Abstract
The pandemic of new coronavirus disease COVID-19 is threatening our health, economy and life style. Collaborations across countries and sectors as a One Health World could be a milestone.
We propose a general protocol, for setting timely active random surveillance of COVID-19, at the human community level, with systematic repeated detection efforts. Strengths and limitations are discussed.
If considered applicable by public health, the protocol could evaluate the status of COVID-19 epidemics consistently and objectively.
Keywords: Pandemic, New coronavirus, COVID-19, Active random surveillance
Highlights
-
•
A base protocol is proposed for setting active random surveillance of COVID-19.
-
•
The protocol is based on widely known veterinary surveillance methodologies.
-
•
By applying the protocol, COVID-19 epidemics could be consistently assessed.
-
•
The protocol could be a milestone in the battle against the pandemic.
-
•
Authorities could consider if the protocol is applicable for public health.
1. Background
During December 2019, a new human disease (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in China [1], and developed into a pandemic in three months.
Health systems worldwide are trying to contain and resolve the outbreaks in the shortest time. Responses appear to be variegated across the globe and several countries reporting internationally, appear to be following a surveillance-control approach based on: a) case identification b) tracings and c) daily reporting of: infected, recovered and deaths, at area and country level [1,2]. The surveillance of clinical cases and traced contacts is particularly useful, especially at the beginning of an outbreak to limit further spread.
Once a new disease spreads within a susceptible population, two surveillance outputs are important to inform actions of risk mitigation and disease control (Fig. 1), i.e. the “true” prevalence (TP) of infected individuals (both symptomatic and not) and, if no cases are found, the confidence in “freedom” (PFree). The TP is the proportion of truly infected that is estimated considering the use of imperfect tests [3]. The PFree instead, is the confidence that disease is absent in a population, or if present, it is below a (beforehand) decided hypothetical cut-off prevalence Pu. In other words, the latter represents the cut-off at which the population would be considered as infected, if at least one positive is found out of those tested . The concept of “freedom” from disease has already had several applications in the veterinary field [[4], [5], [6], [7], [8], [9], [10]] and more recently, also in public health [11].
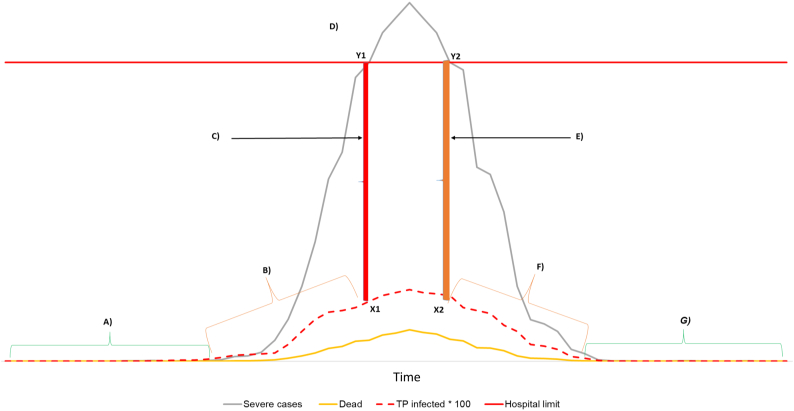
Fig. 1.
Hypothetical representation of phases (A-G) of surveillance and control of COVID-19 at human population level. The dashed red curve represents the true prevalence (TP) of infected people that could be estimated by active random surveillance to inform about the “true” population infection status in real time. A) Surveillance surveys are addressed to initial detection of disease, and if no cases are found, the aimed confidence in freedom PFree could be reached (confidence to be below design prevalence Pu). B) Disease is known to be circulating in the population and random surveys are addressed to estimate the TP and avoid reaching the threshold prevalence (ThreTP, X1), above which the health care system could go under pressure due to high number of severely ill persons. C) Critical situation starts at the hospitals. Red vertical bar connecting ThreTP = X1 and hospitalization limit Y1 of severely ill people (red horizontal bar). D) Critical situation due to TP > ThreTP and application of “draconian” measures. E) Critical situation reduces at the hospitals due to TP = ThreTP (orange bar Y2-X2). F) Disease is still known to be circulating in the population and random surveys are addressed to estimate the TP to monitor the situation before relaxing restrictions. G) Surveillance surveys are addressed at early detection of eventual relapses of disease, and if no cases are found, the aimed confidence in freedom PFree is confirmed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The aim of this article is to propose a general protocol for setting surveys based on random sampling (as frequently used in veterinary surveillance), to the public health settings to assess TP and PFree in a systematic and objective manner. For more information on the justification of this proposal, see accompanying paper [12].
2. Materials and methods
2.1. Possible general protocol for setting a random surveillance component for COVID-19
In the veterinary field, active random surveillance surveys are often used to estimate the TP and PFree, and to decide where and when control actions could be prioritized. Similar outputs are needed to make decisions in public health regarding the COVID-19 epidemic.
The use of repeated randomized surveys at the beginning of the outbreak (Fig. 1) could be applied to substantiate freedom (if the pathogen has not been detected in the area) (Fig. 1 phase A). If detection occurs, the TP can be estimated to understand the level of infection in the population (Fig. 1 phase B).
This can inform actions that can minimize the risk that the estimated TP reaches the threshold prevalence ThreTP (Fig. 1, phase C) at which the related number of hospitalized people could be too high, so that the health care system could be excessively stressed (Fig. 1, phase D). The decision-maker could also choose to use any limit of the TP's confidence interval rather than the estimated TP, depending on risk-appetite and room for uncertainty in the health system. Some examples of how to define threshold prevalences, as well as setting (or comparing) survey prevalence results in the public health, are in [13,14].
During phases D and E, estimating the TP would allow evaluating the eventual effect of “draconian” measures (e.g. strict quarantine, closing borders etc.).
In Phase F, estimating the TP of infected would allow monitoring there are no relapses into phase D if restrictions are relaxed; while in phase G, the output of repeated surveys informs when to relax measures, reassure the public and move resources to areas where urgency is higher.
A general description of how the protocol could be conducted in real time, is shown in Fig. 2. Firstly, the sample sizes (n1 and n2) are calculated, to potentially address both purposes of: estimating TP (n1) and if no cases are found, reaching aimed confidence in freedom, PFree (n2).
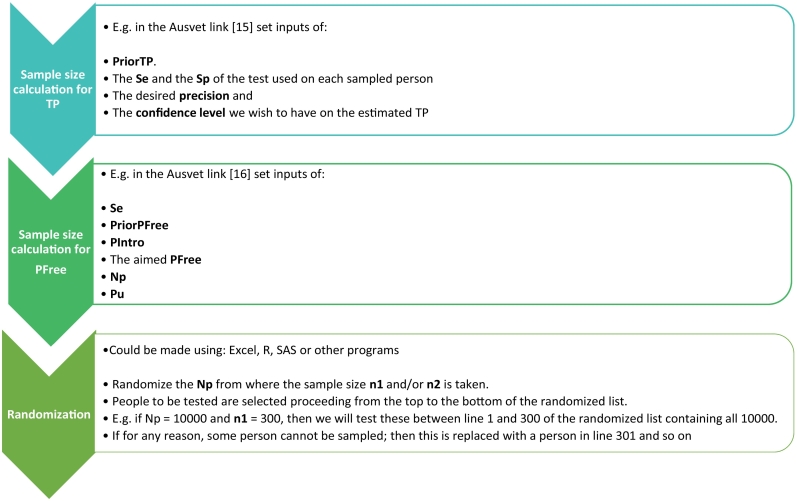
Fig. 2.
General protocol for surveillance of COVID-19 based on repeated surveys: n1 = sample size to estimate true prevalence. TP = True prevalence of infected people within the target population Np. The same protocol, with adapted inputs, can be used to estimate TP of antibody positive people, if an antibody test is applied too; n2 = sample size to reach the aimed confidence in freedom; PFree = aimed confidence in freedom from disease at the area/population level, if no cases are found through the survey. Np = overall target population in the area, from where the n1 and/or n2 are sampled. ThreTP = threshold true prevalence at which the health care system goes under pressure.
Next, a randomized sample size of the target population Np, is selected to ensure that the obtained TP or PFree are statistically reliable across repeated random surveys.
Due to the high transmission rate of SARS-CoV-2, repeating the survey and its interpretation on weekly or fortnightly (Fig. 2) could be sufficient. The frequency of the surveys would depend on the resources available and on the management needs of public health authorities for improved decision making, prioritizations and anticipating response needs (i.e., hospital beds, ventilators, medical staff on location or on call etc.). Obviously, the more frequent the surveying the more updated and timely is the information it provides.
The data analysis and interpretation of results (Fig. 2) gives the “picture” (with related uncertainties) of the epidemiological phase of the outbreak (Fig. 1).
Then, authorities can use the outputs to decide if more actions and resources are needed to reduce the TP in the area, or if this could be considered as “free” with confidence PFree.
In the sections below and in the supplementary material, some practical examples are provided to illustrate the general principles and application of the protocol.
2.1.1. Example of sample size calculation to estimate the TP
Software programs (e.g. R, Excel, SAS, STATA etc) as well as links from the web [e.g. [[15], [16], [17], [18], [19], [20]]] may be applicable, to calculate sample sizes for estimating TP or for reaching the aimed PFree.
For example, the sample size (n1) to estimate the TP could be calculated by using the Ausvet link [15]; which is one of the widely used veterinary links we are most familiar with. In that case the inputs needed are: the desired precision, the confidence level, the sensitivity (Se) and the specificity (Sp) of the test used and the true prevalence assumed (a priori) before the survey is carried out (here called PriorTP) (Fig. 3). By adapting the mentioned inputs, the TP of antibody positive people could be assessed as well.
Fig. 3.
Detailed description example, for sample size calculation and randomization processes for repeated surveys. PriorTP = The true prevalence (TP) assumed before the survey is made. Se = test sensitivity. Sp = test specificity. PIntro = Probability of introduction into the targeted population Np (e.g. during 1 or more sampling days or during time elapsed between two consecutive surveys). PFree = The aimed confidence in freedom to be substantiated by the survey, if all samples (n2) result negative (confidence TP < Pu); Np = The size of the population within the targeted area and considered for the randomization process. Pu = The design prevalence of infected people, at which we can have at least one positive result out of n2 sampled, with the test used. N.B. If both n1 and n2 are calculated, by using the biggest of the two sample sizes, would prepare for both outputs, namely TP (if at least one person is positive) and PFree if all sampled persons result negative. Also notice that, usually, the specificity (Sp) is assumed 100% [[4], [5]] when PFree is assessed.
For the first survey, the PriorTP is derived from the opinion of experts in public health or by using apparent prevalences (AP) as reported from other infected areas to date. In the latter case, it must be considered that AP estimates could be biased, if not based on simple random sampling and if not corrected for imperfect test performance. When the sample size calculation is repeated for each survey, already at the second survey, the PriorPT would be set using the TP estimated from survey one. In the third survey, the PriorPT could be set using the TP from the previous survey and so on.
2.1.2. Example of sample size calculation to reach the aimed confidence in freedom (PFree)
The sample size (n2) to achieve the target confidence in freedom (PFree) can be calculated, for example using the Ausvet link [16] (Fig. 3). Confidence in freedom is only applicable for areas where disease is expected being absent (TP around 0% and below the hypothetical design prevalence Pu) (Fig. 1, Steps A or G).
The Se could be set according to information on test's performance from literature, or according to experts knowledge. The Sp could be assumed 100% [4,5] if false positves are ruled out by follow up tests, or if positives at the first test are directly managed as "truly" positives (e.g. quarantined).
The confidence in freedom before the first survey is made (called here PriorPFree) could be set = 50% [4,5]. Whereas in the following testing, the PFree from the previous survey could be used as PriorPFree.
The probability of introduction into the target population during the survey (or between two consecutive surveys), is here called PIntro and for example, it could be set to 50% or around 0%. In the latter case it would be assumed that during the surveying period (e.g. during a single day of sampling) the probability of disease introduction into the targeted population is negligible (e.g. if movement between areas is not allowed). If possible, it is suggested taking all the samples at the same time. Setting PIntro = 50% would be more conservative than using 0%, because it would include the potential movement of infected people into the target population.
PriorPFree and PIntro values of 50% could be considered “neutral” (as “tossing a coin”).
The required target confidence in freedom (PFree) could be agreed within the task force unit. In the veterinary field, values of 95 or 99% are usually considered sufficient to substantiate freedom from disease at population level [[8], [9], [10]].
2.1.3. Example of randomization process
An example of how to use the randomization process after calculating the sample sizes (n1, n2) is shown in Fig. 3. If available, different software (e.g. Excel, SAS, R) and techniques from other working sectors are equally applicable for this purpose; as long as the user understands the input parameters and the uncertainty they carry on the outputs. Also, when the Np is defined, it could be assumed that after a while and especially before control measures are implemented, the virus could be more or less widespread within the target population. This assumption seems reasonable, if considering that the virus has spread worldwide within just a few months, and considering the syndromic data reported to date from large infected areas and cities where several thousands of cases occurred within just a few weeks from first detection.
3. Discussion
Although the principles of the surveying protocol are derived from the animal health realm, where they have had extensive use [e.g. [[4], [5], [6], [7], [8], [9], [10]]], a similar approach in surveillance of the human population could be applicable [11], and requires public health leadership. At a first glance, the application of the proposed principles and protocol (Fig. 1, Fig. 2, Fig. 3), could appear as unfeasible; because it could require overpassing some social, logistic and economical barriers; as never seen before. Nevertheless, we must also consider that this is a particular situation, which is challenging the health and economy as rarely seen in history. If the virus has been able to adapt across species, the human society should be able adapting quickly too, by working across countries, sectors and communities. The protocol is not a strict guideline with which everybody must comply. Rather, is a proposal for potential solutions and it is only a “starting” point open to improvements and adaptations across different capacities and realities. In Table 1, are resumed some considerations on: strengths, limitations, and possible solutions for the protocol applications.
Table 1.
Strengths, limitations and possible solutions to consider for applying the proposed COVID-19 surveillance and control protocol.
| Strengths |
|---|
|
|
|
|
|
|
|
| Limitations and possible solutions |
|
|
|
|
|
Funding statement
No funding was received for this work.
Authors statement
Alessandro Foddai developed the idea, designed the protocol, carried out the literature review, and drafted the manuscript. Johanne Ellis-Iversen contributed to the idea and to the protocol setting, carried out part of the literature review and revised the manuscript. Juan Lubroth gave his critical opinion, supported the revision of the manuscript and the generalization of its concept from a disease control/management perspective.
Declaration of Competing Interest
None of the authors have any conflict of interests
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2020.100129.
Appendix A. Supplementary data
Example of protocol´s application.
References
- 1.Wu Z., McGoogan G.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020:E1–E4. doi: 10.1001/jama.2020.264. [DOI] [PubMed] [Google Scholar]
- 2.Wold Health Organization (WHO) Coronavirus, Situation Updates. 2020. https://www.who.int/health-topics/coronavirus accessed on 18 March 2020.
- 3.Moreno-Torres K., Wolfe B., Saville W., Garabed R. Estimating Neospora caninum prevalence in wildlife populations using Bayesian inference. Ecol. Evol. 2016;6(7):2216–2225. doi: 10.1002/ece3.2050. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin P.A.J., Cameron A.R., Greiner M. Demonstrating freedom from disease using multiple complex data sources 1: a new methodology based on scenario trees. Prev. Vet. Med. 2007;79:71–97. doi: 10.1016/j.prevetmed.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Martin P.A.J., Cameron A.R., Barfod K., Sergeant E.S.G., Greiner M. Demonstrating freedom from disease using multiple complex data sources 2: case study-classical swine fever in Denmark. Prev. Vet. Med. 2007;79:98–115. doi: 10.1016/j.prevetmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Cameron A.R. The consequences of risk-based surveillance: developing output-based standards for surveillance to demonstrate freedom from disease. Prev. Vet. Med. 2012;105:280–286. doi: 10.1016/j.prevetmed.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Cameron A., Njeumi F., Chibeu D., Martin T. A Manual for Veterinarians on The Design And Analysis of Surveillance For Demonstration of Freedom From Disease. Food and Agriculture Organization of the United Nations; Rome: 2014. Risk based disease surveillance; pp. 1–215.http://www.fao.org/3/a-i4205e.pdf Available at: [Google Scholar]
- 8.Foddai A., Nielsen L.R., Willeberg P., Alban L. Comparison of output-based approaches used to substantiate bovine tuberculosis free status in Danish cattle herds. Prev. Vet. Med. 2015;121:21–29. doi: 10.1016/j.prevetmed.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Foddai A., Stockmarr A., Boklund A. Evaluation of temporal surveillance system sensitivity and freedom from bovine viral diarrhea in Danish dairy herds using scenario tree modelling. BMC Vet. Res. 2016;12:1–12. doi: 10.1186/s12917-016-0744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foddai A., Floyd T., McGiven J., Grace K., Evans S. Evaluation of the English bovine brucellosis surveillance system considering probability of disease introduction and non-random sampling. Prev. Vet. Med. 2020;176:1–14. doi: 10.1016/j.prevetmed.2020.104927. [DOI] [PubMed] [Google Scholar]
- 11.Michael E., Smith M.E., Katabarwa M.N., Byamukama E., Griswold E., Habomugisha P. Substantiating freedom from parasitic infection by combining transmission model predictions with disease surveys. Nat. Commun. 2018;9(4324):1–13. doi: 10.1038/s41467-018-06657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foddai A., Lindberg A., Lubroth J., Ellis-Iversen J. Surveillance to improve evidence for community control decisions during the COVID-19 pandemic – opening the animal epidemic toolbox for public health. One Health. 2020 doi: 10.1016/j.onehlt.2020.100130. In Prep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilukha O.O., Blanton C. Interpreting Results of Field Surveys Using Probability Calculators. 2020. https://www.ennonline.net/fex/39/results
- 14.WHO, CDC, UNAIDS, FHI 360 . For Populations at Risk of HIV. 2017. Global HVI Strategic Information working group. Biobehavioural survey guidelines; pp. 1–515.https://apps.who.int/iris/bitstream/handle/10665/258924/9789241513012-eng.pdf?sequence=1&ua=1 accessed on 24 March 2020. [Google Scholar]
- 15.https://epitools.ausvet.com.au/prevalencess accessed on 24 March 2020.
- 16.https://epitools.ausvet.com.au/freedomssthree accessed on 24 March 2020.
- 17.https://www.who.int/ncds/surveillance/steps/resources/EpiInfo/en/ accessed on 24 March 2020.
- 18.https://www.surveysystem.com/sscalc.htm accessed on 24 March 2020.
- 19.https://www.abs.gov.au/websitedbs/D3310114.nsf/home/Sample+Size+Calculator accessed on 24 March 2020.
- 20.https://epitools.fp7-risksur.eu/tools/index?toolId=46 accessed on 24 March 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example of protocol´s application.