Abstract
It is important to identify viruses in animals because most infectious diseases in humans are caused by viruses of zoonotic origin. African green monkey is a widely used non-human primate model in biomedical investigations. In this study, total RNAs were extracted from stool samples of 10 African green monkeys with diarrhea. High-throughput sequencing was used to characterize viromes. PCR and Sanger sequencing were used to determine the full genome sequences. Great viral diversity was observed. The dominant viruses were enteroviruses and picobirnaviruses. Six enterovirus genomes and a picobirnavirus RNA-dependent RNA polymerase sequence were characterized. Five enteroviruses belonged to two putative new genotypes of species Enterovirus J. One enterovirus belonged to EV-A92. The picobirnavirus RNA-dependent RNA polymerase sequence had the highest nucleotide similarity (93.48%) with human picobirnavirus isolate GPBV6C2. The present study helped to identify the potential zoonotic viruses in African green monkeys. Further investigations are required to elucidate their pathogenic roles in animals and humans.
Keywords: Virome, African green monkey, Enterovirus, Picobirnavirus
Highlights
-
•
Great viral diversity was observed from stool samples of African green monkeys in China
-
•
Six novel enterovirus genomes and a picobirnavirus RdRp sequence were characterized
-
•
Two new genotypes of species Enterovirus J that had not been reported before were assigned
-
•
The picobirnavirus RdRp sequence had 93.48% nucleotide similarity with human picobirnavirus isolate GPBV6C2
1. Introduction
Viruses are considered the most abundant and diverse organisms. They infect almost every other organism in nature and cause diseases ranging from self-limiting diseases to acute fatal diseases (Pallansch et al., 2013; Herrington et al., 2015). A great number of human infectious diseases are caused by viruses that evolved from zoonotic viruses such as SARS-Cov from bats (Hu et al., 2017), ebola virus from bats (Biek et al., 2006) and HIV from non-human primates (NHPs) (Sharp and Hahn, 2010; Chahroudi et al., 2012). It is important to identify viruses in animals to provide basic information for their spread to humans and their relationships with pathogenic infections.
African green monkey is a widely used NHP model in biomedical investigations because of its similarities to humans in terms of anatomy, immunology and physiology (Jasinska et al., 2013). Ward et al. (2008) used adult male African green monkeys as a preclinical pharmacokinetic model to validate their utility for predicting human pharmacokinetics using a set of marketed compounds. African green monkey was considered a viral reservoir which might spread viral pathogens to humans (Chahroudi et al., 2012). Some research investigated certain viruses in African green monkeys. Oberste et al. (2013) investigated three picornavirus genera (Enterovirus, Parechovirus, and Sapelovirus) in African green monkeys at the Dazark Zoo of Bangladesh, but none was detected. Gallagher et al. (2017) detected simian picobirnaviruses in fecal samples from diarrhea-free African green monkeys of the Caribbean region. However, little research has reported the virome in stools of African green monkeys.
In this study, we collected stools from 10 African green monkeys with diarrhea in a captive colony in China and then detected the gut RNA virome using high-throughput sequencing technology. Great diversity of virus families were identified. Enterovirus and picobirnavirus reads accounted for the vast majority of the mammalian virus reads. They were identified and further characterized. This knowledge may provide evidence of the causes of diarrhea and lead to effective strategies for preventing the transmission of viruses from African green monkeys.
2. Materials and methods
2.1. Specimen collection
Twenty-two African green monkeys from a captive colony at an animal research center of Beijing in China were found to have contracted diarrhea. Ten stool samples from separate African green monkeys with diarrhea were collected and stored at −70 °C until further high-throughput sequencing analysis.
2.2. RNA extraction
The stool samples were diluted with phosphate-buffered saline (Thermo Fisher Scientific, USA) at a w/v ratio of 20% and crushed with a homogeniser at a speed of 50 Hz for 10 s. The supernatants were collected after centrifugation at 12,000 rpm for 10 min and then filtered through 0.22-μm filters (Merck, Germany). Total RNAs were extracted from 400 μl of the clarified supernatants using a High Pure Viral RNA Kit (Roche, Switzerland) in accordance with the manufacturer's instructions.
2.3. High-throughput sequencing
RNA sequencing libraries were constructed using Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific). High-throughput sequencing was then performed using an Ion Torrent S5 sequencer (Thermo Fisher Scientific). The raw data were filtered to remove low-quality reads using FASTP (Chen et al., 2018). Filtered reads were normalized using BBNorm software (Biowulf, USA) and then compared with the local virus databases established from NCBI using BLAST (Cameron et al., 2006) to discover virus-related sequences. Then, mammalian virus reads were extracted for analysis.
2.4. Genome sequencing
Reads similar to certain virus were abstracted and assembled to contigs using Newbler software v2.9 (Roche). Consensus sequences were constructed with viral reads mapped to the most similar sequences by BLAST using CLC Genomic Workbench v.9.0 (Qiagen Bioinformatics, Germany). The gaps between the sequence contigs were determined using PCR and Sanger sequencing. A cDNA amplification kit for 5′ and 3′ rapid amplification of cDNA ends (TaKaRa, Japan) was used to determine the full terminal sequences in accordance with the manufacturer's instructions. Sequences were validated by mapping the original high-throughput sequencing data with genome sequences in this study as references using CLC Genomic Workbench v.9.0. The verified high-throughput sequencing depth and coverage of each sequence were shown in supplementary material Table S1.
2.5. Genome analysis
The online BLASTn and BLASTp methods (Cameron et al., 2006) were used to compare the viral nucleotide or amino acid sequences with all of the currently published sequences in the NCBI database. The complete genomic sequences were annotated using the RAST (Rapid Annotation using Subsystem Technology) server (Aziz et al., 2008). The pairwise nucleotide identities of enteroviruses were calculated using Clustal Omega (Madeira et al., 2019).
2.6. Phylogenetic analysis
Sequences were aligned using MAFFT version 7 (Katoh and Standley, 2013). MEGA 7 (Kumar et al., 2016) was used to generate maximum-likelihood phylogenetic trees with the best calculate model. Bootstrap values were calculated from 1000 replicates.
2.7. GenBank accession numbers
The GenBank accession numbers for the complete genomes of novel enterovirus strains CHN/BJ/2018-1J, CHN/BJ/2018-8J, CHN/BJ/2018-9J, CHN/BJ/2018-10J, CHN/BJ/2018-2J, and CHN/BJ/2018-1A in this study are MN427526, MN427527, MN427528, MN865120, MN865119 and MN427525, respectively. The GenBank accession number for the nucleotide sequence of picobirnavirus strain SPBV-3 RNA-dependent RNA polymerase (RdRp) is MN871976. The GenBank accession numbers for the reference sequences enterovirus J strain 1631, human enterovirus 92 strain RJG7, human picobirnavirus RdRp gene isolate GPBV6C2, picobirnavirus strain PBV/Simian/KNA/08873/2015 RdRp sequence are AF326766.2, EF667344.1, AB517732.1, KY053140.1.
3. Results
3.1. Overview of viral sequences
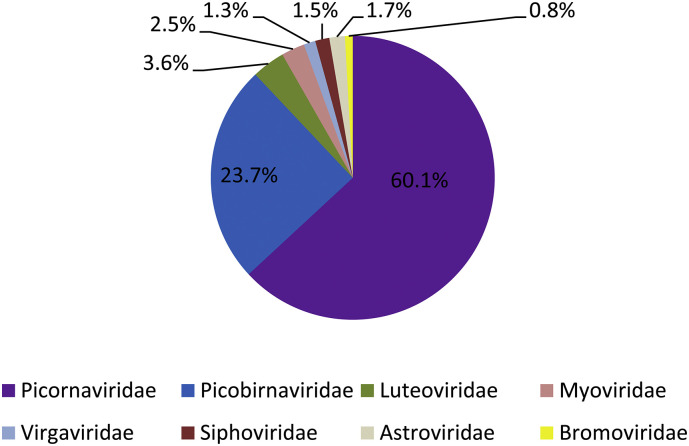
Great viral diversity was detected from stool samples of African green monkeys in this study. Reads in most abundant viral families are depicted in Fig. 1 . The most abundant virus families were Picornaviridae and Picobirnaviridae. The diverse reads related to these two families occupied 83.8% (60.1% and 23.7%, respectively) of the total viral reads. Sequences of plant virus families (Luteoviridae, Virgaviridae, Bromoviridae) and bacteriophage families (Myoviridae, Siphoviridae) were not further analyzed. Reads in mammalian virus families (Picornaviridae, Picobirnaviridae, Astroviridae) were further characterized.
Fig. 1.
Overview of the most abundant viral families among all virus reads identified in stool samples of African green monkeys in this study. Families with fewer than 100 reads were excluded. Virus families are indicated by the color code on the bottom. Percentages of reads in different viral families are indicated in the pie graph. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Further analysis of these mammalian virus reads are shown in Table 1 . Enterovirus, picobirnavirus, sapelovirus and bastrovirus sequences were detected. Enterovirus reads were found in nine African green monkeys, accounting for almost 100% of the mammalian virus reads in five samples, while 0.2–6.4% in the other four samples. Picobirnavirus reads were found in seven African green monkeys. They accounted for 84.8%–95.6% of the mammalian virus reads in five animals, while only 0.2% and 0.7% in the other two animals. The remaining putative mammalian virus reads were related to sapelovirus or bastrovirus, accounting for 0.2%–7.6% of the mammalian virus reads in different samples. 36 sapelovirus reads were found mainly in one green monkey with high abundance of enterovirus reads, and 5–136 bastrovirus reads was found in six green monkeys with picobirnavirus reads. Enterovirus and picobirnavirus sequences, which were the majority in the samples, were further characterized. Attempts to obtain more sequences from sapelovirus and bastrovirus were unsuccessful, probably because of low virus titers in the samples.
Table 1.
Characterisation of the mammalian RNA viral sequence reads in stool samples from African green monkeys with diarrhea.
| ID | Total reads no. of mammalian virus | Name of the match | Total reads no. of a specific virus | Percentage of specific virus reads | Virus strains identified |
|---|---|---|---|---|---|
| AGM01 | 6925 | Enterovirus | 6817 | 98.4% | CHN/BJ/2018-1J |
| Sapelovirus | 36 | 0.5% | CHN/BJ/2018-1A | ||
| Picobirnavirus | 48 | 0.7% | |||
| Bastrovirus | 11 | 0.2% | |||
| Other unclassified | 13 | 0.2% | |||
| AGM02 | 2222 | Enterovirus | 2204 | 99.2% | CHN/BJ/2018-2J |
| Other unclassified | 18 | 0.8% | |||
| AGM03 | 848 | Picobirnavirus | 811 | 95.6% | SPBV-3 |
| Bastrovirus | 37 | 4.4% | |||
| AGM04 | 125 | Enterovirus | 8 | 6.4% | SPBV-4 |
| Picobirnavirus | 106 | 84.8% | |||
| Bastrovirus | 5 | 4.0% | |||
| Other unclassified | 6 | 4.8% | |||
| AGM05 | 1798 | Enterovirus | 4 | 0.2% | SPBV-5 |
| Picobirnavirus | 1657 | 92.2% | |||
| Bastrovirus | 136 | 7.6% | |||
| Other unclassified | 1 | 0.1% | |||
| AGM06 | 784 | Enterovirus | 7 | 0.9% | SPBV-6 |
| Picobirnavirus | 732 | 93.4% | |||
| Bastrovirus | 45 | 5.7% | |||
| AGM07 | 924 | Enterovirus | 2 | 0.2% | SPBV-7 |
| Picobirnavirus | 855 | 92.5% | |||
| Bastrovirus | 66 | 7.1% | |||
| Other unclassified | 1 | 0.1% | |||
| AGM08 | 263 | Enterovirus | 260 | 98.9% | CHN/BJ/2018-8J |
| Other unclassified | 3 | 1.1% | |||
| AGM09 | 1268 | Enterovirus | 1251 | 98.7% | CHN/BJ/2018-9J |
| Picobirnavirus | 2 | 0.2% | |||
| Other unclassified | 15 | 1.2% | |||
| AGM10 | 70 | Enterovirus | 70 | 100.0% | CHN/BJ/2018-10J |
3.2. Enterovirus
Six complete enterovirus genomes were detected from five samples with abundant enterovirus reads. The six genome lengths ranged from 7319 bp to 7370 bp. The annotated results from RAST illustrated that all six genomes had only one large typical enterovirus open-reading frame encoding all of the structural proteins and non-structural proteins. All the six strains were enteroviruses that had not been reported before. Four strains belonged to a putative new genotype of species Enterovirus J, another strain belonged to another putative new genotype of species Enterovirus J, and the other strain belonged to EV-A92.
Four enterovirus strains had high degrees of genome nucleotide similarity (92.05%–97.83%) and polyprotein amino acid identity (96.17%–99.4%) with each other. Their VP1 sequences exhibited 91.06%–99.07% nucleotide similarity and 94.08%–99.65% amino acid identity. They were tentatively named CHN/BJ/2018-1J, CHN/BJ/2018-8J, CHN/BJ/2018-9J and CHN/BJ/2018-10J, abbreviated as 1J, 8J, 9J, 10J in this article, respectively. Online BLASTp analysis of their polyproteins showed that they had the highest identity (72.58%–73.09%) with strain 1631 of species Enterovius J.
The enterovirus strain CHN/BJ/2018-2J (abbreviated as 2J) also displayed the greatest similarity to strain 1631, with 72.10% polyprotein amino acid identity. Its genome shared 85.37%–86.41% nucleotide similarity and 92.43%–93.4% amino acid identity with that of strains 1J, 8J, 9J and 10J. VP1 sequence of 2J exhibited relatively low nucleotide similarity (73.17%–74.45%) and amino acid identity (79.79%–81.18%) with those of strains 1J, 8J, 9J and 10J.
The pairwise similarity of strains 1J, 8J, 9J, 10J and 2J with stains of species Enterovius J was calculated using Clustal Omega as presented in Table 2 . VP1 regions of strains 1J, 8J, 9J, 10J and 2J had significantly lower identities with stains of species Enterovius J than observed for other regions.
Table 2.
The nucleotide and deduced amino acid sequence pairwise identities of strains 1J, 8J, 9J, 10J, and 2J with species Enterovirus J.
| Region | Identity of 1J, 8J, 9J, 10J with Enterovirus J (%) |
Identity of 2J with Enterovirus J (%) |
||
|---|---|---|---|---|
| Nucleotide | Amino acid | Nucleotide | Amino acid | |
| Polyprotein | 65.14–67.59 | 70.32–73.53 | 65.79–67.04 | 71.24–73.3 |
| 1A (VP4) | 67.65–71.57 | 73.5–89.71 | 69.61–71.08 | 73.53–89.71 |
| 1B (VP2) | 58.57–65.02 | 61.32–68.31 | 60.36–62.55 | 62.55–65.02 |
| 1C (VP3) | 59.63–62.73 | 57.38–64.41 | 58.79–59.63 | 57.38–62.29 |
| 1D (VP1) | 50–58.16 | 46.35–55.45 | 51.08–59.52 | 46.35–53 |
| P1 | 57.22–61.11 | 57.58–61.06 | 57.58–59.58 | 58.06–59.61 |
| 2A | 64–67.78 | 65.33–72.67 | 65.33–69.33 | 67.33–74.67 |
| 2B | 61.28–69.7 | 72.73–79.8 | 61.28–70.03 | 72.73–78.79 |
| 2C | 70.83–72.66 | 79.27–82.93 | 71.75–72.26 | 79.88–82.01 |
| 3A | 65.52–69.35 | 70.11–73.56 | 67.05–70.11 | 71.26–75.86 |
| 3B | 57.58–68.18 | 72.73–77.27 | 59.09–63.64 | 72.73–77.27 |
| 3C | 70.49–73.04 | 78.14–81.42 | 71.22–72.86 | 79.23–81.97 |
| 3D | 72.29–74.17 | 80.95–87.45 | 73.3–74.1 | 83.77–86.8 |
| 2C+3CD | 71.98–73.52 | 80.78–83.56 | 72.52–73.38 | 82.22–83.44 |
The other enterovirus was tentatively named strain CHN/BJ/2018-1A (abbreviated as 1A). Nucleotide BLASTn analysis of the whole genomic sequence indicated that 1A exhibited the highest sequence similarity of 82.8% (99% coverage) with human enterovirus 92 strain RJG7. BLASTp analysis of the polyprotein amino acid sequence revealed that strains 1A and RJG7 shared 94.7% identity (100% coverage). VP1 sequence of strain 1A exhibited 80.87% nucleotide identity (100% coverage) and 89.23% amino acid identity (100% coverage) with that of strain RJG7.
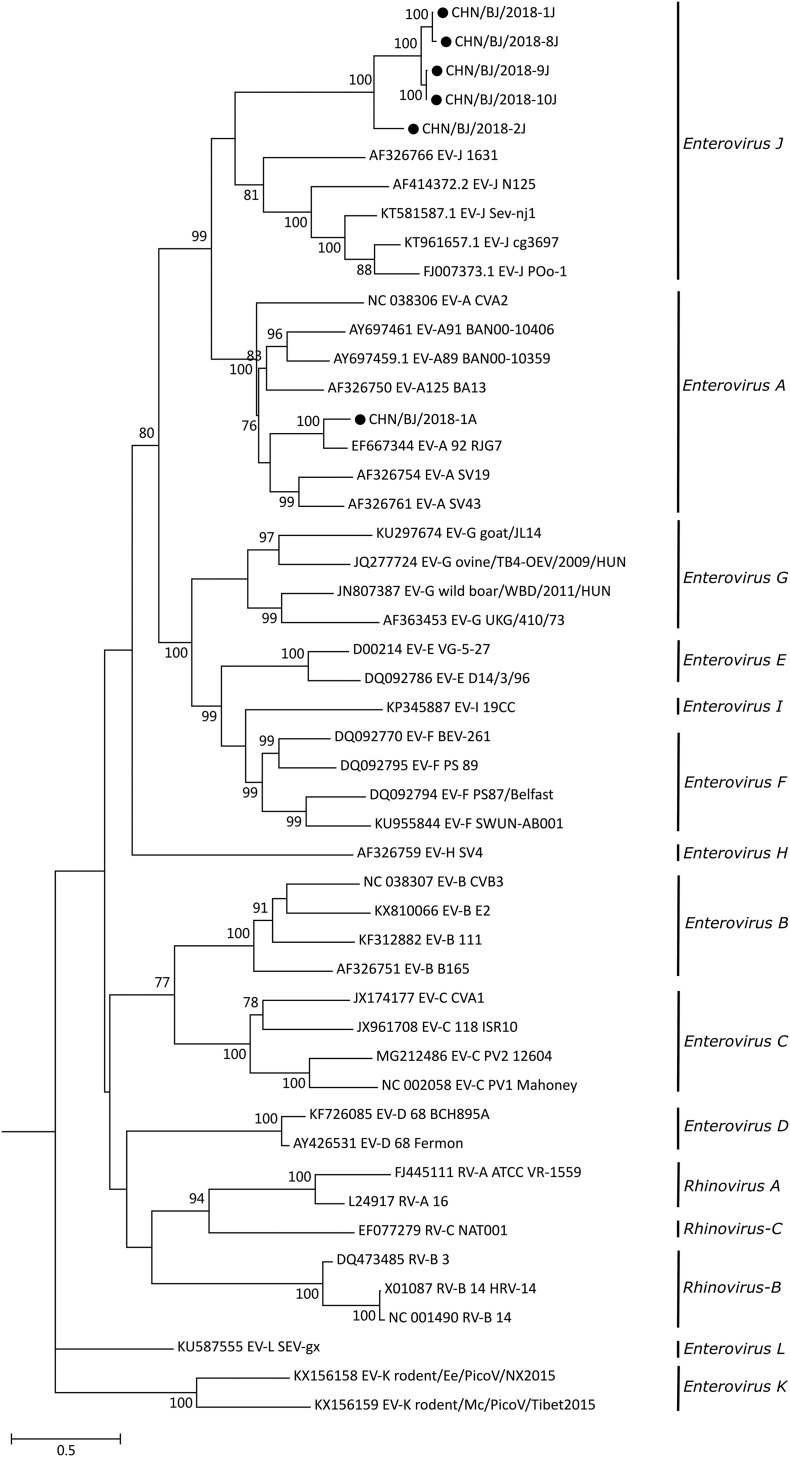
To further confirm the genetic relationships of enterovirus strains in our study with other enteroviruses, a phylogenetic tree (Fig. 2 ) was constructed on the basis of their VP1 nucleotide sequences. The virus strains used in the phylogenetic analysis included the six strains in this study and the typical enterovirus strains listed online (http://www.picornaviridae.com/enterovirus/enterovirus.htm). The maximum-likelihood phylogenetic tree was generated using the GTR+G+I model. Five strains 1J, 8J, 9J, 10J and 2J clustered into a clade with stains of species Enterovius J, indicating that they might be members of species Enterovius J. Strain 1A had the closest genetic relationship with human enterovirus 92 strain RJG7 of species Enterovius A.
Fig. 2.
Phylogenetic tree of enterovirus strains in our study and other enteroviruses based on VP1 nucleotide sequences. The maximum-likelihood method was used to construct the tree with 1000 bootstrap replicates. Bootstrap values are listed at the nodes. The scale represents the evolutionary distance. Strains detected in this study are indicated by black dots.
3.3. Picobirnavirus
Most of the picobirnavirus reads were assigned to RdRp sequences. Mapping these reads from separate samples with human picobirnavirus RdRp gene isolate GPBV6C2 serving as a reference using CLC workbench, five picobirnavirus RdRp complete sequences were assembled from the five samples abundant with picobirnavirus reads. The five picobirnavirus strains were named SPBV-3, SPBV-4, SPBV-5, SPBV-6 and SPBV-7, respectively. These sequences shared 100% sequence identity with each other. Only the sequence of the typical strain SPBV-3 RdRp gene was submitted to GenBank. BLASTn analysis illustrated that the typical picobirnavirus strain SPBV-3 RdRp sequence had the highest nucleotide similarity of 93.48% (100% coverage) with that of human picobirnavirus isolate GPBV6C2. BLASTp analysis of the SPBV-3 RdRp amino acid sequence revealed that it shared the highest similarity of 96% (100% coverage) with that of human picobirnavirus GPBV6C2.
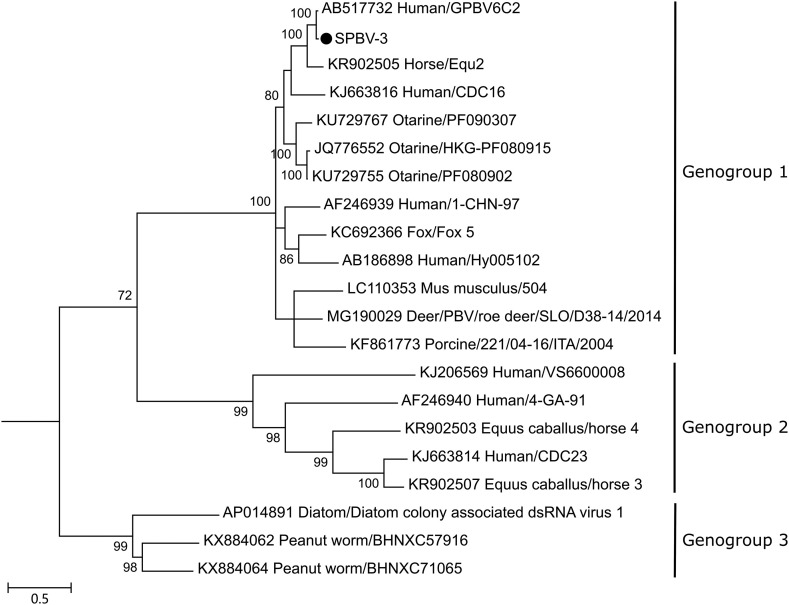
On the basis of sequence diversity in RdRp, picobirnaviruses are classified into three genogroups (Delmas et al., 2019). To further confirm the relationships of SPBV-3 and other picobirnaviruses, a phylogenetic tree (Fig. 3 ) based on RdRp amino acid sequences was constructed. RdRp sequences from SPBV-3, GPBV6C2 and picobirnaviruses listed in the ICTV picobirnavirus report (Delmas et al., 2019) were included in the phylogenetic analysis. The maximum-likelihood phylogenetic tree was generated using the LG+G+I model. SPBV-3 clustered into a clade with picobirnaviruses of genogroup 1 and showed the closest genetic distance with strain GPBV6C2.
Fig. 3.
Phylogenetic tree of picobirnavirus SPBV-3 detected in our study and other picobirnaviruses in ICTV based on the RNA-dependent RNA polymerase amino acid sequences. The maximum-likelihood method was used to construct the tree with 1000 bootstrap replicates. Bootstrap values are listed at the nodes. The scale represents the evolutionary distance. Strain SPBV-3 detected in this study is indicated by a black dot.
4. Discussion
Viral metagenomic analysis conducted in this study revealed the high diversity of viruses in stool samples from African green monkeys with diarrhea in China. Enterovirus and picobirnavirus sequences were highly abundant in the examined samples.
Enterovirus, a genus of Picornaviridae, is one of the most common infectious viruses in humans and animals. Although most enterovirus infections are asymptomatic, some may cause serious or fatal diseases, such as acute flaccid paralysis, aseptic meningitis, hand-foot-and-mouth disease, epidemic pleurodynia, diarrhea and severe respiratory disease (Pallansch et al., 2013; Pons-Salort et al., 2015; Scully et al., 2018). Enteroviruses may circulate between humans and NHPs. Some enterovirus types with a worldwide distribution in humans have been detected in NHPs (Sadeuh-Mba et al., 2014; Mombo et al., 2015). It is therefore extremely important to identify the circulating enteroviruses among NHPs because they might mutate and infect humans, thereby causing new emerging diseases. Recently, a growing number of enteroviruses were reported in NHPs. SEV-gx was identified from stool samples of rhesus macaques (Ao et al., 2016). Mombo (Mombo et al., 2017) detected EV-B112 in a chimpanzee fecal sample and EV-B113 in a mandrill fecal sample. Wang et al. (2016) identified Sev-nj1 from a fecal sample from rhesus. In this study, we detected six novel enteroviruses in stool samples from captive African green monkeys with diarrhea in China. Strain CHN/BJ/2018-1A might be a member of species Enterovirus A, most of which can infect humans. It had the highest similarity with human enterovirus 92 strain RJG7, which was found in an NHP with diarrhea (Oberste et al., 2007). The other five strains had the closest relationships with strain 1631 of species Enterovirus J isolated from Macaca mulatta (Oberste et al., 2007). These six strains were found in five separate monkeys in this study. Coinfection of strain CHN/BJ/2018-1A and CHN/BJ/2018-1J was detected in one African green monkey. Whether the six enteroviruses were pathogenic to African green monkeys should be verified by additional research. Substantial attention and surveillance of these enteroviruses in African green monkeys and people who had contact with these animals may be necessary.
The genus Enterovirus consists of 15 species, namely Enterovirus A-L and Rhinovirus A-C (ICTV Report, 2020). Members of a enterovirus species should share greater than 70% polyprotein amino acid sequence identity, greater than 60% amino acid identity in region P1 and greater than 70% amino acid identity in region 2C plus 3CD (ICTV Report, 2020). Enterovirus genotype was currently defined as sharing greater than 75% nucleotide similarity and greater than 85% amino acid identity in the VP1 coding region (Lukashev et al., 2018). Four strains 1J, 8J, 9J, 10J shared 70.32%–73.53% polyprotein amino acid identity, 57.58%–61.06% amino acid identity in P1 and 80.78%–83.56% amino acid identity in region 2C plus 3CD (Table 2) with strains of species Enterovirus J. We suggested that they should be classified into species Enterovirus J. VP1 sequences of these four strains and that of species Enterovirus J shared 50%–58.16% nucleotide similarity and 46.35%–55.45% amino acid identity (Table 2). This met the criteria for being grouped into different genotypes. Therefore, strains 1J, 8J, 9J, 10J formed a new enterovirus genotype. Strain 2J and strains of species Enterovirus J also met the species demarcation criteria, with 71.24%–73.3% polyprotein amino acid identity, 58.06–59.61% amino acid identity in P1, 82.22%–83.44% amino acid identity in 2C plus 3CD (Table 2). It should be classified into species Enterovirus J. VP1 sequence of strain 2J exhibited low similarity with that of species Enterovirus J (51.08%–59.52% nucleotide similarity, 46.35%–53% amino acid identity) (Table 2) and strains 1J, 8J, 9J, 10J (73.17%–74.45% nucleotide similarity, 79.79%–81.18% amino acid identity). Therefore, strain 2J represented another new genotype. Strain 1A had high VP1 similarity (80.87% nucleotide similarity, 89.23% amino acid identity) with human enterovirus 92 strain RJG7, thus, it should be classified into genotype 92 of species Enterovirus A.
Picobirnaviruses are small, non-enveloped viruses with a genome of two unrelated linear dsRNA segments. Picobirnaviruses have been detected in the faeces of humans and a wide range of animals worldwide, but their pathogenicity remains unknown (Malik et al., 2014; Delmas et al., 2019). In this study, picobirnavirus sequences were detected from seven out of ten stool samples of green monkeys with diarrhea, suggesting a high picobirnavirus infection ratio in African green monkeys. Picobirnavirus strain PBV/Simian/KNA/08873/2015 RdRp sequence detected from an African green monkey in the Caribbean region (Gallagher et al., 2017) had 69.93% nucleotide similarity (92% coverage) and 63.27% amino acid identity (100% coverage) with that of the picobirnavirus in this study, illustrating the great diversity among picobirnaviruses in African green monkeys. The high nucleotide similarity (93%) of picobirnavirus RdRp detected in this study with that of human picobirnavirus GPBV6C2 found in a stool specimen from a patient with diarrhea indicated that African green monkey could be a reservoir of human picobirnavirus. The possibility that human picobirnaviruses could circulate among African green monkeys and humans could not be eliminated. Picobirnavirus strains in this study had a 100% identity, while enteroviruses showed diversity among the same animal colony. These viruses might have different evolutionary histories or dynamics, which is worthy of further investigations.
In brief, we investigated the RNA virome in stool samples of African green monkeys with diarrhea in China, and identified great viral diversity. Enterovirus and picobirnavirus reads accounted for the majority of mammalian virus sequences. Full genomic characterisation of six enterovirus strains and a picobirnavirus RdRp sequence were depicted. Two new genotypes of enteroviruses were defined. The enteroviruses and picobirnavirus identified in this study are worthy of further investigation to clarify their pathogenicity, and surveillance is needed to determine their transmission to humans or other animals.
The following are the supplementary data related to this article.
The verified high-throughput sequencing depth and coverage of each sequence in this study.
Funding
This research was funded by the National Key Research and Development Program of China (grant number: 2018YFA0903000), National Natural Science Foundation of China (grant number: 81672001), and China Mega-Project on Infectious Disease Prevention (grant numbers: 2018ZX10305410-001 and 2018ZX10711001-003-001).
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- Ao Y.Y., Yu J.M., Zhang C.Y., Xin Y.Y., Li L.L., Duan Z.J. Identification of a novel enterovirus species in rhesus macaque in China. Sci. Rep. 2016;6:28526. doi: 10.1038/srep28526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Walsh P.D., Leroy E.M., Real L.A. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog. 2006;2 doi: 10.1371/journal.ppat.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M., Williams Hugh E., Cannane Adam. A deterministic finite automaton for faster protein hit detection in BLAST. J. Comput. Biol. 2006;13:965–978. doi: 10.1089/cmb.2006.13.965. [DOI] [PubMed] [Google Scholar]
- Chahroudi A., Bosinger S.E., Vanderford T.H., Paiardini M., Silvestri G. Natural SIV hosts: showing AIDS the door. Science (80-.) 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Attoui H., Ghosh S., Malik Y.S., Mundt E., Vakharia V.N., Ictv Report C. ICTV virus taxonomy profile: Picobirnaviridae. J. Gen. Virol. 2019;100:133–134. doi: 10.1099/jgv.0.001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher C.A., Navarro R., Cruz K., Aung M.S., Ng A., Bajak E., Beierschmitt A., Lawrence M., Dore K.M., Ketzis J., Malik Y.S., Kobayashi N., Ghosh S. Detection of picobirnaviruses in vervet monkeys (Chlorocebus sabaeus): molecular characterization of complete genomic segment-2. Virus Res. 2017;230:13–18. doi: 10.1016/j.virusres.2016.12.021. [DOI] [PubMed] [Google Scholar]
- Herrington C.S, Coates P.J, Duprex W.P. Viruses and disease: emerging concepts for prevention, diagnosis and treatment. J Pathol. 2015;235(2):149–152. doi: 10.1002/path.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zeng L.-P., Yang X.-L., Ge X.-Y., Zhang W., Li B., Xie J.-Z., Shen X.-R., Zhang Y.-Z., Wang N., Luo D.-S., Zheng X.-S., Wang M.-N., Daszak P., Wang L.-F., Cui J., Shi Z.-L. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:1–27. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV Report for Genus Enterovirus. 2019. https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae/681/genus-enterovirus (accessed 2 February 2020)
- Jasinska A.J., Schmitt C.A., Service S.K., Cantor R.M., Dewar K., Jentsch J.D., Kaplan J.R., Turner T.R., Warren W.C., Weinstock G.M., Woods R.P., Freimer N.B. Systems biology of the vervet monkey. ILAR J. 2013;54:122–143. doi: 10.1093/ilar/ilt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev A.N., Vakulenko Y.A., Turbabina N.A., Deviatkin A.A., Drexler J.F. Molecular epidemiology and phylogenetics of human enteroviruses: is there a forest behind the trees? Rev. Med. Virol. 2018;28 doi: 10.1002/rmv.2002. [DOI] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y.S., Kumar N., Sharma K., Dhama K., Shabbir M.Z., Ganesh B., Kobayashi N., Banyai K. Epidemiology, phylogeny, and evolution of emerging enteric Picobirnaviruses of animal origin and their relationship to human strains. Biomed. Res. Int. 2014;2014:780752. doi: 10.1155/2014/780752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombo I.M., Berthet N., Lukashev A.N. First Detection of an Enterovirus C99 in a Captive Chimpanzee with Acute Flaccid Paralysis, from the Tchimpounga Chimpanzee Rehabilitation Center, Republic of Congo. PLoS One. 2015;10(8):e0136700. doi: 10.1371/journal.pone.0136700. Published 2015 Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombo I.M., Lukashev A.N., Bleicker T., Brunink S., Berthet N., Maganga G.D., Durand P., Arnathau C., Boundenga L., Ngoubangoye B., Boue V., Liegeois F., Ollomo B., Prugnolle F., Drexler J.F., Drosten C., Renaud F., Rougeron V., Leroy E. African non-human primates host diverse enteroviruses. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M.S., Maher K., Pallansch M.A. Complete genome sequences for nine simian enteroviruses. J. Gen. Virol. 2007;88:3360–3372. doi: 10.1099/vir.0.83124-0. [DOI] [PubMed] [Google Scholar]
- Oberste M.S., Feeroz M.M., Maher K., Nix W.A., Engel G.A., Begum S., Hasan K.M., Oh G., Pallansch M.A., Jones-Engel L. Naturally acquired picornavirus infections in primates at the Dhaka zoo. J. Virol. 2013;87:572–580. doi: 10.1128/JVI.00838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallansch M.A., Whitton J.L., Oberste M.S., Howley P.M. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Knipe D.M., editor. Fields Virology. 6th ed. Wolters Kluwer; Philadelphia: 2013. pp. 490–530. [Google Scholar]
- Pons-Salort M., Parker E.P., Grassly N.C. The epidemiology of non-polio enteroviruses: recent advances and outstanding questions. Curr. Opin. Infect. Dis. 2015;28:479–487. doi: 10.1097/QCO.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeuh-Mba S.A., Bessaud M., Joffret M.-L., Endegue Zanga M.-C., Balanant J., Mpoudi Ngole E., Njouom R., Reynes J.-M., Delpeyroux F., Rousset D. Characterization of enteroviruses from non-human primates in Cameroon revealed virus types widespread in humans along with candidate new types and species. PLoS Negl. Trop. Dis. 2014;8:1–12. doi: 10.1371/journal.pntd.0003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.J., Basnet S., Wrangham R.W., Muller M.N., Otali E., Hyeroba D., Grindle K.A., Pappas T.E., Thompson M.E., Machanda Z., Watters K.E., Palmenberg A.C., Gern J.E., Goldberg T.L. Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerg. Infect. Dis. 2018;24:267–274. doi: 10.3201/eid2402.170778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. Vol. 365. 2010. The Evolution of HIV-1 and the Origin of AIDS; pp. 2487–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shao S., Wang H., Shen Q., Yang S., Zhang W. An enterovirus from a captive primate in China. Springerplus. 2016;5:1281. doi: 10.1186/s40064-016-2966-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward K.W., Coon D.J., Magiera D., Bhadresa S., Nisbett E., Lawrence M.S. Exploration of the African green monkey as a preclinical pharmacokinetic model: intravenous pharmacokinetic parameters. Drug Metab. Dispos. 2008;36:715–720. doi: 10.1124/dmd.107.019315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The verified high-throughput sequencing depth and coverage of each sequence in this study.