint.pdf?sfvrsn=4c22e36_2 [Google Scholar]
- 13.World Health Organization Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) 2020. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov
- 14.World Health Organization Novel coronavirus (2019-nCoV) - situation report - 10 - 30 January 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200130-sitrep-10-ncov.pdf?sfvrsn=d0b2e480_2
- 15.Ince Y., Yasa C., Metin M., Sonmez M., Meram E., Benkli B. Crimean-Congo hemorrhagic fever infections reported by ProMED. Int J Infect Dis : IJID : official publication of the International Society for Infectious Diseases. 2014;26:44–46. doi: 10.1016/j.ijid.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Madoff L.C. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis : an official publication of the Infectious Diseases Society of America. 2004;39:227–232. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- 17.Madoff L.C., Woodall J.P. The internet and the global monitoring of emerging diseases: lessons from the first 10 years of ProMED-mail. Arch Med Res. 2005;36:724–730. doi: 10.1016/j.arcmed.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Rolland C., Lazarus C., Giese C., Monate B., Travert A.S., Salomon J. Early detection of public health emergencies of international concern through undiagnosed disease reports in ProMED-mail. Emerg Infect Dis. 2020;26:336–339. doi: 10.3201/eid2602.191043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrion M., Madoff L.C. ProMED-mail: 22 years of digital surveillance of emerging infectious diseases. Int Health. 2017;9:177–183. doi: 10.1093/inthealth/ihx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai A.N., Anyoha A., Madoff L.C., Lassmann B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int J Infect Dis. 2019;84:48–53. doi: 10.1016/j.ijid.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ProMED About ProMED-mail. 2014. https://promedmail.org/about-promed/
- 22.ProMED About ProMED-mail. 2020. https://promedmail.org/about-promed/
- 23.Nasner-Posso K.M., Cruz-Calderon S., Montufar-Andrade F.E., Dance D.A., Rodriguez-Morales A.J. Human melioidosis reported by ProMED. Int J Infect Dis. 2015;35:103–106. doi: 10.1016/j.ijid.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollack M.P., Pringle C., Madoff L.C., Memish Z.A. Latest outbreak news from ProMED-mail: novel coronavirus -- Middle East. Int J Infect Dis : IJID : official publication of the International Society for Infectious Diseases. 2013;17:e143–e144. doi: 10.1016/j.ijid.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonilla-Aldana D.K., Quintero-Rada K., Montoya-Posada J.P., Ramirez S., Paniz-Mondolfi A., Rabaan A. SARS-CoV, MERS-CoV and now the 2019-novel CoV: have we investigated enough about coronaviruses? - a bibliometric analysis. Trav Med Infect Dis. 2020:101566. doi: 10.1016/j.tmaid.2020.101566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan J.F.-W., Yuan S., Kok K.-H., To K.K.-W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020 doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020 doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallego V., Berberian G., Lloveras S., Verbanaz S., Chaves T.S., Orduna T. The 2014 FIFA World Cup: communicable disease risks and advice for visitors to Brazil--a review from the Latin American Society for Travel Medicine (SLAMVI) Trav Med Infect Dis. 2014;12:208–218. doi: 10.1016/j.tmaid.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Ji W., Wang W., Zhao X., Zai J., Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Med Virol. 2020 doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau S.K.P., Li K.S.M., Luk H.K.H., He Z., Teng J.L.L., Yuen K.Y. Middle East respiratory syndrome coronavirus antibodies in bactrian and hybrid camels from dubai. mSphere. 2020;5 doi: 10.1128/mSphere.00898-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S.L., Saif L. Emerging viruses without borders: the Wuhan coronavirus. Viruses. 2020;12 doi: 10.3390/v12020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubin E.J., Baden L.R., Morrissey S., Campion E.W. Medical journals and the 2019-nCoV outbreak. N Engl J Med. 2020 doi: 10.1056/NEJMe2001329. [DOI] [PubMed] [Google Scholar]
- 36.The L. Emerging understandings of 2019-nCoV. Lancet. 2020;395:311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veljkovic V., Vergara-Alert J., Segalés J., Paessler S. Use of the informational spectrum methodology for rapid biological analysis of the novel coronavirus 2019-nCoV: prediction of potential receptor, natural reservoir, tropism and therapeutic/vaccine target [version 2; peer review: awaiting peer review] F1000Research. 2020;9 doi: 10.12688/f1000research.22149.1. [DOI] [PMC free article] [PubMed] [Google Scholar]