Abstract
Autophagy, an evolutional conserved lysosomal degradation process, has been implicated to play an important role in cellular defense against a variety of microbial infection. Interestingly, numerous studies found that some pathogens, especially positive-single-strand RNA viruses, actually hijacked autophagy machinery to promote virus infection within host cells, facilitating different stages of viral life cycle, from replication, assembly to egress. Enterovirus, a genus of positive-strand RNA virus, can cause various human diseases and is one of main public health threat globally, yet no effective clinical intervention is available for enterovirus infection. Here we summarized recent literature on how enteroviruses regulate and utilize autophagy process to facilitate their propagation in the host cells. The studies on the interplay between enterovirus and autophagy not only shed light on the molecular mechanisms underlying how enterovirus hijacks cellular components and pathway for its own benefits, but also provide therapeutic option against enterovirus infection.
Keywords: Autophagy, Enterovirus, Replication organelles
1. Introduction
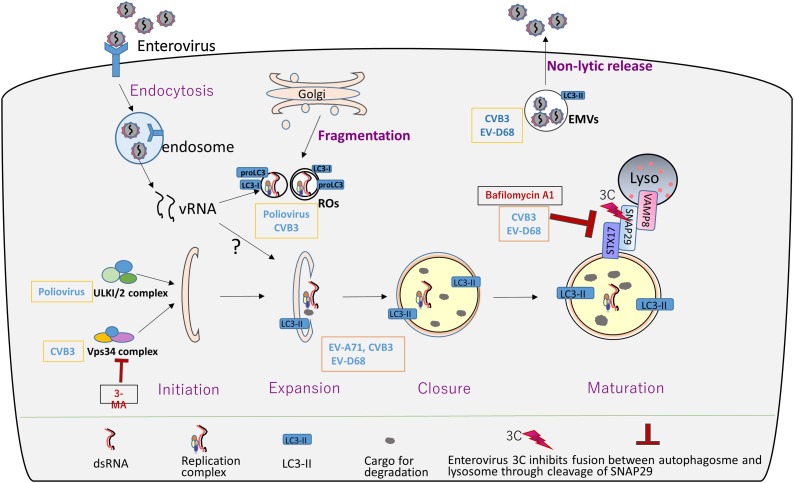
Autophagy is a process of “self-eating”, an essential cellular pathway that maintains hemostasis by degrading misfolded protein, damaged organelles, or invading pathogens [1,2]. It is widely found in eukaryotic organisms, ranging from yeast to mammals, and plays an important role in various physiological and pathophysiological process, including cell growth, tumor formation and intracellular microbial infection [1,3]. Thus, autophagy pathway is a potential therapeutic target for many diseases, including tumor, diabetes, neurodegenerative disease, and pathogenic infection. It is thought that autophagy functions as host defense against viral infection via the uptake of pathogens for degradation. However, increasing evidences show that many positive RNA viruses, including flavivirus and enterovirus, hijack the autophagy machinery to promote their propagation, from genome RNA replication, viral assembly to progeny virions egress [[4], [5], [6], [7], [8]]. In this review, we will discuss recent reports on how enteroviruses utilize autophagy machinery to propagate and avoid lysosomal capture and clearance (Fig. 1 ).
Fig. 1.
Overview of Enterovirus’ subversion of autophagy pathway. In enterovirus infections, the life cycle of enterovirus begins with recognition and binding to receptors on plasma membrane, followed by receptor-mediated endocytosis which results in virus uncoating and release of viral genome RNA (vRNA) into cytoplasm induced by pH change in endosomal compartment. The vRNA was primarily used as mRNA template to translate and synthesis viral proteins which was required for subsequent genome replication. It is reported that poliovirus utilizes ULK1 complex and CVB3 requires Vps34 complex to initiate autophagy. EV-A71, CVB3 and EV-D68 were beneficial from virus-induced formation of autophagosomes to promote viral genome replication. During late autophagy, autophagosomes fuse with lysosomes to form autolysosomes to degrade cargos engulfed by autophagosomes. However, in CVB3 and EV-D68 infection, viral protease 3C cleaves SNAP29, leading to blockage of auto-lysosome formation and autophagic flux. Poliovirus and CVB3 were capable of recruiting either proLC3 or LC3-I to replication organelles (ROs) (single-membrane or double-membrane) to promote viral replication in an autophagy-independent manner. Besides, CVB3 and EV-D68 manipulate extracellular membrane vesicles (EMVs) to promote non-lytic release of infectious progeny virions. 3-MA is an early-stage autophagy inhibitor that blocked PI3K3C/Vps34, while bafilomycin A1 is a late-stage autophagy inhibitor that inhibit fusion of autophagosome with lysosome.
2. Autophagy
Autophagy can be divided into the following three types: macro-autophagy, micro-autophagy and chaperone-mediated autophagy (CMA) [9]. Microautophagy is a process that lysosome directly engulfs cytoplasmic components for degradation, while CMA is highly selective, which can only degrade soluble protein, not organelles, in the cytoplasm. Macroautophagy, on the other hand, refers to the formation of a double membrane structure within a cell, which degrades both organelles and proteins in either selective or non-selective manner. Macroautophagy, referring as autophagy hereafter, can be generally divided into four stages: initiation and nucleation of phagophore, elongation and closure to form autophagosome, fusion of autophagosome with lysosome to form autolysosome, and degradation of engulfing cargos inside autolysosome [10,11].
3. Initiation and nucleation of phagophore
Phagophore biogenesis is initiated when a serine/threonine kinase ULK1, also known as ATG1 in yeast, forms a complex with autophagy-related protein 13 (ATG13), autophagy-related protein 101 (ATG101), and RB1 Inducible Coiled-Coil 1 (RB1CC1, also known as FIP200) [12]. The kinase activity of ULK1 complex is required for the recruitment of phosphatidylinositol 3-kinase (PI3KC3) complex to the site of phagophore nucleation [13]. PI3KC3, also known as vacuolar protein sorting 34 (VPS34) that converts phosphatidylinositol (PI) into phosphatidylinositol-3-phosphate (PI3P), forms a complex with other proteins, including general vesicular transport factor p115, Beclin-1, and ATG14 L in PI3KC3 complex I or UV radiation resistance-associated gene protein (UVRAG) in PI3KC3 complex II [[14], [15], [16]]. The PI3P produced by the class III PI3K complex I is essential for recruitment of PI3P-binding proteins of the WIPI family and its effectors that are required for expansion and subsequent sealing of the isolation membrane [17,18]. One of essential upstream regulators of ULK1 is mTOR kinase (The mammalian target of rapamycin), which inhibits the initiation of autophagy in the presence of growth factors and adequate nutrients [19,20]. Rapamycin binds to 12 kDa FK506-binding protein (FKBP12) to form a complex, which binds to mTOR to inhibit its activity, thereby activating the initiation of autophagy [[21], [22], [23]]. Upon nutrient stimulation, mTOR complexes 1 (mTORC1), containing mTOR, raptor, mammalian lethal with SEC13 protein 8 (MLST8), PRAS40 and DEPTOR, directly phosphorylates Ulk1 to inhibit autophagy [[24], [25], [26]]. The mTOR pathway has been reported to regulate more than 20 autophagy-related genes, which are assembled into functional complex to initiate autophagy [17,27,28]. Notably, although ULK1 complex is required for nutrient-dependent mTORC1-associated autophagy, it also plays a key role in mTOR-independent autophagy [25,29].
4. Phagophore elongation and closure
The Atg8 family, known as LC3 in mammals, contains at least 6 members: microtubule-associated proteins 1 light chain subfamily (MAP1LC3A, MAP1LC3B, MAP1LC3C) and the γ-aminobutyric acid receptor-associated protein (GABARAP) subfamily (GABARAP, GABARAPL1 and GABARAPL2) [30]. The proLC3 proteins are firstly subjected to a post-translation modification at the C-terminus by the cysteine protease autophagy-related protein 4 (ATG4), including ATG4A-D, and the modified cytosolic LC3-I is then conjugated to the lipid phosphatidylethanolamine (PE) on phagophore membranes by a complex ubiquitin-like E3 ligase system [31]. Briefly, the E1-like enzyme autophagy-related protein 7 (ATG7) activates the C-terminal glycine residue of LC3-I, transfers it to the E2-like enzyme autophagy-related protein 3 (ATG3), and finally with the help of E3-like enzyme ATG12-ATG5-ATG16L1 complexes, LC3-I fuses with PE to become the lapidated LC3-II, which is tightly attached to phagophore membranes [15]. Moreover, LC3 also plays an essential role in cargo recruitment in selective autophagy, via interaction with cargo receptors, e.g. neighbor of BRCA1 gene NBR1, nuclear dot protein 52 (NDP52), optineurin (OPTN), and sequestosome-1 (SQSTM1), which contain LC3-interacting regions (LIRs) [32,33]. The polyubiquitin-binding protein, SQSTM1, also known as p62, which contains both LC3-interacting region (LIR) and ubiquitin binding domain, recruits the cargo mediated by ubiquitin–p62–LC3 interactions [34,35]. The continuously recruitment of ATG8s and cargos to phagophore leads to expansion of phagophore, followed by closure, maturation and formation of double-membrane autophagosomes. However, currently, the mechanisms involved in the sealing of the phagophore membrane is still unclear [17,18]. During the maturation of autophagosome, some autophagy-related proteins (e.g. ULK1 and ATG16L1) that localize on outer membrane of autophagosome, are cleaved and removed from autophagosome [27].
5. Autophagosome-lysosome fusion and cargo degradation
Afterwards, autophagosome and lysosome move together to get closer enough for tethering and fusion to become autolysosome. Besides, autophagosome also fuses with late endosome to form amphisome [36]. Plenty of tethering factors are involved in this process, which can mainly be divided into 3 categories: the homotypic fusion and protein sorting (HOPS) complex, small GTPases of the Ras-related (Rab) proteins and SNARE proteins [37,38]. It has been shown that the SNARE protein syntaxin 17 (STX17) is recruited to membrane of mature autophagosomes upon autophagy induction, and STX17 subsequently interacts with HOPS complex (VPS11, VPS16, VPS18, VPS33A, VPS39 and VPS41) [39]. The HOPS complex also interacts with Rab7, which localizes on late endosome and lysosome, mediated by Rab7 effectors, PLEKHM1 and RILP [40,41]. Thus, the HOPS complex is essential for fusion of autophagosome with lysosome. It is reported that STX17 forms a complex with another two SNARE proteins, synaptosomal associated protein 29 (SNAP29) and vesicle-associated membrane protein 8 (VAMP8), to facilitate the fusion between autophagosomes and lysosomes. Digestion of the inner autophagosome membrane by lysosomal enzymes and release of the cargos of the autophagosome to the lumen of the lysosome lead to formation of complete autolysosome [37]. Degradation of these components not only provides nutrients to maintain various cellular functions, but also plays an important role in the process of innate immunity [2].
6. Methods for monitoring autophagy
To monitor autophagy activities within mammalian cells, which is a highly dynamic cellular process, plenty of methods have been adopted in the past twenty years. These techniques have been systematically introduced and summarized [[42], [43], [44], [45]]. The methods mentioned in the articles that we summarized in this review will be selected and introduced below.
Electron microscopy is the most traditional and reliable method for monitoring autophagy. Autophagosome is a double-membrane compartment engulfing undigested cytoplasmic components, while the autolysosome is a single-membrane vesicle which is occasionally difficult to be distinguished from other single-membrane organelles, e.g. endocytic compartments [43]. As electronic microscopy requires proficient technique and expertise to identify different organelles, it is not an ideal method study autophagy, especially for those who are new in this field.
As mentioned above, when autophagy occurs, nascent cytoplasmic proLC3 is processed by Atg4 into LC3-I and then conjugated with phosphatidylethanolamine (PE) to become LC3-II by a complex ubiquitin-like E3 ligase system, which bounds to autophagosome [46,47]. Thus, LC3 is a most widely used marker for autophagic membrane. On SDS-PAGE, it usually shows two bands in LC3 immunoblotting: LC3-II (apparent mobility, 16 kD) migrates faster than LC3-I (apparent mobility, 18 kD) [48]. Quantification of LC3-II level or LC3-II/LC3-I ratio by western-blot provides valuable insight on autophagic process. Usually, the upregulated level of LC3-II represents increased autophagic activity. However, either blocking fusion of autophagosome and lysosome or inhibiting the protease activity in lysosome can also lead to accumulation of LC3-II [42,49]. Therefore, though measurement of LC3-II/LC3-I by western blot is quantitative and relatively convenient, it is usually combined with other techniques to monitor autophagic activities [43,50].
LC3 is often fused with a fluorescent protein, e.g. GFP and RFP. In GFP-LC3 puncta formation assay which counts the average number of punctate structures per cell by fluorescence microscopy, green punctate structures primarily represent autophagosomes [48,51]. Since GFP can be easily quenched in acidic pH, membrane-bound LC3 autolysosome is almost absent under fluorescence microscopy. In contrast, RFP is more stable in acidic organelles, thus RFP-LC3 can be detected in both autophagosome and autolysosome. Another useful probe to monitor autophagic flux has been developed based on the difference in nature of GFP and RFP, which fuses tandem fluorescent-tag mRFP-GFP with LC3 to generate tfLC3 construct [52]. In tfLC3 expressing cells, autophagosomes can be visualized as yellow puncta as both mRFP and GFP exist, while autolysosome exhibits red only signal due to the stability of mRFP in acidic compartment. Both yellow and red puncta are increased during autophagic flux, whereas the accumulation of yellow puncta only indicates that fusion between autophagosome and autolysosome is blocked, or protease activity is inhibited in autolysosome [52].
7. Enterovirus: pathogenesis and life cycle
Enterovirus, belonging to the family Picornaviridae, is a genus of small, non-enveloped, single-stranded positive-sense ss(+)RNA viruses, and includes 12 species, from Enterovirus species A to L and Rhinovirus species A to C [53]. Among them, Enterovirus A71 (EV-A71), Enterovirus D68 (EV-D68), Coxsackievirus A16 (CVA16), Coxsackievirus A10 (CVA10) and Coxsackievirus A6 (CVA6) are most common causative agents responsible for outbreak of human hand, foot and mouth disease (HFMD), most often affecting children [54,55]. Though HFMD is self-limiting in most of the cases, the increasing morbidity and neurological sequelae caused by these viruses have been a huge burden of public health in recent years across the world, especially in Asia-Pacific region [[56], [57], [58], [59]]. The clinical manifestation of HFMD patients ranges from mild fever, blisters on the limbs and in the mouth, and gastrointestinal symptoms, e.g. nausea and diarrhea, to severe neurological symptoms which might lead to life-long paralysis [54,55]. The most well-known enterovirus, poliovirus, was one of the most horrible pathogens that caused poliomyelitis in last century, affecting numerous of children each year, with 1%–2% of the infected patients showing lifelong paralysis sequelae [60]. Luckily, since the launch of “Global Polio Eradication Initiative” initiated by World Health Assembly (WHA) in 1988, global incidence of poliomyelitis caused by poliovirus infection has been dramatically reduced to nearly 1%; through broad epidemiological surveillance and highly effective vaccination program, poliomyelitis caused by poliovirus has been reported by only three countries, Afghanistan, Pakistan, and Nigeria, across the world in recent years [[61], [62], [63]]. Another important human enterovirus is Coxsackievirus B3 (CVB3), which has been the major causative agent of viral myocarditis associated with heart dysfunction, also commonly affecting children and young adults [64,65]. CVB3-induced myocarditis can lead to dilated cardiomyopathy (DCM), which is a common cause of heart transplantation as it is the only feasible choice to overcome DCM currently [66,67].
The life cycle of enterovirus begins with recognition and binding to receptors on plasma membrane, followed by receptor-mediated endocytosis which results in virus uncoating and release of viral genome RNA (vRNA) into cytoplasm induced by pH change in endosomal compartment [68,69]. Subsequently, vRNA serves as a primary template to translate into a large polyprotein that are cleaved by viral proteases 2Apro, 3Cpro and 3CDpro into ten proteins, including four capsid proteins (VP1, VP2, VP3 and VP4), two viral proteases (2A, and 3C), a RNA-dependent-RNA-polymerase (3D), two proteins involved in RNA synthesis (2B, and 2C), and a primer of initiation of RNA synthesis (3AB) [68,69].
Then, RNA genome replication, mediated by the RNA-dependent RNA polymerase 3Dpol and other host factors, takes place in virus-induced membranous compartments, called replication organelles (ROs) [70]. Like other positive-sense RNA viruses, e.g. Dengue Virus (DENV), West Nile Virus (WNV), Hepatitis C Virus (HCV), Flock House Virus (FHV) and Severe Acute Respiratory Syndrome (SARS)-Coronavirus, enterovirus can subvert host protein network and lipid landscape to rebuild membranous structures as ROs, which provide ideal sites for generating RNA genome and escaping from host defense induced by replication intermediates, double-stranded RNA (dsRNA) [[71], [72], [73], [74]]. It has been shown that the ROs of enterovirus are localized to cis-Golgi membranes, e.g. poliovirus [75], trans-Golgi network, e.g. CVB3 [76], or endoplasmic reticulum–Golgi interface, e.g. human rhinovirus [77], or highly dynamic secretory pathway organelles. Upon enterovirus infection, the secretory pathway is hijacked and remodeled, in which small Ras-family GTPase Arf1 and its guanine nucleotide exchange factor GBF1 are recruited to the replication complex, and phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) are subsequently lured to the complex, where it produces phosphatidylinositol-4-phosphate (PI4P) lipids [[77], [78], [79]]. Together with viral proteins, these host factors rebuild a distinct elaborate membranous microenvironment to support viral RNA genome replication. Autophagosome, which is an important component of the intracellular membranous structures with highly dynamic activity, has been suggested to provide scaffold to recruit replication complex for several positive-stranded RNA viruses. Based on two-dimensional electron microscopy (EM) studies, ROs of enterovirus contains both single-membrane and double-membrane structures [80]. It is unclear that whether this double-membrane structures are originated from autophagic structures or not. Currently, whether autophagic compartments directly function as replication organelles or indirectly correlate with viral replication are still debatable, as too limited evidences have been provided to draw a conclusion.
Viral RNA amplification begins with synthesis of a negative-strand copy from viral genome to generate a dsRNA intermediate. The negative-strand RNA then serves as template to generate more positive-strand RNAs, which either serve as mRNA templates for viral protein synthesis and replication or are incorporated into structural capsid proteins for package and further process into mature infectious progeny virions [68]. In the end, the newly assembled virions egress through either lytic or non-lytic release of membrane-bound structures or via cell lysis [81,82].
8. Interplay of autophagy and enterovirus
8.1. Enterovirus 71 (EV-A71)
Several studies revealed that EV-A71 induced autophagy to offer a favorable micro-environment for its propagation within infected host. Accumulation of GFP-LC3 puncta was observed in EV-A71 infected RD and SKN-SH cells in a time-dependent manner, which is consistent with LC3 immunoblotting result, showing increased lipid-bound LC3-II level in infected cells [83]. The autophagosome-like structures were also detected in both SK-N-SH cells and neuron cells isolated from EV-A71 infected mice under transmission electron microscopy [83]. Subsequently, EV-A71 structural protein VP1 and nonstructural protein 2C were found to be co-localized with autophagosome marker LC3 and late endosome protein, mannose 6-phosphate receptor (M6PR), and LC3-II conversion was induced in EV-A71 infected SK-N-SH cells and in the brain tissues of in EV-A71 infected suckling mice [84]. Treatment of cells with 3-MA, an early-stage inhibitor that blocks PI3K activity, or bafilomycin A1 or saikosaponin D, which are late stage autophagy inhibitors that block autophagosome-lysosome fusion, suppressed viral production, while induction of autophagy by tamoxifen, rapamycin and serum starvation promoted viral yields [83,85]. Likewise, 3-MA alleviated the clinical symptoms and decreased the viral titer in the brain tissues isolated from the EV-A71 infected mice [84]. These data suggest that EV-A71 induces the formation of autophagosomes and benefits form it.
EV-A71-induced autophagic activity was shown to be regulated by inhibiting the expression level of a microRNA, miR-30a, which binds to 3’-UTR of Beclin 1 to inhibit autophagy, resulting in decrease of EV-A71 replication [86]. The interplay between EV-A71-induced autophagy and apoptosis was also explored [87]. Not surprisingly, inhibition of autophagy and lysosomal protease impaired EV-A71 replication, resulting in decreased apoptotic activity; while blockage of apoptosis led to upregulated autophagic activity with increasing conversion of LC3-I to LC3-II [87]. In addition, both autophagy and apoptosis were found to be involved in the release of progeny virus [87].
Although all these results indicate that EV-A71 induces the formation of autophagosomes to facilitate its replication, it is unknown whether EV-A71 induces autophagic flux, since no reports have actually shown that the number of autolysosome is increased in EV-A71 infected cells. In addition, the detailed mechanisms of how EV-A71 manipulates autophagic machinery and in which stage does EV-A71 benefit form autophagy remain elusive.
8.2. Coxsackievirus B3 (CVB3)
Several research groups reported that CVB3 usurps autophagosome to support its replication both in vivo and in vitro [[88], [89], [90], [91], [92], [93]]. CVB3 infection induced the formation of double-membranous autophagosome-like vesicle in infected Hela and HEK293A cells under electron microscopy [91]. Compared to uninfected HEK293A cells, increased GFP-LC3 puncta and LC3-II/LC3-I ratio were detected in CVB3 infected cells [91]. Inhibition of autophagy pathway by treatment with 3-MA and knockdown of several essential autophagy-related genes, e.g. ATG7, Beclin-1, or PIK3C3, reduced viral production of CVB3. In contrast, nutrient deprivation (HBSS incubation) or mTOR inhibitor (rapamycin) promoted CVB3 propagation [91,93]. In the in vivo study, autophagy-like vesicles were also observed in the pancreatic acinar cells and heart isolated from CVB3 infected mice [89,90].
However, SQSTM1 was accumulated in the infected cells and large autophagy-related vesicles were formed in the infected cells, suggesting that the late stages of autophagy, either fusion of autophagosome with lysosome or degradation in autolysosome, is blocked in CVB3 infected cells [90]. Likewise, autophagic flux was inhibited in CVB3 infected HEK293 and Hela cells [88,89]. In CVB3 infected cells, GFP and RFP signals were co-localized and accumulated, suggesting that fusion of autophagosome with lysosome is blocked. The inhibition of autophagic flux was mediated through the modification of soluble N-ethylmaleimide-sensitive factor activating protein receptor (SNARE) proteins. In addition, the level of STX17 (a SNARE of the autophagosome) was decreased following CVB3 infection and overexpression of STX17 rescued the formation of autolysosome, part of lysosomal function, and reduced cell death associated with CVB3 infection [88]. Furthermore, SNAP29 and Pleckstrin homology domain-containing family M member 1 (PLEKHM1), two important proteins regulating fusion of autophagosome and lysosome, were cleaved by proteinase 3C (a nonstructural protein of CVB3), thereby leading to dysfunction of autolysosome formation and degradation [89]. Knockdown of SNAP29 or PLEKHM1 promoted CVB3 intracellular viral yields [89]. Taken together, these data suggest that CVB3-induced disruption of SNAP29 and PLEKHM1 arrests autophagy flux, leading to the increase of LC3-bound membrane structures which may offer sites for viral replication and package.
Most recently, Shi et al. found that SQSTM1 was cleaved by viral protease 2Apro following CVB3 infection in Hela cells [94]. The cleaved SQSTM1 was no longer recognized and engulfed into autophagosome, thereby compromising its ubiquitous function. In addition, the truncated SQSTM1 failed to activate the NFKB pathway, indicating that CVB3 exploits this strategy to avoid host defense. Interestingly, Alirezaei et al. reported that CVB3 also utilized LC3 in either autophagy-dependent or -independent manner in infected mice to support viral replication [95]. The authors used three recombinant CVB3 viruses encoding either wildtype or mutated autophagy-related genes (proLC3, proLC3G120A, and ATG4BC74A) to compare the replication efficacy in mice. They found that proLC3 virus induced the accumulation of lipid-bound LC3-II and replicated efficiently in an autophagy-dependent manner. Whereas the proLC3G120A virus with the mutated LC3 at glycine residue failed to attach to PE, instead, it interacted with the ER-resident protein SEL1L and used it as the alternative membranous structures for CVB3 replication. Interestingly, the ATG4BC74A virus, in which mutated ATG4B fails to process proLC3 into LC3-I and leads to accumulation of proLC3 proteins, was also capable of replicating efficiently. Therefore, CVB3 utilizes unprocessed LC3, LC3-I or PE-bound LC3 flexibly to facilitate viral replication, suggesting that CVB3 is capable of exploiting autophagy-related proteins in both autophagy-dependent and -independent manner. Nevertheless, these results indicate that CVB3 is quite versatile, using various strategies to manipulate autophagy-related proteins and host membranous structures to rebuild replication scaffold both in vivo and in vitro, and to avoid host defense.
Besides manipulating autophagy components to promote viral replication, CVB3 is found to take advantage of autophagic components as original resource of extracellular membrane vesicles (EMVs) to facilitate its dissemination within host cells [81,89]. Robinson et al. found that extracellular microvesicles (EMVs) isolated from CVB3-infected cells contained infectious CVB3 virions, indicating that CVB3 may be releases from host cell through EMVs in addition to cell lysis [81]. Recently, Mohamud et al. also found that LC3 and viral structural protein VP1 were co-existed in EMVs isolated from CVB3-infected cells, indicating that egress of CVB3 progeny virions may be achieved through secretory vesicles originated from autophagic membrane [89]. Altogether, these findings indicate that CVB3 exploits autophagy machinery to promote both viral replication and viral release.
8.3. Poliovirus
Poliovirus has been reported to benefit from virus induced autophagy according to several research [5,[96], [97], [98], [99]]. Double-membrane autophagy-like vesicles can be observed in poliovirus infected COS-1 cells under electron microscopy [96]. It is speculated that these replication vesicles are originated from Endoplasmic Reticulum [96]. However, Belov et al. found that GM130 (cis-Golgi marker) was fragmented into multiple puncta-like structures upon poliovirus infection, and showed co-localization with viral nonstructural protein 3A, suggesting that the poliovirus ROs membrane are most likely derived from cis-Golgi membranes [75]. They also found that single-membrane compartments were evoked at early stage of poliovirus infection, while plenty of double-membrane structures were accumulated at the end of replication cycle [75]. The authors proposed that formation of late double-membrane vesicles in infected cells is attributed to engulfment of early single-membrane chambers [75]. Similar to EV-A71 and CVB3, poliovirus infection was found to recruit LC3 to viral replication complex, which was also co-localized with LAMP1 (autolysosome/lysosome marker) [5]. Likewise, knockdown of autophagy-related proteins (LC3 and ATG12), or treatment with 3-MA decreased poliovirus viral load, while activation of autophagy by tamoxifen or rapamycin promoted viral production [5]. Moreover, Taylor and Kirkegaard found that nonstructural viral protein 2BC and 3A directly modified LC3 and recruited it to cellular membrane, suggesting that poliovirus manipulates autophagy in a non-canonical manner [100]. Surprisingly, Richards et al. recently found that poliovirus dsRNA does not co-localized with LC3 in poliovirus infected Hela cells, hinting that autophagosome might not be the direct origins of reorganized membrane [101].
Corona Velazquez et al. found that depletion of several important conventional members of autophagic complex, such as ULK1, ULK2 and RB1CC1/FIP200, failed to affect poliovirus production [97]. Moreover, they found that SQSTM1 was cleaved and digested in a non-canonical manner in poliovirus infected cells [97], similar to the aforementioned CVB3 infected cells [94]. However, Abernathy et al. recently reported that depletion of autophagy-related protein 9 (ATG9), ULK1, FIP200 and LC3B undermined poliovirus replication, while knockout of VPS34, Beclin-1 and ATG5 did not affect viral production [99]. They further demonstrated that poliovirus recruited the non-lipid-bound form of LC3 to the remodeled membranes in GFP-LC3-G120A expressing cells, indicating that the canonical PE-linked LC3, an essential form in autophagosome formation, is not required for poliovirus replication [99]. Although the detailed mechanism of how poliovirus utilizes autophagy network reported from different research teams is not be perfectly consistent, these data do suggest that poliovirus manipulates a non-canonical autophagy pathway to facilitate its propagation, and poliovirus-induced reorganized membranes might not be originated from the autophagosomes.
8.4. Other enteroviruses
Foot-and-mouth disease virus (FMDV) has been reported to benefit from induced autophagy [102,103]. O'Donnell et al. reported that GFP-LC3 puncta were accumulated and co-localized with non-structural viral proteins 2B, 2C and 3A in FMDV-infected MCF-10A cells. In addition, co-localizations of viral structural protein VP1 with Atg5, and GFP-LC3 with LAMP-1 were detected in FMDV-infected cells [102]. As expected, inhibition of autophagy by either 3-MA or small RNA interfering (LC3 or ATG12) suppressed viral production, while activation of autophagy through rapamycin treatment promoted viral yield [102]. Subsequently, Berryman et al. found that FMDV induced autophagosome formation in an ATG5-dependent manner in infected CHO cells [103]. However, treatment of wortamanin, an inhibitor of PI3K3C/VPS34, did not inhibit GFP-LC3 puncta induced by FMDV infection, suggesting that FMDV induces autophagy in a PI3K-independent manner [102]. Unlike other enteroviruses, FMDV seems to induce autophagy at early stage of infection, as both UV-inactivated virus and empty FMDV capsids could induce autophagosome formation [102]. Another special phenomenon observed in FMDV infected cells is that viral structural protein VP1 but not nonstructural proteins (3A and RNA-polymerase 3D) co-localizes with LC3, indicating that autophagosome is involved in early stage of viral life cycle before genome replication. Taken together, these findings suggest that FMDV exploits autophagy in a non-canonical manner during cell entry to promote infection, instead of providing membranes for replication.
Enterovirus D68 (EV-D68) also benefits from induced autophagy in host cells, as inhibition of autophagy by depletion of E1-like enzyme ATG7 or treatment with bafilomycin A1 undermined viral yield, while induction of autophagy by starvation promoted EV-D68 propagation [104]. Similar to the aforementioned CVB3 infection [89,94], accumulation of GFP-LC3 puncta and cleavage of SNAP29 and SQSTM1 by EV-D68 3C protease were detected in EV-D68 infected Hela cells [104]. The cleaved SNAP29, which reportedly regulated formation of autolysosome, facilitated EV-D68 in early infection but was later cleaved. In addition, SNAP47 whose role is unknown in autophagy pathway, was suggested to play a role in regulating EV-D68 induced autophagy and viral release [104]. These findings suggest that EV-D68 benefits from early stage of autophagy and blocks autophagic flux through cleavage of important factors which regulate autolysosome formation, thereby remodeling autophagic trafficking to facilitate viral release.
9. Conclusion
Following infection of enterovirus, intracellular membranes are rearranged to facilitate the concentrated viral vesicles to locate on secretory organelles, providing a superior environment for RNA genome replication [[71], [72], [73], [74],77,78]. In addition, these remodeled membrane compartments help to isolate viral RNAs from cytoplasmic RNases, preventing being degraded and avoiding from detection by cytosolic RNA sensors to mitigate immune responses. However, the origin of the reorganized membranes remains elusive. Though most of the research work we summarized in this review show that enteroviruses, e.g. CVB3 [[88], [89], [90], [91], [92], [93]], EV-A71 [[83], [84], [85]] and poliovirus, manipulate autophagy network to facilitate viral replication, no direct evidences show that viral RNA replication actually takes place in autophagosome or autolysosome. From our point of view, using dsRNA (replication intermediate) antibody to label replication organelles is more ideal and specific than using viral protein immunostaining. According to the study of Richards et al., poliovirus dsRNA was not co-localized with GFP-LC3, hinting that reorganized replication compartments are not originated from autophagic membrane [101]. In early stage of poliovirus infection, only single-membrane compartments that support viral replication were detected, whereas double-membrane vesicles were only seen later in the poliovirus life cycle [75]. It is reasonable to speculate that poliovirus might first induce the accumulation of single-membrane vesicles as ROs for genome replication, and some of the accumulated ROs later might then be engulfed to form autophagosomes. It is also possible that the newly assembled viruses are engulfed by autophagosomes later in poliovirus life cycle.
Along this line, it has been reported that depletion of VPS34 complex, which is an important autophagy complex that responsible for generating PI3P in mammalian cells, did not affect poliovirus replication [99]. Similarly, it has been shown that wortamanin, an inhibitor of PI3K3C/VPS34, failed to block GFP-LC3 puncta formation in the poliovirus infected cells [102]. This again suggest that enterovirus do not directly usurp autophagy compartments as replication sites, as it is known that lipid component of autophgosome/autolysosome is PI3P. Accumulating evidences have actually shown that enterovirus recruits type III phosphatidylinositol 4-kinases (PI4KIIIs) to replication sites, where increased amount of phosphoinositides 4-phosphate (PI4P) is generated; the PI4P is subsequently exchanged for cholesterol at replication organelles [76,77,[105], [106], [107], [108], [109], [110]]. Taken together, these findings hint that enterovirus manipulates autophagy-related proteins to boost viral replication in an autophagy-independent manner.
Conventionally, microbial infection-induced autophagy is supposed to act as immune response, leading to engulfment and clearance of the invading pathogens. However, most of positive-strand RNA viruses, e.g. enteroviruses, are capable of utilizing different components of autophagic network to counter host defense and take advantage of it. Notably, each member of enterovirus may subvert autophagy machinery and benefit from this process via a distinct strategy. More efforts are needed to reveal detailed mechanisms on how enterovirus manipulate autophagy network to achieve efficient propagation. Particularly, it is of great interest to determine whether enterovirus directly uses autophagsome or autolysosome as replication sites, and whether enterovirus blocks the autophagosome-lysosome fusion to avoid lysosomal degradation or for viral release. The deep understanding of the interplay between enterovirus and autophagy will undoubtedly provide new therapeutic targets against enterovirus infection.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgements
We apologize to those whose work could not be cited here due to the space limitation. This work was supported by Hong Kong Research Grant Council (RGC) grants (11101717), NSFC (21778045), CAS-Croucher Funding Scheme, Guangdong-Hong Kong joint innovation Research Scheme (#2016A050503010), and Shenzhen government research grant (JCYJ20160229165235739 and JCYJ 20170413141331470) to JY.
References
- 1.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018:1. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N., Klionsky D.J. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 4.Dreux M., Gastaminza P., Wieland S.F., Chisari F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson W.T., Giddings T.H., Jr, Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y.-R., Lei H.-Y., Liu M.-T., Wang J.-R., Chen S.-H., Jiang-Shieh Y.-F., Lin Y.-S., Yeh T.-M., Liu C.-C., Liu H.-S. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi J., Luo H. Interplay between the cellular autophagy machinery and positive-stranded RNA viruses. Acta Biochim. Biophys. Sin. (Shanghai) 2012;44:375–384. doi: 10.1093/abbs/gms010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- 9.Klionsky D.J. The molecular machinery of autophagy: unanswered questions. J. Cell. Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 12.McAlpine F., Williamson L.E., Tooze S.A., Chan E.Y. Regulation of nutrient-sensitive autophagy by uncoordinated 51-like kinases 1 and 2. Autophagy. 2013;9:361–373. doi: 10.4161/auto.23066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itakura E., Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirth M., Joachim J., Tooze S.A. Elsevier; 2013. Tilte. [Google Scholar]
- 15.Liang C., Feng P., Ku B., Oh B.-H., Jung J.U. UVRAG: a new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 16.Fan W., Nassiri A., Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14 (L) Proc. Natl. Acad. Sci. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson S.R., Simonsen A. Membrane dynamics in autophagosome biogenesis. J. Cell. Sci. 2015;128:193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- 19.Jung C.H., Ro S.-H., Cao J., Otto N.M., Kim D.-H. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 21.Heitman J., Movva N.R., Hall M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 22.Raught B., Gingras A.-C., Sonenberg N. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thoreen C.C., Sabatini D.M. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5:725–726. doi: 10.4161/auto.5.5.8504. [DOI] [PubMed] [Google Scholar]
- 24.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S.-i., Natsume T., Takehana K., Yamada N. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., Rubinsztein D.C. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 28.Hurley J.H., Young L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L.J., Rubinsztein D.C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdollahzadeh I., Schwarten M., Gensch T., Willbold D., Weiergräber O.H. The Atg8 family of proteins—modulating shape and functionality of autophagic membranes. Front. Genet. 2017;8:109. doi: 10.3389/fgene.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 32.Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y.-K., Lee J.-A. Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 2016;49:424. doi: 10.5483/BMBRep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 35.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez‐Wandelmer J., Reggiori F. Amphisomes: out of the autophagosome shadow? EMBO J. 2013;32:3116–3118. doi: 10.1038/emboj.2013.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu L., Chen Y., Tooze S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura S., Yoshimori T. New insights into autophagosome–lysosome fusion. J. Cell. Sci. 2017;130:1209–1216. doi: 10.1242/jcs.196352. [DOI] [PubMed] [Google Scholar]
- 39.Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., Mizushima N. The HOPS complex mediates autophagosome–lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijdeven R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., Neefjes J. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 2016;7:11808. doi: 10.1038/ncomms11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.kleine Balderhaar H.J., Ungermann C. CORVET and HOPS tethering complexes–coordinators of endosome and lysosome fusion. J. Cell. Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Klionsky D.J., Cuervo A.M., Seglen P.O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 45.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Longatti A., Tooze S. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 48.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T.L.C.3. A mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asanuma K., Tanida I., Shirato I., Ueno T., Takahara H., Nishitani T., Kominami E., Tomino Y. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. Faseb J. 2003;17:1165–1167. doi: 10.1096/fj.02-0580fje. [DOI] [PubMed] [Google Scholar]
- 50.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T.L.C.3. GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell. Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 53.Ho B.C., Yang P.C., Yu S.L. MicroRNA and pathogenesis of enterovirus infection. Viruses. 2016;8 doi: 10.3390/v8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aswathyraj S., Arunkumar G., Alidjinou E., Hober D. Hand, foot and mouth disease (HFMD): emerging epidemiology and the need for a vaccine strategy. Med. Microbiol. Immunol. 2016;205:397–407. doi: 10.1007/s00430-016-0465-y. [DOI] [PubMed] [Google Scholar]
- 55.Ventarola D., Bordone L., Silverberg N. Update on hand-foot-and-mouth disease. Clin. Dermatol. 2015;33:340–346. doi: 10.1016/j.clindermatol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Lugo D., Krogstad P. Enteroviruses in the early 21st century: new manifestations and challenges. Curr. Opin. Pediatr. 2016;28:107–113. doi: 10.1097/MOP.0000000000000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- 58.Bian L., Wang Y., Yao X., Mao Q., Xu M., Liang Z. Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev. Anti. Ther. 2015;13:1061–1071. doi: 10.1586/14787210.2015.1058156. [DOI] [PubMed] [Google Scholar]
- 59.Mao Q., Wang Y., Yao X., Bian L., Wu X., Xu M., Liang Z. Coxsackievirus A16: epidemiology, diagnosis, and vaccine. Hum. Vaccin. Immunother. 2014;10:360–367. doi: 10.4161/hv.27087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lévêque N., Semler B.L. A 21st century perspective of poliovirus replication. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arita M. Poliovirus studies during the endgame of the polio eradication program. Jpn. J. Infect. Dis. 2017;70:1–6. doi: 10.7883/yoken.JJID.2016.356. [DOI] [PubMed] [Google Scholar]
- 62.Lopalco P.L. Wild and vaccine-derived poliovirus circulation, and implications for polio eradication. Epidemiol. Infect. 2017;145:413–419. doi: 10.1017/S0950268816002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Racaniello V.R. One hundred years of poliovirus pathogenesis. Virology. 2006;344:9–16. doi: 10.1016/j.virol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 64.Chang H., Li X., Cai Q., Li C., Tian L., Chen J., Xing X., Gan Y., Ouyang W., Yang Z. The PI3K/Akt/mTOR pathway is involved in CVB3-induced autophagy of HeLa cells. Int. J. Mol. Med. 2017;40:182–192. doi: 10.3892/ijmm.2017.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esfandiarei M., McManus B.M. Molecular biology and pathogenesis of viral myocarditis. Annu. Rev. Pathmechdis. Mech. Dis. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 66.Fairweather D., Stafford K.A., Sung Y.K. Update on coxsackievirus B3 myocarditis. Curr. Opin. Rheumatol. 2012;24:401. doi: 10.1097/BOR.0b013e328353372d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Massilamany C., Gangaplara A., Reddy J. Intricacies of cardiac damage in coxsackievirus B3 infection: implications for therapy. Int. J. Cardiol. 2014;177:330–339. doi: 10.1016/j.ijcard.2014.09.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baggen J., Thibaut H., Strating J.R., van Kuppeveld F.J. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018:1. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 69.Garmaroudi F.S., Marchant D., Hendry R., Luo H., Yang D., Ye X., Shi J., McManus B.M. Coxsackievirus B3 replication and pathogenesis. Future Microbiol. 2015;10:629–653. doi: 10.2217/fmb.15.5. [DOI] [PubMed] [Google Scholar]
- 70.van der Schaar H.M., Dorobantu C.M., Albulescu L., Strating J.R., van Kuppeveld F.J. Fat (al) attraction: picornaviruses usurp lipid transfer at membrane contact sites to create replication organelles. Trends Microbiol. 2016;24:535–546. doi: 10.1016/j.tim.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linden Lvd., Wolthers K.C., van Kuppeveld F.J. Replication and inhibitors of enteroviruses and parechoviruses. Viruses. 2015;7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 73.Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sasvari Z., Nagy P.D. Making of viral replication organelles by remodeling interior membranes. Viruses. 2010;2:2436–2442. doi: 10.3390/v2112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belov G.A., Nair V., Hansen B.T., Hoyt F.H., Fischer E.R., Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu N.-Y., Ilnytska O., Belov G., Santiana M., Chen Y.-H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roulin P.S., Lötzerich M., Torta F., Tanner L.B., van Kuppeveld F.J., Wenk M.R., Greber U.F. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe. 2014;16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 78.Hsu N.Y., Ilnytska O., Belov G., Santiana M., Chen Y.H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., Cameron C.E., Ehrenfeld E., van Kuppeveld F.J., Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belov G.A., Altan-Bonnet N., Kovtunovych G., Jackson C.L., Lippincott-Schwartz J., Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van der Schaar H., Melia C., van Bruggen J., Strating J., van Geenen M., Koster A., Bárcena M., van Kuppeveld F. Illuminating the sites of enterovirus replication in living cells by using a split-GFP-tagged viral protein. mSphere. 2016;1:e00104–00116. doi: 10.1128/mSphere.00104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson S.M., Tsueng G., Sin J., Mangale V., Rahawi S., McIntyre L.L., Williams W., Kha N., Cruz C., Hancock B.M., Nguyen D.P., Sayen M.R., Hilton B.J., Doran K.S., Segall A.M., Wolkowicz R., Cornell C.T., Whitton J.L., Gottlieb R.A., Feuer R. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lai J.K., Sam I.C., Chan Y.F. The autophagic machinery in enterovirus infection. Viruses. 2016;8 doi: 10.3390/v8020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang S.C., Chang C.L., Wang P.S., Tsai Y., Liu H.S. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 2009;81:1241–1252. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee Y.-R., Wang P.-S., Wang J.-R., Liu H.-S. Enterovirus 71-induced autophagy increases viral replication and pathogenesis in a suckling mouse model. J. Biomed. Sci. 2014;21:80. doi: 10.1186/s12929-014-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li C., Huang L., Sun W., Chen Y., He M.L., Yue J., Ballard H. Saikosaponin D suppresses enterovirus A71 infection by inhibiting autophagy. Signal Transduct. Target. Ther. 2019;4:4. doi: 10.1038/s41392-019-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu Y., Xu W., Chen D., Feng C., Zhang L., Wang X., Lv X., Zheng N., Jin Y., Wu Z. Enterovirus 71 induces autophagy by regulating has-miR-30a expression to promote viral replication. Antiviral Res. 2015;124:43–53. doi: 10.1016/j.antiviral.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 87.Xi X., Zhang X., Wang B., Wang T., Wang J., Huang H., Wang J., Jin Q., Zhao Z. The interplays between autophagy and apoptosis induced by enterovirus 71. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Tian L., Yang Y., Li C., Chen J., Li Z., Li X., Li S., Wu F., Hu Z., Yang Z. The cytotoxicity of coxsackievirus B3 is associated with a blockage of autophagic flux mediated by reduced syntaxin 17 expression. Cell Death Dis. 2018;9:242. doi: 10.1038/s41419-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohamud Y., Shi J., Qu J., Poon T., Xue Y.C., Deng H., Zhang J., Luo H. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018;22:3292–3303. doi: 10.1016/j.celrep.2018.02.090. [DOI] [PubMed] [Google Scholar]
- 90.Kemball C.C., Alirezaei M., Flynn C.T., Wood M.R., Harkins S., Kiosses W.B., Whitton J.L. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J. Virol. 2010;84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong J., Zhang J., Si X., Gao G., Mao I., McManus B.M., Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang H., Li X., Cai Q., Li C., Tian L., Chen J., Xing X., Gan Y., Ouyang W., Yang Z. The PI3K/Akt/mTOR pathway is involved in CVB3-induced autophagy of HeLa cells. Int. J. Mol. Med. 2017;40:182–192. doi: 10.3892/ijmm.2017.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tabor-Godwin J.M., Tsueng G., Sayen M.R., Gottlieb R.A., Feuer R. The role of autophagy during coxsackievirus infection of neural progenitor and stem cells. Autophagy. 2012;8:938–953. doi: 10.4161/auto.19781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi J., Wong J., Piesik P., Fung G., Zhang J., Jagdeo J., Li X., Jan E., Luo H. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy. 2013;9:1591–1603. doi: 10.4161/auto.26059. [DOI] [PubMed] [Google Scholar]
- 95.Alirezaei M., Flynn C.T., Wood M.R., Harkins S., Whitton J.L. Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo. Autophagy. 2015;11:1389–1407. doi: 10.1080/15548627.2015.1063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suhy D.A., Giddings T.H., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Corona Velazquez A., Corona A.K., Klein K.A., Jackson W.T. Poliovirus induces autophagic signaling independent of the ULK1 complex. Autophagy. 2018;14:1201–1213. doi: 10.1080/15548627.2018.1458805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bird S.W., Maynard N.D., Covert M.W., Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc. Natl. Acad. Sci. U. S. A. 2014;111:13081–13086. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abernathy E., Mateo R., Majzoub K., van Buuren N., Bird S.W., Carette J.E., Kirkegaard K. Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.2006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor M.P., Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 2007;81:12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richards A.L., Soares-Martins J.A., Riddell G.T., Jackson W.T. Generation of unique poliovirus RNA replication organelles. MBio. 2014;5:e00833–00813. doi: 10.1128/mBio.00833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O’Donnell V., Pacheco J.M., LaRocco M., Burrage T., Jackson W., Rodriguez L.L., Borca M.V., Baxt B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology. 2011;410:142–150. doi: 10.1016/j.virol.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berryman S., Brooks E., Burman A., Hawes P., Roberts R., Netherton C., Monaghan P., Whelband M., Cottam E., Elazar Z., Jackson T., Wileman T. Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J. Virol. 2012;86:12940–12953. doi: 10.1128/JVI.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Corona A.K., Saulsbery H.M., Corona Velazquez A.F., Jackson W.T. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018;22:3304–3314. doi: 10.1016/j.celrep.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van der Schaar H.M., Leyssen P., Thibaut H.J., De Palma A., van der Linden L., Lanke K.H., Lacroix C., Verbeken E., Conrath K., MacLeod A.M. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIβ. Antimicrob. Agents Chemother. 2013;57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spickler C., Lippens J., Laberge M.-K., Desmeules S., Bellavance É., Garneau M., Guo T., Hucke O., Leyssen P., Neyts J. Phosphatidylinositol 4-kinase III beta is essential for replication of human rhinovirus and its inhibition causes a lethal phenotype in vivo. Antimicrob. Agents Chemother. 2013;57:3358–3368. doi: 10.1128/AAC.00303-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ishikawa-Sasaki K., Sasaki J., Taniguchi K. A complex comprising phosphatidylinositol 4-kinase IIIβ, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex. J. Virol. 2014;88:6586–6598. doi: 10.1128/JVI.00208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Delang L., Paeshuyse J., Neyts J. The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem. Pharmacol. 2012;84:1400–1408. doi: 10.1016/j.bcp.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arita M. Phosphatidylinositol‐4 kinase III beta and oxysterol‐binding protein accumulate unesterified cholesterol on poliovirus‐induced membrane structure. Microbiol. Immunol. 2014;58:239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]
- 110.Altan-Bonnet N., Balla T. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 2012;37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]