Abstract
In this study, the transcriptional response of grouper to Singapore grouper iridovirus (SGIV) stimulation was characterized using RNA sequencing. Transcriptome sequencing of three test groups in the grouper was performed using the Illumina MiSeq platform. The three test groups were a control group, which was injected with PBS buffer; a high-susceptible (HS) group, which died shortly after the SGIV injection; and a high-resistance (HR) group, which survived the SGIV injection. In total, 38,253 unigenes were generated. When the HS group was compared with the control group, 885 unigenes were upregulated and 487 unigenes were downregulated. When the HR and control groups were compared, 1114 unigenes were upregulated and 420 were downregulated, and when the HR and HS groups were compared, 1010 unigenes were upregulated and 375 were downregulated. In the KEGG analysis, two immune-related pathways, the p53 and peroxisome proliferator-activated receptor pathways, were detected with highly significant enrichment. In addition, 7465 microsatellites and 22,1569 candidate single nucleotide polymorphisms were identified from our transcriptome data. The results suggested several pathways that are associated with traits of disease susceptibility or disease resistance, and provided extensive information about novel gene sequences, gene expression profiles, and genetic markers. This may contribute to vaccine research and a breeding program against SGIV infection in grouper.
Keywords: Orange-spotted grouper, Singapore grouper iridovirus (SGIV), Transcriptome, Disease resistance, Signaling pathway
Highlights
-
•
This study suggested several pathways associated with traits of disease susceptibility or disease resistance in grouper.
-
•
This study offered information about novel gene sequences, gene expression profiles, and genetic markers in grouper.
-
•
This may contribute to vaccine research and a breeding program against SGIV infection in grouper.
1. Introduction
The orange-spotted grouper (Epinephelus coioides) is one of the most important economical marine aquaculture species in many Asia countries. Moreover, because of its high nutritional properties and appealing taste, the grouper possesses a high market value in China. However, in recent years, along with the fast development of intensive large-scale aquaculture, the incidence of disease outbreaks has increased dramatically. Bacterial and viral infectious diseases have caused serious damage to the grouper aquaculture industry. Singapore grouper iridovirus (SGIV), a novel member of the Iridovirus pathogens, can cause >90% mortality in cultured grouper (Qin et al., 2003). Moreover, surviving groupers harboring these pathogens can transmit them to other fish horizontally in the same water environment, leading to more serious economic losses in grouper aquaculture. To date, the molecular mechanisms of SGIV pathogenesis and virus–host interactions are still largely unknown, which has been preventing the development of effective anti-viral strategies.
RNA-sequencing (RNA-Seq) using next-generation sequencing is one of the most useful methods to survey the character of a transcriptome because it offers a great deal of data on gene expression. Recently, transcriptome profiling using next-generation sequencing technologies have provided new insights into immune responses against various pathogenic infections in many aquaculture animals, such as the zebrafish (Danio rerio) (Zhang et al., 2013), common carp (Cyprinus carpio) (Li et al., 2015), large yellow croaker (Pseudosciaena crocea) (Mu et al., 2014), turbot (Scophthalmus maximus) (Pereiro et al., 2012), and Japanese sea bass (Lateolabrax japonicus) (Xiang et al., 2010). These studies used a high-throughput sequencing approach to identify numerous novel genes and expression profiles relating to the immune response.
In our previous study, we used 454 pyrosequencing technology to analyze the difference in the transcriptomes between the control and SGIV group (Huang et al., 2011a,b). The study provided important information on the host responses to viral infection. For further understanding of the immune mechanism and disease resistance mechanism of grouper to SGIV, we performed an analysis of three transcriptomes in this study: a control group, grouper that were highly susceptible (HS group) to SGIV and grouper that were highly resistant (HR group). These databases suggested several candidate pathways and genes that were associated with traits of disease susceptibility or disease, as well as offering extensive information about novel gene sequences, gene expression profiles, and genetic markers. The result of this study may contribute to vaccine research and breeding programs for resistance against SGIV infection in groupers.
2. Materials and methods
2.1. Experimental fish and immune challenge
Orange-spotted groupers used in the experiment were reared at an aquaculture farm in Huizhou City, Guangdong Province. A total of 300 randomly selected fish (weight: 50 ± 10 g) were acclimated for one week in filtered seawater with constant aeration (salinity, ∼30‰; temperature, ∼25 °C; pH, ∼7.9). They were fed with commercial grouper diet three times a day, and half of the seawater was changed every day to maintain quality. Before the injection experiment, genomic DNA extracted from the immune tissues (the mixture of liver, spleen, and head-kidney tissues) of three randomly selected individuals were amplified with specific primers of SGIV genes to ensure that those samples were not infected.
SGIV was originally isolated from diseased grouper, and SGIV propagation was performed as described previously (Hegde et al., 2002; Qin et al., 2003).
Based on our previous studies (Yang et al., 2016a, 2016b), the pathogen challenge experiment was implemented according to the following steps: 240 fish were intraperitoneally injected with 100 μL SGIV suspension (Concentration: 1 × 106 TCID50/mL), and another 30 fish were intraperitoneally injected with 100 μL saline (concentration: 0.9%) as the control group. After the treatments, all the samples were sent to seawater tanks and then reared for 2 weeks. The mortality data in all groups were obtained daily over 2 weeks.
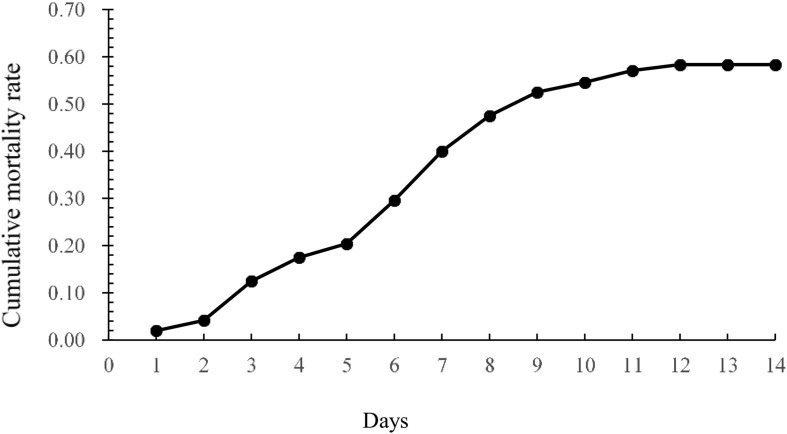
According to the mortality data, 30 fish died during the first 72 h and were considered as the HS group, and 100 fish were still alive at the end of the challenge experiment; these were considered to be the HR group. The cumulative mortality rate during the injection test is shown in Fig. 1 . During the whole period, no fish died in the control group.
Fig. 1.
The cumulative mortality rate during the injection test.
From a total of nine fish, three samples for each group were randomly selected. For each fish, total RNA from liver, spleen, and head-kidney was extracted separately and mixed as one RNA pool. Then, nine RNA pools were used for transcriptome library construction.
2.2. RNA extraction and library preparation for transcriptome sequencing
Total RNA was extracted from the immune tissues using Trizol Reagent (Takara, Tokyo, Japan) according to the manufacturer's instructions. The RNA degradation and contamination of each sample was monitored on 1.5% agarose gels. RNA concentration and integrity were further measured using a Qubit RNA Assay Kit in a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA), separately. Only RNA samples of high quality were used for constructing the cDNA libraries.
One RNA pool contained a mixture of RNA from liver, spleen, and kidney from one fish. Approximately 1.5 μg RNA was used as input material; 0.5 μg RNA per tissue (liver, spleen, or kidney). Then, nine RNA pools, three for the control group, three for the HS group, and three for the HR group, were produced and sequenced by the Sagene Biotechnology Company (Guangzhou, China) using the Illumina Genome Analyser II platform.
Briefly, mRNA was isolated from total RNA using poly-T oligo-attached magnetic beads, and then the purified mRNA was cleaved into small fragments using divalent cations, under elevated temperature. First-strand cDNA was synthesized with a random hexamer primer and M-MuLV Reverse Transcriptase. The second-strand cDNA was then synthesized and adapters were ligated to the products. The cDNA fragments of 150–200 bp were selected preferentially and purified. The size-selected and adaptor-ligated cDNA were then subjected to PCR amplification for 15 cycles. Finally, PCR products were purified and library quality was assessed on the Illumina HiSeq 2500 platform.
2.3. Transcriptome assembly and gene function annotation
Raw reads were filtered from total reads. The filter steps were: (1) remove reads containing the adapter, (2) remove reads containing ploy-N, and (3) remove low quality reads with a percentage of nucleotides with SQ ≤ 20, higher than 50%. Then reads with quality scores >20 were selected for downstream analyses. Transcriptome assembly of those selected reads was carried out using Trinity software (Grabherr et al., 2011), and the main parameters used in the assembly process were: min contig length, 100; min glue, 3; path reinforcement distance, 85; and min k-mer cov, 3. Then, the contigs were generated using Trinity software and were further processed by TGICL V2.1 software (Pertea et al., 2003) to remove redundant sequences and perform additional assembly. Finally, the resulting unique transcripts were termed unigenes.
Gene function was annotated based on four databases: Nr (all non-redundant GenBank CSampleB translations), KEGG (database resource for understanding high-level functions and utilities of the biological system), Swiss-Prot (a manually annotated and reviewed protein sequence database), and KOG/COG (The Clusters of Orthologous Groups of proteins [COGs] database). Firstly, unigenes were ‘blasted’ (processed) on these four databases using Blastx. Then, functional annotation information for these unigenes was obtained, based on similar protein sequences.
2.4. Quantification of gene expression levels and differential expression analysis
Gene expression levels in tissues (liver, spleen, and head-kidney tissues) from nine fish were estimated using RNA-Seq by Expectation-Maximization, based on fragments per kilobase, per million reads (Li and Dewey, 2011). Differential expression analysis was performed using the DESeq R package with default parameters (Wang et al., 2010). The p values (q-values) were adjusted using the Benjamini and Hochberg's approach (Avagyan, 2009). Genes with fold changes of expression level >2 and q-value <0.05 were assigned as differentially expressed.
2.5. GO and KEGG pathway enrichment analyses
Go enrichment analysis of the differentially expressed genes (DEGs) was conducted using the software GOseq V1.16.2 (Young et al., 2010). The statistical enrichment of DEGs in KEGG pathways was tested using the software KOBAS (Mao et al., 2005), with a p value < 0.05.
2.6. Simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) detection
SSRs from the transcriptome data were detected using Microsatellite (MISA, http://pgrc.ipk-gatersleben.de/misa/) (Thiel et al., 2003). The parameters, based on those used in similar studies, were adjusted to identify mono-, di-, tri-, tetra-, penta- and hexa-nucleotide motifs with a minimum of 8, 6, 5, 5, 4, and 4 repeats, respectively (Wei et al., 2014; Moraortiz et al., 2016). The candidate SNPs were analyzed with SOAP software using parameters set as -ut-QI-L90 (Li et al., 2009).
2.7. Gene expression validation using real-time PCR
To validate the gene expression level of RNA-seq, ten DEGs were randomly selected to perform real-time PCR. Specific primers were designed using Primer 5; the sequences of primers are listed in Table 1 . The β-actin gene was used as a reference gene. Primer specificity was ascertained using the following steps: PCR amplification, sequencing of PCR products, and Blast analysis in the NCBI database.
Table 1.
Genes and primer sequences used in the validation of gene expression.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Beta-action | TACGAGCTGCCTGACGGACA | GGCTGTGATCTCCTTCTGCA |
| Alcohol dehydrogenase 1 (ADH1) | CTGGAATGCGTCGGAAAT | GGGTCGGACAGCAACATC |
| Fatty aldehyde dehydrogenase (ALDH3A2) | GCCCTCTACGTCTTCTCG | AGCCCATACCACTGTTCC |
| Cathepsin F(CTSF) | GGGGTATTAGTGCTTTAGTTG | CACGCTGTGCTGATGGTC |
| 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase (CYP8B1) | TTTGCCTATGTGCCTTGT | TGGCAGGTTGATGGTGTA |
| N-oxide-forming 2 (FMO2) | AGTTGTTGAGCCTGGTGA | CAGAGGAAAGAGGGTGGT |
| N-oxide-forming 5 (FMO5) | TTTGTTCTCGCCAATCCA | GGGTCATCCGTCAGGTTT |
| Thyroid hormone receptor beta (THRB) | GGTCAGCAGGTATTTAGG | ATTCTTGGGTATGTCTTCA |
| Toll-like receptor 5 (TLR5) | ACTTCCGAGTTTAGATTAC | GCAGAGTCACAAGGGTAT |
| UDP-glucuronosyltransferase 1-1 (UGT1A1) | ACCCTCCCGTCAAAGAAT | TTGGAGCAGTGGAATGACGAAC |
| UDP-glucuronosyltransferase 2A1 (UGT2A1) | CTCTACCCAACATCTCACA | CTTCCTCTTCTACGCACT |
Another 18 fish samples were selected from test samples, six samples from the control group (collected on day14 after infection), six samples from the HS group (collected on day 14 after infection), and six samples from the HR group (collected on day three after infection). As described in the methods (Sections 2.1, 2.2), the RNA sample mix (RNA extracted from liver, spleen, and head-kidney) from each fish was obtained and reversed transcribed to cDNA, and used as a template in the validation experiments.
The cDNA used as a template for real-time PCR was synthesized using a cDNA Synthesis Kit (TOYOBO, Japan) following the manufacturer's instructions. Real-time PCR was performed on a Roche LightCycler 480 Real-time PCR system (Roche, Switzerland) using a SYBR Green qPCR Mix kit (Toyobo, Osaka, Japan). PCR was carried out in a total volume of 10 μL containing 5 μL of 2*SYBR Green PCR buffer, 0.5 μL of each primer (10 μM), 1 μL cDNA, and 3 μL RNase-free H2O.
Each reaction was performed in triplicate under the following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s. Gene expression level was compared and converted to fold differences by the 2−△△CT method (Livak and Schmittgen, 2001). The standard error of the mean (SEM) was used to report the data. One-way analysis of variance was performed using SPSS 21.0 software (SPSS Inc., Chicago, IL, USA). A P value < 0.05 was considered statistically significant.
3. Results
3.1. Statistics of transcriptome sequencing and assembly
Three cDNA libraries representing the control, HS, and HR groups were constructed. A total of 398,682,010 raw reads were generated from these three groups. The control group contained 132,430,556 clean reads, the HS group contained 132,430,556 clean reads, and the HR group contained 132,810,280 clean reads. The Q30 was higher than 96% and the GC content was ∼49% (Supplemental Table 1).
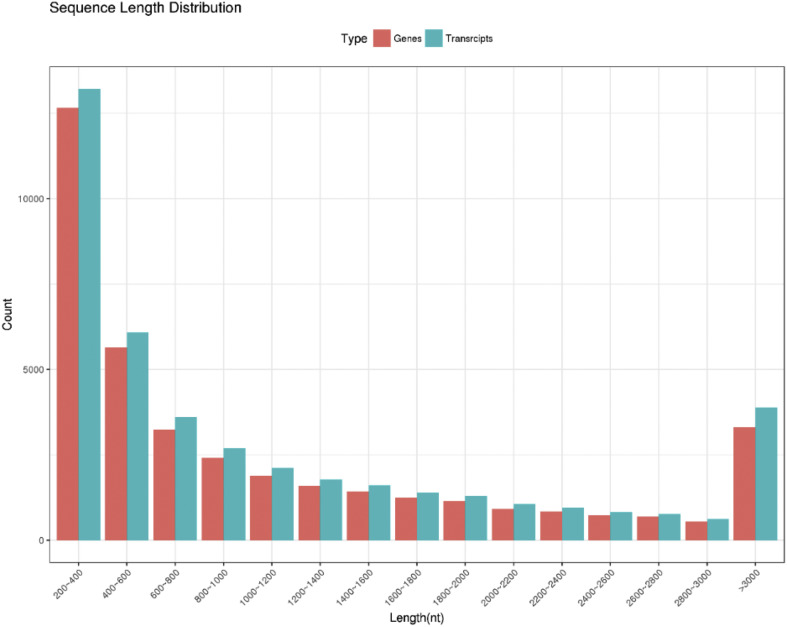
After discarding ambiguous nucleotides, short reads, and low-quality reads, clean reads were selected to be assembled into sequences. Then 38,253 unigenes and 41,866 transcripts were generated by using Trinity software (Supplemental Table 2). The length distribution of assembled unigenes is shown in Fig. 2 . The figure revealed that most of the unigenes ranged from 200 to 3000 bp, and ∼5% of unigenes were >3000 bp. These results demonstrated that the transcriptome data was high-quality, and the unigenes could be used for subsequent analysis. The sequence data of unigenes were deposited in the Sequence Read Archives (SRA) at NCBI under accession number SRP149880.
Fig. 2.
Length distribution of the assembled genes and transcripts.
3.2. Functional analysis of the transcriptome
To evaluate the function of the unigenes, 38,253 unigene sequences were aligned with seven public databases. The results showed that 23,497 (61.42%) unigenes were annotated in at least one database, and 10,801 genes (28.23%) were annotated in all four databases. The annotation success rate of unigenes was 23,467 in NR (61.34%), 13,548 in KEGG (35.41%), 19,550 in SwissProt (51.10%), and 15,543 in KOG (40.63%) (Supplemental Table 3).
3.3. Identification and analysis of DEGs
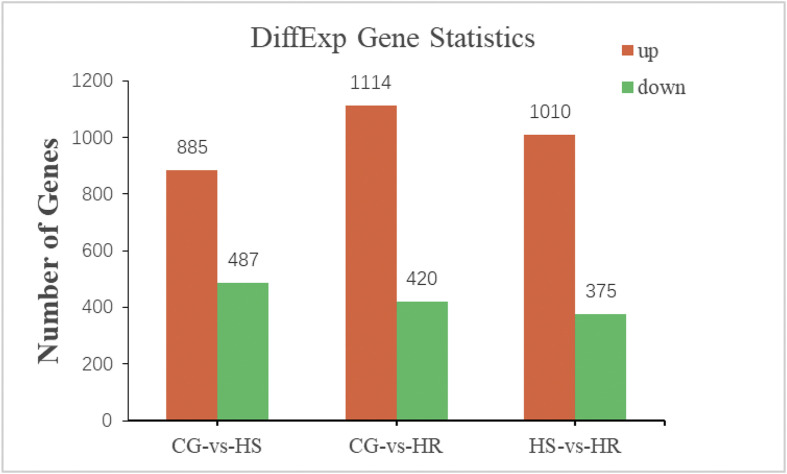
A false discovery rate) cutoff ≤0.05 and log2Ratio ≥1 was used as the threshold to select DEGs. When compared with the control group, the number of DEGs in the HR group (1,534) was higher than that in the HS group (1,372), and there were 1385 DEGs between the HR and HS groups (Fig. 3 ). In comparison with the unigene expression profile of the control group, 885 unigenes were upregulated and 487 unigenes were downregulated in the HS group, and 1114 unigenes were upregulated and 420 unigenes were downregulated in the HR group. When the HR group was compared with the HS group, there were 1010 upregulated and 375 downregulated genes in the HR group (Fig. 3). The details of changes of DEGs between different groups are listed in Supplemental Tables 4, 5, and 6.
Fig. 3.
Differentially expressed genes between groups.
To further analyze the DEGs between the SGIV-challenged and non-challenged individuals, the DEGs from the HS and control groups were further annotated with the GO and KEGG databases.
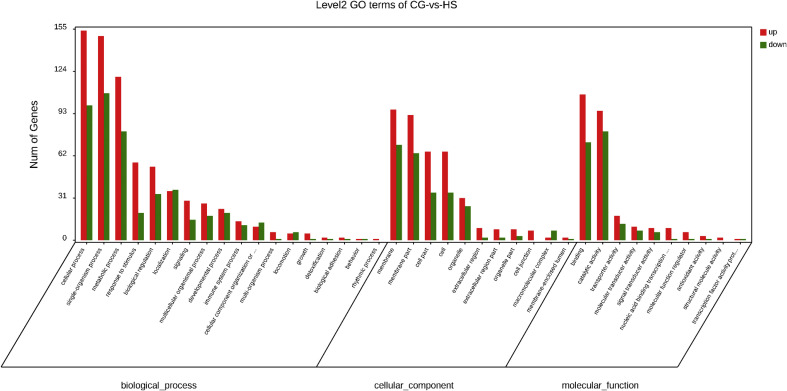
In the GO enrichment analysis between the HS and control group, all DEGs were classified into three GO categories: biological process, cellular component, and molecular function. The biological process was the most prevalent category, followed by cellular component, and molecular function. The biological process category included 18 subcategories, whereas cellular component and molecular function each contained 11 and 10 subcategories, respectively. Among the various categories of biological process, the frequency of the top three clusters were single-organism process (18.80%), cellular process (18.44%), and metabolic process (14.58%) (Fig. 4 ). In the GO enrichment analysis between the HR and control group and between the HS and HR group, the most prevalent categories and the top three clusters of the various categories were in accordance with the above results (Supplemental Figures 1 and 2).
Fig. 4.
Enriched GO terms in the differentially expressed genes (DEGs) between the control group and highly susceptible (HS) group.
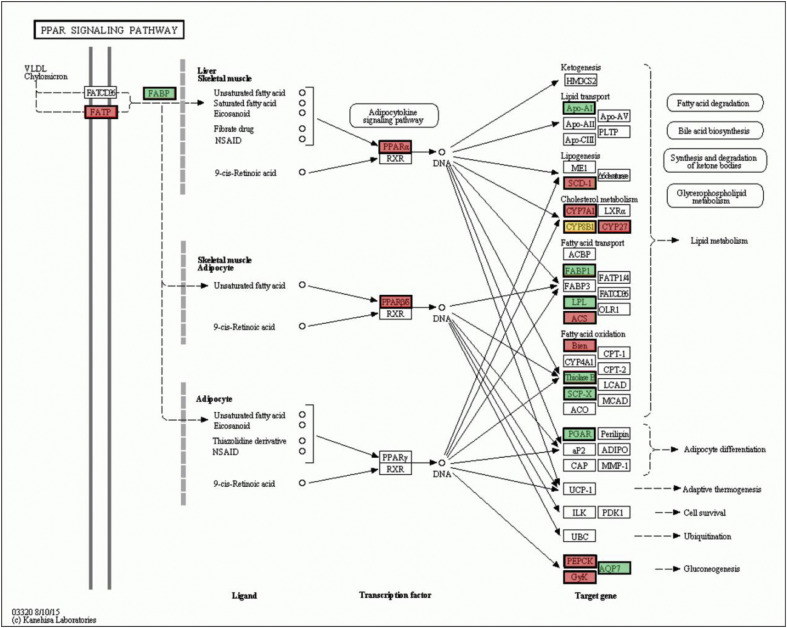
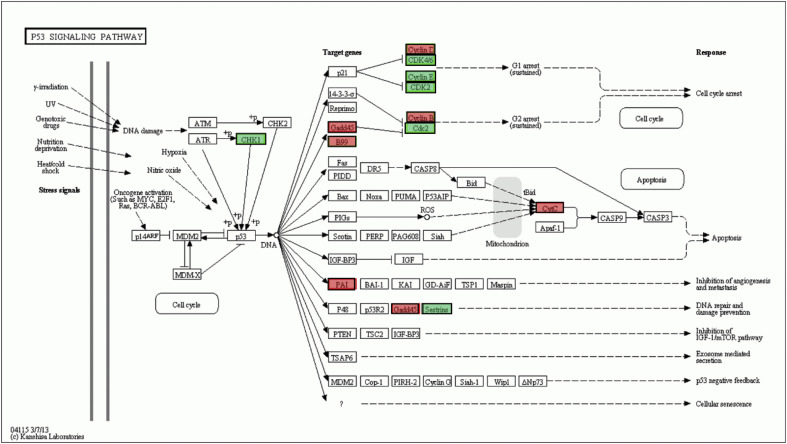
In the KEGG analysis of any pairwise comparison group, most KEGG pathways with highly significant enrichment were associated with metabolism, such as tyrosine metabolism, pyrimidine metabolism, and carbon metabolism. Besides these metabolism pathways, two immune-related pathways, the p53 pathway and the peroxisome proliferator-activated receptor (PPAR) pathway, were also detected with highly significant enrichment (Supplemental Tables 7, 8 and 9). When the HS and control groups were compared, there were nine genes enriched in the p53 pathway (P = 0.003), and also nine genes enriched in the PPAR pathway (P = 0.045) (Supplemental Table 7). Among the DEGs in the p53 pathway, Gadd45, CyclinD, and CyclinB were associated with cell cycle arrest, and among them, Gadd45 was upregulated, while CyclinD and CyclinB were downregulated; PIGs and Cytc genes, which are associated with apoptosis, were upregulated; PAI, which is associated with inhibition of angiogenesis and metastasis, was upregulated; and Gadd45 and Sestrins, which are associated with DNA repair and damage prevention, were also upregulated (Fig. 5 ). In the PPAR pathway, seven genes, PPAR-α, CYP7A1, LPL, ACS, PGAR, PEPCK, and GYK, were upregulated, and two genes, CYP8B1, Bien, were downregulated. Especially, the PPAR-α gene, a crucial regulatory gene in PPAR pathway, was upregulated (Fig. 6 ).
Fig. 5.
The enriched DGEs in the PPAR signaling pathway There were three pairwise comparison groups (CG vs HS, CG vs HR and HS vs HR) in our study, and there were three kinds of DEGs from the different comparison groups. And it is hard to show the downregulated or upregulated information in one figure. For instance, the A gene was included in both the DEGs of CG vs HS group, and the CG vs HR group, and it was upregulated in the HS group when comparing with CG and HS, while downregulated in the HR group when comparing the CG and HR. Hence, we only show the group information of enriched DEGs with colors: Green, DGSs existing in only one pairwise comparison group; Red, DEGs existing in any two pairwise comparison groups; Yellow, DGSs existing in all three pairwise comparison groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Enriched DGEs in the p53 signaling pathway The DGSs included in the CG vs HS groups, and CG vs HR groups are shown. Green, DGSs existing in only one pairwise comparison group; Red, DEGs existing in any two pairwise comparison groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
When the HR and control group were compared, there were 12 genes enriched in the p53 pathway with a P value of 0.004 (P < 0.05), and also 12 genes enriched in the PPAR pathway (P = 0.001) (Supplemental Table 8). Among the genes in the p53 pathway, the CHK1 gene, with a function associated with p53 phosphorylation, was downregulated; CyclinD, CDK4/6, CyclinE, CDK2, CyclinB, Cdc2, and B99, which are associated with cell cycle arrest, were down-regulated, while Gadd45, which is associated with both cell cycle arrest and DNA repair and damage prevention, was upregulated; the Cytc gene, which is associated with apoptosis, was upregulated; the PAI gene, which is associated with inhibition of angiogenesis and metastasis, was upregulated (Fig. 5). In the PPAR pathway, 11 genes, FATP, PPAR-α, Apo-AI, CYP7A1, CYP8B1, CYP27, PPAR-β, SCP-X, PEPCK, GyK, and AQP7, were upregulated, and only one gene, SCD-1, was downregulated. Interestingly, besides PPAR-α, PPAR-β was also upregulated (Fig. 6).
When the HR and HS group were compared, the PPAR pathway was significantly enriched (P = 0.001), while the p53 pathway was not (P = 0.97) (Supplemental Table 9). There were only two genes enriched in the p53 pathway, the Sestrins gene, which is associated with DNA repair and damage prevention, was downregulated and Cyclin G, which is associated with p53 negative feedback, was upregulated. In the PPAR pathway of the HR group, FATP, FABP, CYP8B1, CYP27, PPAR-β, FABP1, Bien, and Thiclase were upregulated, while SCD-1 and ACS were downregulated (Fig. 6).
3.4. SSR and SNP analysis
A total of 7465 microsatellites from 41,866 sequences were identified using Microsatellite (MISA, http://pgrc.ipk-gatersleben.de/misa/) (Thiel et al., 2003). The percentage of dinucleotide, trinucleotide, mononucleotide, pentanucleotide, and hexanucleotide SSRs was 64.97%, 46.70%, 10.71%, 3.09%, and 2.03%, respectively (Supplemental Fig. 3).
In addition, 72,853 candidate SNPs were identified from our transcriptome data. Among all SNPs, 50,538 were transitions and 35,020 were transversions. The top four mutation types were C-T, G-A, A-G, and T-C (Supplemental Fig. 4).
3.5. Validation of RNA sequencing via real-time PCR
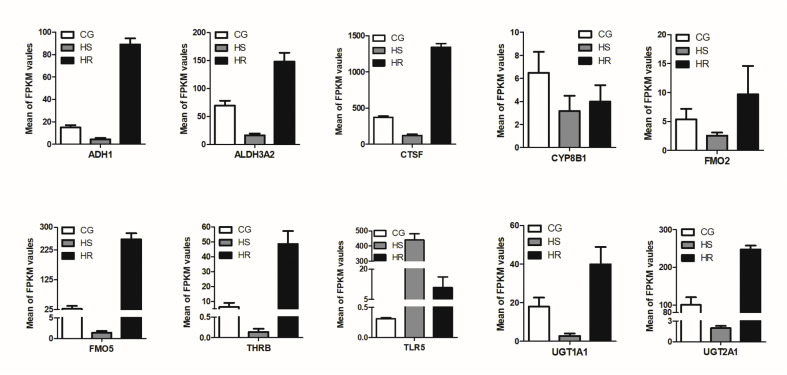
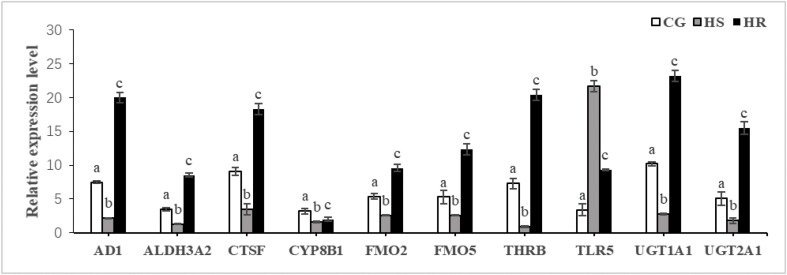
To validate the gene expression levels of RNA-Seq, real-time PCR was further conducted for 10 randomly selected genes: ADH1, ALDH3A2, CTSF, CYP8B1, FMO2, FMO5, THRB, TLR5, UGT1A1, and UGT2A1. The results showed that except for a slight difference in fold values between different groups, the expression patterns of these randomly selected genes were consistent with those from our transcriptome data (Fig. 7, Fig. 8 ). This suggested that our Illumina RNA-Seq data were reliable.
Fig. 7.
Expression levels of 10 randomly-selected DEGs in the transcriptome data CG, the control group; HS, high susceptibility group; HR, high-resistance group; FPKM, fragments per kilobase of transcript per million mapped reads.
Fig. 8.
Validation of differentially expressed genes (DEGs) by qPCR CG, the control group; HS, high susceptibility group; HR, high-resistance group. Different letters on the graph denote significant statistical difference (P < 0.05).
4. Discussion
Grouper is one of the most important economical aquaculture species in various Asian countries (Pierre et al., 2008). In recent years, due to high-intensive farming and climate change, bacterial and viral infections have spread more easily and widely, especially members of the Iridoviridae. SGIV, a member of the Iridoviruses, often leads to a highly lethal and serious systemic disease in groupers, causing tremendous economic losses (Qin et al., 2001; Gibson-Kueh et al., 2003). Therefore, an investigation on the molecular response to SGIV infection and SGIV disease-resistant is required to develop methods of prevention such as vaccines and disease-resistant breeding. In the present study, we performed a transcriptome analysis of the main immune tissue (spleen, liver, and head kidney) in control and SGIV susceptible and resistant orange-spotted grouper samples using an RNA-Seq platform, aiming to obtain more insights into the molecular mechanism of the grouper immune response to SGIV. We identified 38,253 unigenes and observed the DEGs related to immune responses. A large number of SSR and SNP markers in the transcripts were also identified, and these data provide a valuable genetic resource for the further development of molecular markers and genetic breeding studies.
For KEGG pathway analysis, all significantly enriched pathways were clustered into 18 KEGG classes. Ten classes belonged to metabolic systems, such as the metabolism of cofactors and vitamins, amino acid metabolism, lipid metabolism, carbohydrate metabolism, energy metabolism, xenobiotic biodegradation and metabolism, global and overview, biosynthesis of other secondary metabolites, nucleotide metabolism, and metabolism of other amino acids. Two classes belonged to organismal systems, such as the endocrine system and immune system. Three classes belonged to genetic information processing systems, such as translation folding, sorting and degradation, and replication and repair. Two classes belonged to cellular processing systems, such as cell growth and death, transport, and catabolism. One class belonged to environmental information processing systems, such as signaling molecules and interactions. These results indicated that SGIV not only impacted on metabolic and immune systems in the grouper, but it also affected the endocrine system, genetic information processing systems, cellular processing systems, and environmental information processing systems.
Among all the highly significant enrichment pathways, two pathways, the p53 pathway and the PPAR pathway, play important roles in the innate antiviral immune response in grouper. The p53 pathway is an important signal pathway that regulates the growth and development of cells, and also plays a role in modulating the cell cycle, cellular senescence, and apoptosis. In the immune system, apoptosis is essential for the maintenance of homeostasis. Several studies have shown that apoptotic cell death occurs in a wide range of viral infections (Clarke and Tyler, 2009; Kinpara et al., 2013), and this virus-induced apoptosis is thought to be related to the activation of the c-Jun N-terminal kinase, NF-κB, and p53 pathways (Huang et al., 2011a,b; Kinpara et al., 2013). In our previous study, we found that SGIV could induce typical apoptosis in teleost cells (Huang et al., 2011a,b). In the present study, the p53 pathway was enriched significantly in the two SGIV challenged groups, while the c-Jun N-terminal kinase and NF-κB pathways were not. We speculate that the p53 pathway may play a more important role in the SGIV-induced apoptosis than the other two pathways. However, the molecular mechanism of SGIV-induced apoptosis is still ambiguous, our DEG data associated with the p53 pathway may provide a reference for further research.
PPARs are a type of ligand-independent transactivation factor. There are three PPAR subtypes, PPAR-α, PPAR-β, and PPAR-γ, which belong to the superfamily of nuclear receptors. PPARs not only regulate lipid metabolism and glucose homeostasis but also play an important role in inflammatory control (Chinetti et al., 2000; Regnier and Sargis, 2014; Jenna et al., 2015).
In mammals, all PPARs can be activated by fatty acids. They are expressed in many tissues and mediate lipid catabolism by regulating the transcription of target genes encoding enzymes involved in peroxisomal and mitochondrial beta-oxidation of fatty acids (Kersten et al., 2001; Reddy and Hashimoto, 2001; Hansmannel et al., 2003; Mandard et al., 2004).
In addition, PPARs were reported to play a regulatory role in innate and adaptive immunity (Clark, 2002), which exerts an anti-inflammatory activity in various cell types by inhibiting the expression of some inflammatory genes, including the cytokine genes, AP-1, and NF-κB (Delerive et al., 2001). Following antigen infection, an increase of NF-κB expression may elicit an inflammatory response, while the PPARs could be responsible for regulating the host inflammatory responses and maintaining the immune homeostasis (Kelly et al., 2004). In mammalian macrophages, a high expression level of the PPAR-γ gene was detected and its expression was dramatically upregulated during inflammatory responses and induced by IL-4 and other immune-regulatory molecules (Ricote et al., 1998a, 1998b; Daynes and Jones, 2002). Furthermore, PPAR-γ could also directly inhibit the activity of other pro-inflammatory transcription factors, such as the activator protein-1 and NF-κB, in a ligand-dependent way, and thereby negatively regulate inflammatory responses (Ricote et al., 1998a, 1998b). In endothelial cells and hepatocytes, levels of inflammatory targets, such as vascular cell adhesion molecule-1 and serum amyloid A, were increased in the absence of PPAR-α, and the response to inflammatory stimuli was prolonged (Devchand et al., 1996; Ziouzenkova et al., 2003; Han et al., 2006). All these studies indicate that PPAR-α is very important in the immune system of an organism.
In teleosts, studies on PPARs are limited. PPAR sequences and tissue distribution were reported in several species such as Megalobrama amblycephala (Zhao et al., 2011; Li et al., 2013) and Salmo salar (Andersen et al., 2000). In Atlantic salmon, researchers found that the diversity of PPAR-γ alleles influenced susceptibility to disease (Sundvold et al., 2010). In orange-spotted grouper, studies indicated that PPAR-γ transcript expression showed a significant increase following Vibrio challenge (Luo et al., 2015). However, the potential role of teleost PPARs in the immune defense system is still not clear. The data of our study may provide references for further research of PPAR pathways in fish.
For validation tests, ten genes were randomly selected. Their functions are: (1) ADH1, which metabolizes a wide variety of substrates including ethanol, hydroxysteroids, and lipid peroxidation products; (2) ALDH3A2, which primarily metabolizes fatty aldehydes to fatty acids; (3) Cathepsin F, which is a member of the multigene family of lysosomal cysteine proteinases, that has regulatory roles in several homeostasis processes (Lee et al., 2013); (4) CYP8B1, which plays an important role in the synthesis of cholesterol and bile acids (Lorbek et al., 2012); (5) FMO2; (6) FMO5, which are members of the flavin-containing monooxygenase (FMO) gene family, which oxidize a variety of soft nucleophilic substrates (Mccombie et al., 1996); (7) THRB, which is a thyroid hormone receptor (THR) that mediates thyroid hormone action; (8) UGT1A1, which is an enzyme on the glucuronidation pathway; studies have indicated that polymorphisms in its promoter are associated with various diseases and drug responses (Shin et al., 2015); (9) TLR5. Which encodes a member of the toll-like receptor (TLR) family playing fundamental roles in pathogen recognition and activation of innate immune responses; and (10) UGT2A1, which encodes an enzyme in the glucuronidation pathway; it transforms small lipophilic molecules such as steroids and drugs into water-soluble, excretable metabolites. These genes will be useful in elucidating infection response mechanisms of groupers against SGIV infection.
The transcriptome data in our study provide valuable gene information data associated with infection susceptibility/resistance traits in the grouper. This information could be used for the development/identification of disease-resistant molecular markers or elucidation of molecular mechanisms involved in grouper susceptibility/resistance traits. The significantly enriched KEGG pathways also offer new insights into iridovirus-host interactions, e.g. how SGIV impacts grouper metabolic, immune and endocrine systems, how antiviral immune responses proceed, and how to contain cellular homeostasis during SGIV infection.
5. Conclusions
In this study, the transcriptional responses of grouper to SGIV stimulation were characterized using RNA sequencing. Three transcriptome databases, a control group, and groups highly susceptible or resistant to SGIV, were obtained and analyzed. In total, 38,254 unigenes were generated. By comparing the unigene expression profile of the control group, 885 unigenes were found to be upregulated and 487 unigenes were downregulated in the HS group, and 1114 unigenes were upregulated and 420 unigenes were downregulated in the HR group. When the HR and HS groups were compared, there were 1010 upregulated and 375 downregulated genes in the HR group. In the GO enrichment analysis; the most prevalent categories of the three pairwise comparison groups were biological processes, and the top three clusters of the various categories were single-organism processes, cellular processes, and metabolic processes. In KEGG analysis, two immune-related pathways, the p53 and PPAR pathways, had highly significant enrichment. In addition, 7465 microsatellites and 72,853 candidate SNPs were identified from our transcriptome data. The results of this study suggested several pathways that were associated with traits of disease susceptibility or disease resistance, as well as offering extensive information about novel gene sequences, gene expression profiles, and genetic markers. This research may contribute to vaccine research data and to resistant breeding programs against SGIV infection in grouper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 41706144, 41806151, 41606183, and 31330082], the National Key R&D Program of China [grant number 2017YFC1404504], and the China Agriculture Research System [grant number CARS-47-G16]. We thank the support of the Young Science and Technology Talents Training Fund of South China Agricultural University and the open fund of Guangxi Key Laboratory of Marine Environmental Science [GXKLHY18-03]. We also thank the Sagene Biotechnology Company (Guangzhou, China) for technical support and last, we thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dci.2018.09.003.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Andersen Ø., Eijsink V.G.H., Thomassen M. Multiple variants of the peroxisome proliferator-activated receptor ( PPAR ) γ are expressed in the liver of Atlantic salmon ( Salmo salar ) Gene. 2000;255:411–418. doi: 10.1016/s0378-1119(00)00350-4. [DOI] [PubMed] [Google Scholar]
- Avagyan V. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Fruchart J.C., Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- Clarke P., Tyler K.L. Apoptosis in animal models of virus-induced disease. Nat. Rev. Microbiol. 2009;7:144–155. doi: 10.1038/nrmicro2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.B. The role of PPARs in inflammation and immunity. J. Leukoc. Biol. 2002;71:388–400. [PubMed] [Google Scholar]
- Daynes R.A., Jones D.C. Emerging roles of PPARS in inflammation and immunity. Nat. Rev. Immunol. 2002;2:748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- Delerive P., Fruchart J.C., Staels B. Peroxisome proliferator-activated receptors in inflammation control. J. Endocrinol. 2001;169:453. doi: 10.1677/joe.0.1690453. [DOI] [PubMed] [Google Scholar]
- Devchand P.R., Keller H., Peters J.M., Vazquez M., Gonzalez F.J., Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Gibson-Kueh S., Netto P., Ngoh-Lim G.H., Chang S.F., Ho L.L., Qin Q.W. The pathology of systemic iridoviral disease in fish. J. Comp. Pathol. 2003;129:111–119. doi: 10.1016/s0021-9975(03)00010-0. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C.Y., Chiba T., Campbell J.S., Fausto N., Chaisson M., Orasanu G. Reciprocal and coordinate regulation of serum amyloid A versus apolipoprotein A-I and paraoxonase-1 by inflammation in murine hepatocytes. Arterioscler. Thromb. Vasc. Biol. 2006;26:1806. doi: 10.1161/01.ATV.0000227472.70734.ad. [DOI] [PubMed] [Google Scholar]
- Hansmannel F., Clemencet M.J.C.C., Osumi T., Latruffe N., Nicolas F.V. Functional characterization of a peroxisome proliferator response-element located in the intron 3 of rat peroxisomal thiolase B gene. Biochem. Biophys. Res. Commun. 2003;311:149–155. doi: 10.1016/j.bbrc.2003.09.185. [DOI] [PubMed] [Google Scholar]
- Hegde A., Chen C.L., Qin Q.W., Lam T.J., Sin Y.M. Characterization, pathogenicity and neutralization studies of a nervous necrosis virus isolated from grouper, Epinephelus tauvina, in Singapore. Aquaculture. 2002;213:55–72. [Google Scholar]
- Huang X., Huang Y., Ouyang Z., Xu L., Yang Y., Cui H. Singapore grouper iridovirus, a large DNA virus, induces nonapoptotic cell death by a cell type dependent fashion and evokes ERK signaling. Apoptosis Int. J. Program. Cell Death. 2011;16:831–845. doi: 10.1007/s10495-011-0616-y. [DOI] [PubMed] [Google Scholar]
- Huang Y., Huang X., Yan Y., Cai J., Ouyang Z., Cui H. Transcriptome analysis of orange-spotted grouper (Epinephelus coioides) spleen in response to Singapore grouper iridovirus. BMC Genom. 2011;12:556. doi: 10.1186/1471-2164-12-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenna C., Winter M.J., Anke L., Rob C., Owen S.F., Tyler C.R. Effects of the lipid regulating drug clofibric acid on PPARα-regulated gene transcript levels in common carp (Cyprinus carpio) at pharmacological and environmental exposure levels. Aquat. Toxicol. 2015;161:127–137. doi: 10.1016/j.aquatox.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D., Campbell J.I., King T.P., Grant G., Jansson E.A., Coutts A.G. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat. Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Kersten S., Mandard S., Escher P., Gonzalez F.J., Tafuri S., Desvergne B. The peroxisome proliferator-activated receptor alpha regulates amino acid metabolism. FASEB (Fed. Am. Soc. Exp. Biol.) J. 2001;15:1971–1978. doi: 10.1096/fj.01-0147com. [DOI] [PubMed] [Google Scholar]
- Kinpara S., Kijiyama M., Takamori A., Hasegawa A., Sasada A., Masuda T. Interferon-α (IFN-α) suppresses HTLV-1 gene expression and cell cycling, while IFN-α combined with zidovudin induces p53 signaling and apoptosis in HTLV-1-infected cells. Retrovirology. 2013;10:52. doi: 10.1186/1742-4690-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Lee Y.M., Yang H., Noh J.K., Kim H.C., Park C.J. Expression analysis of Cathepsin F during embryogenesis and early developmental stage in olive flounder (Paralichthys olivaceus) Dev. Reprod. 2013;17:221–229. doi: 10.12717/DR.2013.17.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhao Y., Liu Z., Gao C., Yan F., Liu B. De novo assembly and characterization of the spleen transcriptome of common carp (Cyprinus carpio) using Illumina paired-end sequencing. Fish Shellfish Immunol. 2015;44:420–429. doi: 10.1016/j.fsi.2015.03.014. [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Fang X., Yang H., Wang J., Kristiansen K. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Gul Y., Wang W., Qian X., Zhao Y. PPARγ, an important gene related to lipid metabolism and immunity in Megalobrama amblycephala: cloning, characterization and transcription analysis by GeNorm. Gene. 2013;512:321–330. doi: 10.1016/j.gene.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorbek G., Lewinska M., Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids. FEBS J. 2012;279:1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- Luo S., Huang Y., Xie F., Huang X., Liu Y., Wang W. Molecular cloning, characterization and expression analysis of PPAR gamma in the orange-spotted grouper (Epinephelus coioides) after the Vibrio alginolyticus challenge. Fish Shellfish Immunol. 2015;43:310–324. doi: 10.1016/j.fsi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- Mandard S., Müller M., Kersten S. Peroxisome proliferator-activated receptor α target genes. Cell. Molecular Life Sci. Cmls. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccombie R.R., Dolphin C.T., Povey S., Phillips I.R., Shephard E.A. Localization of human flavin-containing monooxygenase genes FMO2 and FMO5 to chromosome 1q. Genomics. 1996;34:426–429. doi: 10.1006/geno.1996.0308. [DOI] [PubMed] [Google Scholar]
- Moraortiz M., Swain M.T., Vickers M.J., Hegarty M.J., Kelly R., Smith L.M.J. De-novo transcriptome assembly for gene identification, analysis, annotation, and molecular marker discovery in Onobrychis viciifolia. BMC Genom. 2016;17:756. doi: 10.1186/s12864-016-3083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y., Li M., Ding F., Ding Y., Ao J., Hu S. De novo characterization of the spleen transcriptome of the large yellow croaker (Pseudosciaena crocea) and analysis of the immune relevant genes and pathways involved in the antiviral response. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Pereiro P., Balseiro P., Romero A., Dios S., Forn-Cuni G., Fuste B. High-throughput sequence analysis of Turbot (Scophthalmus maximus) transcriptome using 454-Pyrosequencing for the discovery of antiviral immune genes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S., Gaillard S., Prévot-D'Alvise N., Aubert J., Rostaing-Capaillon O., Leung-Tack D. Grouper aquaculture: Asian success and Mediterranean trials. Aquat. Conserv. Mar. Freshw. Ecosyst. 2008;18:297–308. [Google Scholar]
- Qin Q.W., Lam T.J., Sin Y.M., Shen H., Chang S.F., Ngoh G.H. Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. J. Virol Meth. 2001;98:17–24. doi: 10.1016/s0166-0934(01)00350-0. [DOI] [PubMed] [Google Scholar]
- Qin Q.W., Chang S.F., Ngoh-Lim G.H., Gibson-Kueh S., Shi C., Lam T.J. Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Dis. Aquat. Org. 2003;53:1–9. doi: 10.3354/dao053001. [DOI] [PubMed] [Google Scholar]
- Reddy J.K., Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- Regnier S.M., Sargis R.M. Adipocytes under assault: environmental disruption of adipose physiology. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2014;1842:520–533. doi: 10.1016/j.bbadis.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Ricote M., Huang J., Fajas L., Li A., Welch J., Najib J. Expression of the peroxisome proliferator-activated receptor γ (PPARγ) in human atherosclerosis and regulation in macrophages by colony stimulating factors and oxidized low density lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7614–7619. doi: 10.1073/pnas.95.13.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.J., Kim J.Y., Cheong H.S., Na H.S., Shin H.D., Chung M.W. Functional study of haplotypes in UGT1A1 promoter to find a novel genetic variants leading to reduced gene expression. Ther. Drug Monit. 2015;37:369. doi: 10.1097/FTD.0000000000000154. [DOI] [PubMed] [Google Scholar]
- Sundvold H., Ruyter B., Ostbye T.K., Moen T. Identification of a novel allele of peroxisome proliferator-activated receptor gamma (PPARG) and its association with resistance to Aeromonas salmonicida in Atlantic salmon (Salmo salar) Fish Shellfish Immunol. 2010;28:394–400. doi: 10.1016/j.fsi.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Thiel T., Michalek W., Varshney R.K., Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor. Appl. Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wei L., Li S., Liu S., He A., Wang D., Wang J. Transcriptome analysis of Houttuynia cordata Thunb. by Illumina Paired-End RNA sequencing and SSR Marker Discovery. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L., Ding H., Dong W., Zhang Y., Shao J. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genom. 2010;11:472. doi: 10.1186/1471-2164-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wei J., Li P., Wei S., Huang Y., Qin Q. MHC polymorphism and disease resistance to Singapore grouper iridovirus (SGIV) in the orange-spotted grouper, Epinephelus coioides. Chin. Sci. Bull. 2016;61:693–699. [Google Scholar]
- Yang M., Wei J., Li P., Wei S., Huang Y., Qin Q. MHC class IIα polymorphisms and their association with resistance/susceptibility to Singapore grouper iridovirus (SGIV) in orange-spotted grouper, Epinephelus coioides. Aquaculture. 2016;462:10–16. [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Alicia O. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-2-r14. R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Gul Y., Li S., Wang W. Cloning, identification and accurate normalization expression analysis of PPARα gene by GeNorm in Megalobrama amblycephala. Fish Shellfish Immunol. 2011;31:462–468. doi: 10.1016/j.fsi.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Zhang H., Fei C., Wu H., Yang M., Liu Q., Wang Q. Transcriptome profiling reveals Th17-like immune responses induced in Zebrafish Bath vaccinated with a live attenuated Vibrio anguillarum. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziouzenkova O., Perrey S., Asatryan L., Hwang J., Macnaul K.L., Moller D.E. Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: evidence for an antiinflammatory role for lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2730. doi: 10.1073/pnas.0538015100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.