Abstract
Aims
A new human coronavirus (HCoV), which has been designated SARS-CoV-2, began spreading in December 2019 in Wuhan City, China causing pneumonia called COVID-19. The spread of SARS-CoV-2 has been faster than any other coronaviruses that have succeeded in crossing the animal-human barrier. There is concern that this new virus will spread around the world as did the previous two HCoVs—Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS)—each of which caused approximately 800 deaths in the years 2002 and 2012, respectively. Thus far, 11,268 deaths have been reported from the 258,842 confirmed infections in 168 countries.
Main methods
In this study, the RNA-dependent RNA polymerase (RdRp) of the newly emerged coronavirus is modeled, validated, and then targeted using different anti-polymerase drugs currently on the market that have been approved for use against various viruses.
Key findings
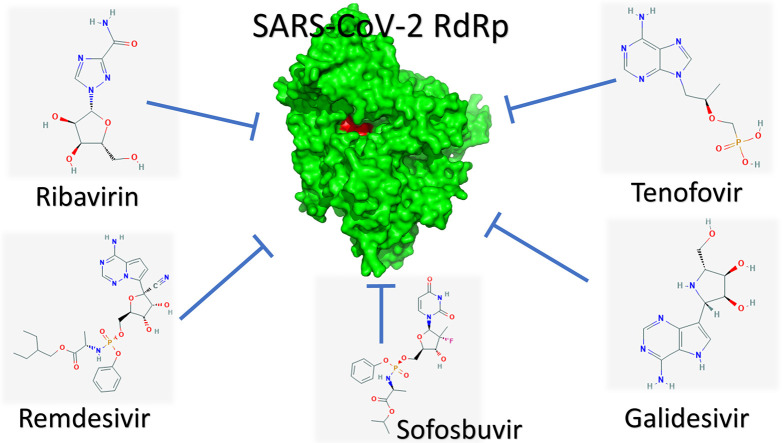
The results suggest the effectiveness of Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir as potent drugs against SARS-CoV-2 since they tightly bind to its RdRp. In addition, the results suggest guanosine derivative (IDX-184), Setrobuvir, and YAK as top seeds for antiviral treatments with high potential to fight the SARS-CoV-2 strain specifically.
Significance
The availability of FDA-approved anti-RdRp drugs can help treat patients and reduce the danger of the mysterious new viral infection COVID-19. The drugs mentioned above can tightly bind to the RdRp of the SARS-CoV-2 strain and thus may be used to treat the disease. No toxicity measurements are required for these drugs since they were previously tested prior to their approval by the FDA.
Keywords: COVID-19, SARS-CoV-2, RdRp, Molecular docking, Structural bioinformatics, Drug repurposing
Graphical abstract
1. Introduction
In December 2019, a rapid and widespread outbreak of a novel coronavirus designated COVID-19, emerged in the city of Wuhan, China [[1], [2], [3]]. According to the World Health Organization (WHO) surveillance draft in January 2020, any traveler to Wuhan City in Hubei Province 2 weeks before the onset of the symptoms is suspected to be infected with COVID-19 [1,4]. Additionally, the WHO distributed interim guidance for laboratories that carry out the testing for the newly-emerged outbreak, as well as infection prevention and control guidance [5,6]. The COVID-19 pneumonia is suspected of having originated in a seafood market, with an unknown animal being responsible for the emergence of the novel virus [2]. There are now surveillance borders around the globe, attempting to prevent the spread of the new mysterious coronavirus [7]. Just two months ago, in mid-January, only 41 cases had been confirmed to be COVID-19 positive, leaving one person dead and 7 in critical care [8]. These numbers are continually increasing each day, and the number of confirmed cases at the time of this writing has now exceeded 258,000, with 11,268 confirmed deaths, mostly in China, Europe, and the Islamic Republic of Iran. On 20 January 2020, the National Health Commission of China confirmed human-to-human transmission of the COVID-19 outbreak [8]. Ten days later, the WHO declared COVID-19 to be a Public Health Emergency of International Concern (PHEIC), then COVID-19 is declared as a pandemic 40 days later by the WHO. The symptoms of COVID-19 include fever, malaise, dry cough, shortness of breath, and respiratory distress [2].
Based on its nucleotide sequences, SARS-CoV-2 is a member of Betacoronaviruses, such as the SARS and MERS HCoVs [[9], [10], [11]]. Currently, seven different strains of human coronaviruses (HCoVs) have been reported, including the 229E and NL63 strains of HCoVs (Alphacoronaviruses), and the OC43, HKU1, SARS, MERS, and SARS-CoV-2 HCoVs (Betacoronaviruses) [2,10,12]. The SARS and MERS HCoVs are the most well-known and aggressive strains of coronaviruses, each causing approximately 800 deaths. According to the WHO, the mortality rates for SARS and MERS HCoV are 10% and 36%, respectively. [10,[12], [13], [14], [15]]. The next few months will reveal the mortality of the new virus; a nearly 4.1% mortality rate for a flanking viral spread may mean millions of deaths.
HCoVs are positive-sense, long (30,000 bp), single-stranded RNA viruses. Two groups of proteins characterize HCoVs: structural proteins, such as Spike (S) marking all coronaviruses, Nucleocapsid (N), Matrix (M), and Envelope (E); and non-structural proteins, such as proteases (nsp3 and nsp5) and RdRp (nsp12) [10]. RdRp is a crucial viral enzyme in the life cycle of RNA viruses; hence, it has been targeted in various viral infections, including the hepatitis C virus (HCV), the Zika virus (ZIKV), and coronaviruses (CoVs) [[16], [17], [18], [19], [20], [21], [22], [23], [24]]. The active RdRp site is highly conserved, with two successive and surface-accessible aspartates in a beta-turn structure. [[25], [26], [27]].
In this study, the SARS-CoV-2 RdRp model was built using the SARS RdRp solved structures from the NCBI protein data bank [28]. After model validation, molecular docking was performed to test several direct-acting antiviral (DAA) drugs against SARS-CoV-2 RdRp, including 5 FDA-approved medications used to treat HCV, the human immunodeficiency virus (HIV), and the Ebola virus; 13 compounds in clinical trials against HCV; and two negative control compounds, in addition to the physiological nucleotides GTP, CTP, UTP, and ATP. The results were promising and suggest possible inhibition from the currently available therapeutics against the newly emerged coronavirus [29].
2. Materials and methods
2.1. Sequence alignment and modeling
The newly-emerged SARS-CoV-2 nucleotide gene (NC_045512.2) was retrieved from the National Center for Biotechnology Information (NCBI) nucleotide database [30]. A homology model for the SARS-CoV-2 RdRp was built using the Swiss Model web server [31]. We used the SARS HCoV solved structure (PDB ID: 6NUR, chain A) as a template for building our model since it was the most sequelogous solved structure (97.08% sequence identity) to SARS-CoV-2 RdRp. 6NUR is a SARS HCoV non-structural protein 12 (nsp12) solved structure (cryo-electron microscopy) with 3.1 Å resolution, which was deposited in the protein data bank in 2019 [32]. Two web servers were used to examine the model: the MolProbity web server of Duke University, and the Structure Analysis and Verification Server (SAVES) of the University of California, Los Angeles (UCLA) [33,34]. Various types of software were used to judge the validity of the model, such as PROCHECK [35], Verify 3D [36], PROVE [37], and ERRAT [38], in addition to the Ramachandran plot of the MolProbity web server. After validation, the model was optimized (classical mechanical, using the MM3 force field) with assistance from the computational chemistry workspace SCIGRESS of Fujitsu [20,22,39,40]. Model minimization was performed after the addition of missed hydrogen atoms to prepare for the docking study [41].
2.2. Molecular docking
AutoDock Vina software was utilized in all of the docking experiments, with the optimized SARS-CoV-2 RdRp model as the docking target [42]. In addition, SARS HCoV RdRp (PDB ID: 6NUR) and hepatitis C virus (HCV) non-structural protein 5B (NS5B) RdRp (PDB ID: 2XI3) were used as docking targets for comparison. A total of 24 compounds were tested against SARS-CoV-2, SARS HCoV, and HCV NS5B RdRps, namely, the four physiological nucleotides (GTP, UTP, CTP, and ATP), five approved drugs against different viral RdRps (Galidesivir, Remdesivir, Tenofovir, Sofosbuvir, and Ribavirin), 13 compounds currently in clinical trials against HCV NS5B RdRp (Uprifosbuvir, Setrobuvir, Balaprevir, MK0608, R7128, IDX-184, 2′C-methylcytidine, BMS-986094, YAK, PSI-6130, PSI-6206, R1479, and Valopectibine), and two negative control compounds that did not have affinity toward RdRp (Cinnamaldehyde and Thymoquinone). All of the compounds were prepared to be optimized in their active forms in physiological conditions [27].
After docking, the structures were examined using the Protein-Ligand Interaction Profiler (PLIP) web server (Technical University of Dresden) [43].
3. Results and discussion
3.1. SARS-CoV-2 RdRp modeling
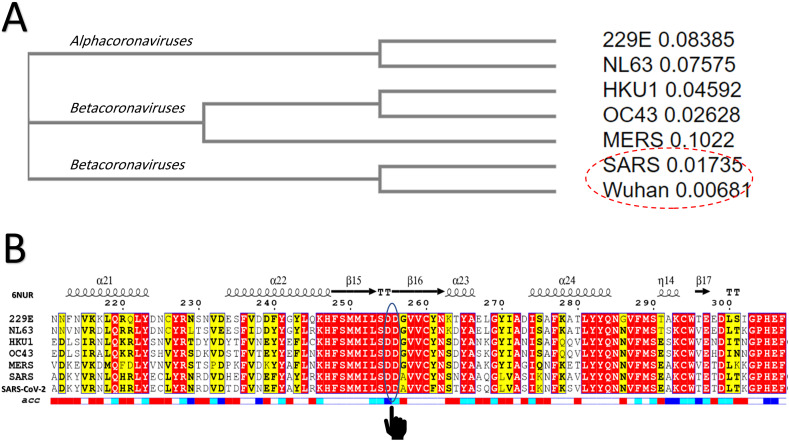
Based on the sequence identity between the SARS-CoV-2 and SARS RdRps, the Swiss model constructed a valid, high-quality model. Fig. 1(A) shows the cladogram representation of the phylogenetic tree for the RdRps of the seven human coronaviruses (229E, NL63, HKU1, OC43, MERS, SARS, and SARS-CoV-2). The distances between each sequence are indicated after the strain name. As shown in the cladogram, the SARS RdRp is the closest strain to the newly emerged SARS-CoV-2 RdRp. The multiple sequence alignment (MSA) of the RdRps from the different strains of HCoV is shown in Fig. 1(B). The SARS HCoV secondary structure (PDB ID: 6NUR) is shown at the top of the MSA, while its water accessibility is found at the bottom (highly accessible residues are in blue, while buried residues are white). The active site residues (D255 and D256) are circled and marked in the MSA. As implied from the MSA, the active site is highly conserved and water accessible in all 7 strains of the human coronaviruses (the red highlighting indicates identical residues, while the yellow highlighting represents conserved residues). The region surrounding the D255 and D256 residues is also conserved and mostly surface accessible in all HCoVs.
Fig. 1.
(A) Phylogenetic analysis of the RdRps of 7 HCoVs (229E, NL63, HKU1, OC43, MERS, SARS, and Wuhan SARS-CoV-2) (cladogram); (B) Multiple sequence alignment of the HCoV RdRp sequences utilizing the Clustal Omega web server and represented by ESpript. Red highlights indicate identical residues while yellow-highlighted residues are less conserved. Secondary structures are described at the top of the MSA for SARS RdRp (PDB ID: 6NUR, chain A), while the surface accessibility is shown at the bottom (blue: highly accessible; white: buried). Active site aspartates (D255 and D256) are marked on the MSA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For SARS-CoV-2 RdRp, the percent sequence identity values against the SARS, MERS, OC43, NL63, 229E, and HKU1 HCoV strains were found to be 90.18%, 56.76%, 55.07%, 48.79%, 48.55%, and 48.16%, respectively. Therefore, SARS HCoV RdRp was determined to be the sequelogous strain to SARS-CoV-2 RdRp, from which we could then build a reliable model. The complete nucleotide genome for SARS-CoV-2 was determined to have sequence identities of 89.12% and 82.34% with bat SARS-like coronavirus isolate bat-SL-CoVZC45 and SARS coronavirus ZS-C, respectively.
The SARS-CoV-2 RdRp model (801 residues) was generated by homology modeling using the Swiss Model web server. The SARS HCoV RdRp (PDB ID: 6NUR, chain A) was employed as a template. The model exhibited a very high (97.08%) sequence identity to the template, suggesting that an excellent model was obtained. Testing of the model validity was mediated by the Ramachandran plot (100% of the residues in the allowed regions, 97.5% in the most favored region), Verify-3D (89.9% of the residues had average 3D-1D scores ≤0.2), and ERRAT (overall quality factor of 95.9%).
3.2. Drugs binding to SARS-CoV-2 RdRp
Prior to testing the ligands against SARS-CoV-2 RdRp, the structures of the small molecules were ensured to be in the optimized active (triphosphate) form. Optimization was performed using the classical MM3 force field, followed by the semi-empirical PM6 force field. After transition state checks, further optimization was performed using the B3LYP functional of density functional theory (DFT) quantum mechanics [41,[44], [45], [46]].
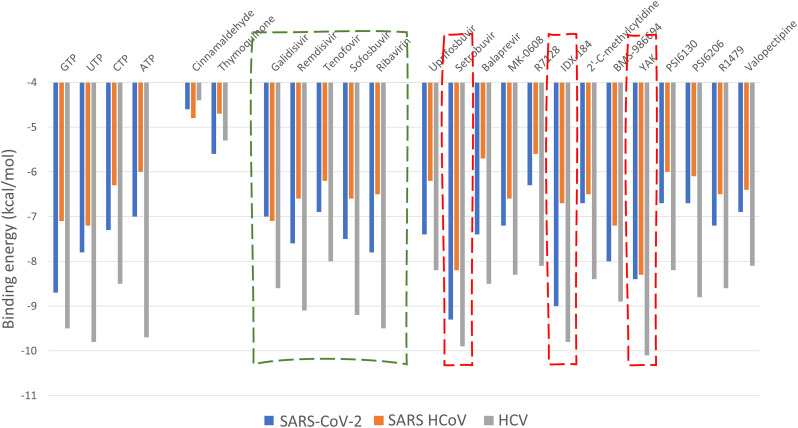
During all of the docking experiments, the active site aspartates (D255 and D256 in the SARS-CoV-2 and SARS RdRp, and D318 and D319 in the HCV NS5B RdRp) were treated as flexible, and the ligands were also flexible. A grid box (30 Å × 30 Å × 30 Å) centered at (142, 139, 150) Å, (141, 139, 149) Å, and (11, 6, 13) Å, for the SARS-CoV-2 RdRp, SARS RdRp, and HCV NS5B RdRp, respectively, was used in the docking experiments by utilizing the AutoDock tools [47]. Fig. 2 shows the docking score values for the SARS-CoV-2 (blue columns), SARS HCoVs (orange columns), and HCV NS5B RdRps (gray columns). The physiological compounds (GTP, UTP, CTP, and ATP) exhibited binding energies for SARS-CoV-2 between −7 (ATP) and −8.7 (GTP) kcal/mol. The two negative control compounds (Cinnamaldehyde and Thymoquinone) displayed low binding energies to all RdRps (−4.4 to −5.6 kcal/mol). The five approved drugs (Galidesivir, Remdesivir, Tenofovir, Sofosbuvir, and Ribavirin) surrounded by the dashed-green box in Fig. 2 were also able to bind the SARS-CoV-2 RdRp, with binding energies of −7.0, −7.6, −6.9, −7.5, and −7.8 kcal/mol, respectively. These drugs were able to bind to the new coronavirus strain RdRp tightly and hence may contradict the polymerase function. In addition, these drugs are potential candidates for inhibiting the RdRps of HCV NS5B (−8.0 to −9.5 kcal/mol) and SARS (−6.2 to −7.1 kcal/mol). Other compounds that are currently in clinical trials can bind to SARS-CoV-2 RdRp, with some showing promising results. Setrobuvir, IDX-184, and YAK (surrounded by red-dashed rectangles in Fig. 2) exhibit firm binding to all of the RdRps (−9.3, −9.0, and −8.4 kcal/mol, respectively for SARS-CoV-2 RdRp). The binding energy values against RdRp for these compounds are better than the native nucleotides. IDX-184 is a guanosine derivative, and it competes for GTP binding with slightly better binding (−9.0 kcal/mol) than GTP (−8.7 kcal/mol). Further analysis of the docking complexes is required to unravel their binding modes with the SARS-CoV-2 RdRp.
Fig. 2.
Bar graph representing the binding energies (in kcal/mol) calculated by AutoDock Vina software for GTP, UTP, CTP, ATP, Cinnamaldehyde, Thymoquinone, Galidesivir, Remdesivir, Tenofovir, Sofosbuvir, Ribavirin, Uprifosbuvir, Setrobuvir, Balaprevir, MK0608, R7128, IDX-184, 2′C-methylcytidine, BMS-986094, YAK, PSI-6130, PSI-6206, R1479, and Valopectibine against SARS-CoV-2 RdRp (blue), SARS HCoV RdRp (orange), and HCV NS5B RdRp (gray). The dashed-red rectangles mark the best three compounds. The dashed green box marks the five drugs (approved by FDA against HCV, EBOV, and HIV). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
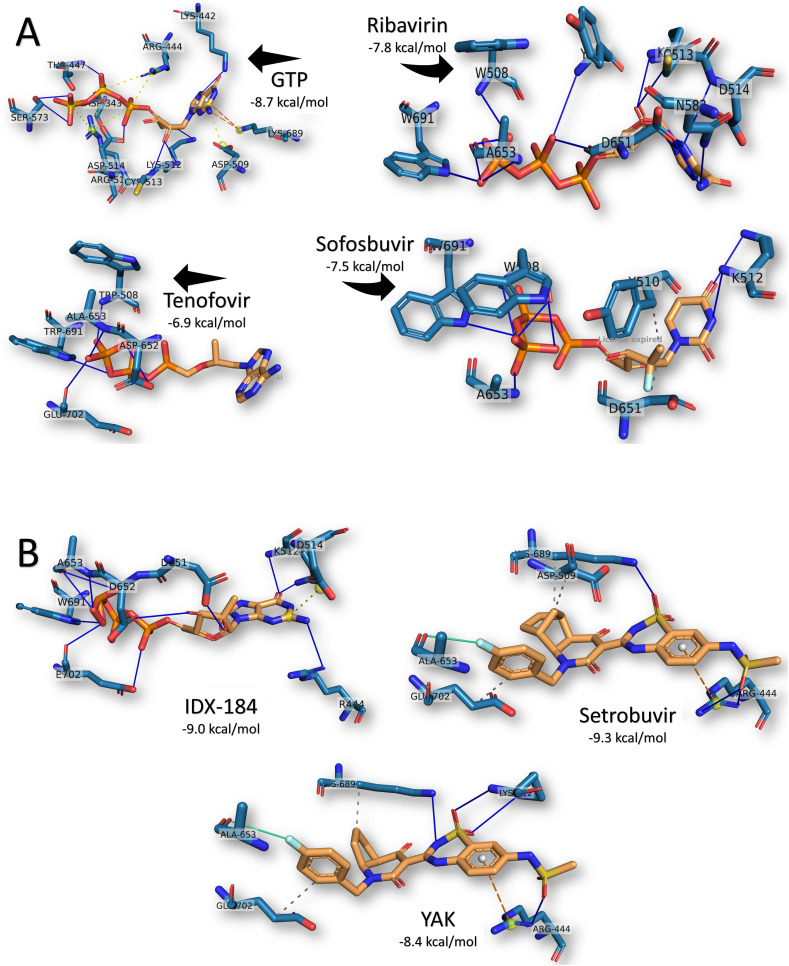
To investigate the possible reasons for the differences in the binding energies, we examined the formed complexes with help from the PLIP web server. Fig. 3(A) shows the formed interactions between the ligands GTP, Ribavirin, Sofosbuvir, and Tenofovir against SARS-CoV-2 RdRp after docking. GTP was found to form 8H-bonds with RdRp residues D343, K442, T447, K512 (2), C513, D514, and S573. Additionally, GTP built six salt bridges with R444 (2), D509, and R515 (3) and two π-cation interaction with K689 from the SARS-CoV-2 RdRp. This vast number of interactions gives the complex its stability with the −8.7 kcal/mol binding energy. This is comparable to the GTP-derivative IDX-184, shown in Fig. 3(B), in which 10H-bonds were formed with the SARS-CoV-2 RdRp residues R444, K512 (2), D651, D652, A653 (3), W691, and E702. On the other hand, two metal interactions were formed between IDX-184 and the active site residue D652 of the RdRp, while only one salt bridge built with D514.
Fig. 3.
Interactions established after docking the (A) GTP, Ribavirin, Sofosbuvir, and Tenofovir and (B) IDX-184, Setrobuvir, and YAK against SARS-CoV-2 RdRp. Drugs are in orange, while cyan lines represent the active protein site pockets. Solid blue lines depict H-bonds, while hydrophobic interactions are gray dashed lines. Additionally, salt bridges, π-cation stacking, and halogen contacts are represented by yellow spheres connected by black dashed lines, orange dashed lines, and cyan lines, respectively. RdRp residues are labeled, and the docking scores are listed under each complex. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For the approved drug Ribavirin, the only interactions established upon docking were the 13H-bonds with W508, Y510, K512, C513, D514, N582, D651 (3), A653 (3), and W691 of the SARS-CoV-2 RdRp. The same pattern was found for Tenofovir, but with a reduced number of H-bonds (5H-bonds with W508, D652, A653, W691, and E702), which was reflected in the binding energy values (−7.8 and −6.9 kcal/mol for Ribavirin and Tenofovir, respectively). On the other hand, Sofosbuvir formed 7H-bonds (W508 (3), K512 (2), A653, and W691) and two hydrophobic contacts (Y510 and D651) with the SARS-CoV-2 RdRp.
Based on the binding energy, the best compounds were discovered to be Setrobuvir (−9.3 kcal/mol), IDX-184 (−9.0 kcal/mol), and YAK (−8.4 kcal/mol). As previously mentioned, IDX-184 exhibited the same interaction pattern as its parent nucleotide (GTP) in binding the RdRp. On the other hand, both Setrobuvir and YAK formed H-bonds, hydrophobic contacts, π-cation contacts, and halogen interactions. Setrobuvir formed 3H-bonds (R444 (2) and K689), three hydrophobic contacts (D509, K689, and E702), a π-cation interaction with R444, and a halogen interaction with A653. YAK had 4H-bonds (R444, K512 (2), and K689), two hydrophobic contacts (K689 and E702), a π-cation interaction, and a halogen interaction with the same residues of Setrobuvir, as seen in Fig. 3(B).
In summary, the five approved drugs (Galidesivir, Remdesivir, Tenofovir, Sofosbuvir, and Ribavirin) can bind to SARS-CoV-2 RdRp, with binding energies comparable to those of native nucleotides (see the graphical abstract). Moreover, the compounds IDX-184, Setrobuvir, and YAK can tightly bind to the newly-emerged coronavirus RdRp, hence contradicting the function of the proteins leading to viral eradication. Additionally, IDX-184 can be used as a seed for new compounds specific against SARS-CoV-2 RdRp active site. Further optimization of the compounds using the high-quality model of the SARS-CoV-2 RdRp may result in a perfect compound able to stop the newly-emerged infection.
4. Conclusions
On 30 January 2020, the WHO declared the COVID-19 pandemic as a Public Health Emergency of International Concern (PHEIC). The newly emerged coronavirus in Wuhan City, Hubei Province, China, is a health concern given that the last outbreaks of this type of virus (SARS and MERS) in the years 2002 and 2012, respectively, each left approximately 800 dead and thousands hospitalized. The present study attempted to test and suggest possible inhibitor drugs, either currently on the market or in clinical trials, to stop the infection immediately. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir showed promising results for use against the newly emerged strain of coronavirus. Besides, the IDX-184, Setrobuvir, and YAK compounds exhibited excellent results for binding SARS-CoV-2 RdRp. We suggest utilizing GTP as a seed to obtain specific inhibitors against SARS-CoV-2 RdRp using its high-quality model.
Acknowledgments
Acknowledgment
Mrs. Kauther M Shaltoot is appreciated for her kind help and support; without her, this work was not possible. Prof. Dr. Wael Elshemey is appreciated for guidance and performing the docking calculations on his computational facility. This work is done during the Junior associate award granted to the author for the period 2016–2021 of the Abdus Salam International Center for Theoretical Physics (ICTP), Trieste, Italy. Additionally, the author wants to thank LetPub for the free language editing of the manuscript as it resembles a work on the newly emerged virus.
Data availability
The docking structures are available upon request from the corresponding author.
Declaration of competing interest
The author declares that there is no competing interest in this work.
References
- 1.Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. Journal of Travel Medicine. 2020;27(2) doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102433. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Organization WH . World Health Organization; 2020. Surveillance Case Definitions for Human Infection With Novel Coronavirus (nCoV): Interim Guidance v1, January 2020. [Google Scholar]
- 5.Organization WH . World Health Organization; 2020. Infection Prevention and Control During Health Care When Novel Coronavirus (nCoV) Infection Is Suspected: Interim Guidance, January 2020. [Google Scholar]
- 6.Organization WH . World Health Organization; 2020. Laboratory Testing of Human Suspected Cases of Novel Coronavirus (nCoV) Infection: Interim Guidance, 10 January 2020. [Google Scholar]
- 7.Parr J. Pneumonia in China: lack of information raises concerns among Hong Kong health workers. BMJ. 2020;368(56) doi: 10.1136/bmj.m56. Published 2020 Jan 8. [DOI] [PubMed] [Google Scholar]
- 8.Yang L. 2020. China Confirms Human-to-Human Transmission of Coronavirus. [Google Scholar]
- 9.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfiky A.A., Mahdy S.M., Elshemey W.M. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J. Med. Virol. 2017;89:1040–1047. doi: 10.1002/jmv.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020 doi: 10.1016/j.jinf.2020.02.026. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO . WHO; 2016. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [Google Scholar]
- 13.Báez-Santos Y.M., Mielech A.M., Deng X., Baker S., Mesecar A.D. Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 2014;88:12511–12527. doi: 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemida M.G., Alnaeem A. Some one health based control strategies for the Middle East respiratory syndrome coronavirus. One Health. 2019;8 doi: 10.1016/j.onehlt.2019.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Organization WH . World Health Organization; 2019. Clinical Management of Severe Acute Respiratory Infection When Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Infection Is Suspected: Interim Guidance. [Google Scholar]
- 16.Elfiky A.A. Zika viral polymerase inhibition using anti-HCV drugs both in market and under clinical trials. J. Med. Virol. 2016;88:2044–2051. doi: 10.1002/jmv.24678. [DOI] [PubMed] [Google Scholar]
- 17.Elfiky A.A. Zika virus: novel guanosine derivatives revealed strong binding and possible inhibition of the polymerase. Futur. Virol. 2017;12:721–728. [Google Scholar]
- 18.Elfiky A.A. Novel guanosine derivatives as anti-HCV NS5b polymerase: a QSAR and molecular docking study. Med. Chem. 2019;15:130–137. doi: 10.2174/1573406414666181015152511. [DOI] [PubMed] [Google Scholar]
- 19.Elfiky A.A., Elshemey W.M. IDX-184 is a superior HCV direct-acting antiviral drug: a QSAR study. Med. Chem. Res. 2016;25:1005–1008. doi: 10.1007/s00044-016-1533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elfiky A.A., Elshemey W.M. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J. Med. Virol. 2018;90:13–18. doi: 10.1002/jmv.24934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elfiky A.A., Elshemey W.M., Gawad W.A., Desoky O.S. Molecular modeling comparison of the performance of NS5b polymerase inhibitor (PSI-7977) on prevalent HCV genotypes. Protein J. 2013;32:75–80. doi: 10.1007/s10930-013-9462-9. [DOI] [PubMed] [Google Scholar]
- 22.Elfiky A.A., Ismail A. Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sci. 2019;238 doi: 10.1016/j.lfs.2019.116958. [DOI] [PubMed] [Google Scholar]
- 23.Elfiky A.A., Ismail A.M. Molecular modeling and docking revealed superiority of IDX-184 as HCV polymerase inhibitor. Futur. Virol. 2017;12:339–347. [Google Scholar]
- 24.Ganesan A., Barakat K. Applications of computer-aided approaches in the development of hepatitis C antiviral agents. Expert Opin. Drug Discovery. 2017;12:407–425. doi: 10.1080/17460441.2017.1291628. [DOI] [PubMed] [Google Scholar]
- 25.Doublie S., Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 26.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elfiky A.A., Ismail A.M. Molecular docking revealed the binding of nucleotide/side inhibitors to Zika viral polymerase solved structures. SAR QSAR Environ. Res. 2018;29:409–418. doi: 10.1080/1062936X.2018.1454981. [DOI] [PubMed] [Google Scholar]
- 28.Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 29.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.NCBI National center of biotechnology informatics (NCBI) database website. 2020. http://www.ncbi.nlm.nih.gov/
- 31.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SAVES . 2020. Structural Analysis and Verification Server Website. [Google Scholar]
- 34.Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski R.A., Rullmann J.A.C., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg D., Lüthy R., Bowie J.U. Methods in Enzymology. Elsevier; 1997. VERIFY3D: assessment of protein models with three-dimensional profiles; pp. 396–404. [DOI] [PubMed] [Google Scholar]
- 37.Joan Pontius JraSJW. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 1996;264:121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- 38.Hooft R.W., Vriend G., Sander C., Abola E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 39.Elfiky AA. The antiviral Sofosbuvir against mucormycosis: an in silico perspective. Future Virology. 2019;14(11):739–744. 0:null. [Google Scholar]
- 40.Summers K.L., Mahrok A.K., Dryden M.D., Stillman M.J. Structural properties of metal-free apometallothioneins. Biochem. Biophys. Res. Commun. 2012;425:485–492. doi: 10.1016/j.bbrc.2012.07.141. [DOI] [PubMed] [Google Scholar]
- 41.Lii J.H., Allinger N.L. Molecular mechanics. The MM3 force field for hydrocarbons. 3. The van der Waals’ potentials and crystal data for aliphatic and aromatic hydrocarbons. J. Am. Chem. Soc. 1989;111:8576–8582. [Google Scholar]
- 42.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 45.Leach A. 2nd edition. Prentice Hall; 2001. Molecular Modelling: Principles and Applications. [Google Scholar]
- 46.Stewart J.J.P. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J. Mol. Model. 2007;13:1173–1213. doi: 10.1007/s00894-007-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The docking structures are available upon request from the corresponding author.