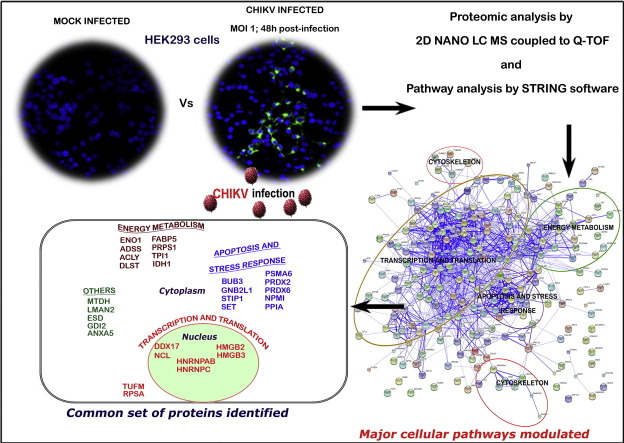
Abstract
Global re-emergence of Chikungunya virus (CHIKV) has renewed the interest in its cellular pathogenesis. We subjected CHIKV-infected Human Embryo Kidney cells (HEK293), a widely used cell-based system for CHIKV infection studies, to a high throughput expression proteomics analysis by Liquid Chromatography–tandem mass spectrometry. A total of 1047 differentially expressed proteins were identified in infected cells, consistently in three biological replicates. Proteins involved in transcription, translation, apoptosis and stress response were the major ones among the 209 proteins that had significant up-regulation. In the set of 45 down-regulated proteins, those involved in carbohydrate and lipid metabolism predominated. A STRING network analysis revealed tight interaction of proteins within the apoptosis, stress response and protein synthesis pathways. We short-listed a common set of 30 proteins that can be implicated in cellular pathology of CHIKV infection by comparing our results and results of earlier CHIKV proteomics studies. Modulation of eight proteins selected from this set was re-confirmed at transcript level. One among them, Nucleophosmin, a nuclear chaperone, showed temporal modulation and cytoplasmic aggregation upon CHIKV infection in double immunofluorescence staining and confocal microscopy. The short-listed cellular proteins will be potential candidates for targeted study of the molecular interactions of CHIKV with host cells.
Biological significance
Chikungunya remained as a neglected tropical disease till its re-emergence in 2005 in the La RéUnion islands and subsequently, in India and many parts of South East Asia. These and the epidemics that followed in subsequent years ran an explosive course leading to extreme morbidity and attributed mortality to this originally benign virus infection. Apart from classical symptoms of acute fever and debilitating polyarthralgia lasting for several weeks, a number of complications were documented. These included aphthous-like ulcers and vesiculo-bullous eruptions on the skin, hepatic involvement, central nervous system complications such as encephalopathy and encephalitis, and transplacental transmission. The disease has recently spread to the Americas with its initial documentation in the Caribbean islands. The Asian genotype of this positive-stranded RNA virus of the Alphavirus genus has been attributed in these outbreaks. However, the disease ran a similar course as the one caused by the East, Central and South African (ECSA) genotype in the other parts of the world. Studies have documented a number of mutations in the re-emerging strains of the virus that enhances mosquito adaptability and modulates virus infectivity. This might support the occurrence of fiery outbreaks in the absence of herd immunity in affected population. Several research groups work to understand the pathogenesis of chikungunya and the mechanisms of complications using cellular and animal models. A few proteomics approaches have been employed earlier to understand the protein level changes in the infected cells. Our present study, which couples a high throughput proteomic analysis and a comparative review of these earlier studies, identifies a few critical molecules as hypothetical candidates that might be important in this infection and for future study.
Keywords: Chikungunya virus, Shotgun proteomics, LC–MS/MS, Differential expression
Graphical abstract
Highlights
-
•
High throughput expression proteomics analysis in HEK293 cells
-
•
Identified four major cellular pathways affected in Chikungunya virus infection
-
•
Short-listed 30 key proteins modulated by a comparative review
-
•
Confirmed modulation of Nucleophosmin and other selected proteins upon infection
1. Introduction
Advances in analytical techniques to explore global proteome changes have found increasing use in host–pathogen interaction studies in infections [1], [2]. Conventional gel-based systems for the purpose use a two-dimensional protein electrophoresis followed by protein identification by mass spectrometry. The newer shotgun proteomics methods have greatly enhanced the spatiotemporal resolution of proteome identification compared to these conventional methods and use liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS) [3]. However, irrespective of these advances, the cross-platform consistency of many proteomics studies in repeatedly identifying the same set of proteins has been rather low, even for identical disease conditions [4]. Variations in experimental design or adopted methodologies can be attributed for these discrepancies. Nevertheless, this has necessitated a comparative approach of multiple studies to reach a meaningful conclusion of the molecules and pathways reliably linked to the pathological process. The present study describes the use of such an approach for understanding the cellular infection process by Chikungunya virus (CHIKV), a recently re-emerged pathogen.
CHIKV has gained global significance with the recent introduction of the virus from the African and Asian countries to the American continents [5]. CHIKV is a positive-sense, single-stranded RNA virus of the genus Alphavirus transmitted to humans by mosquitoes of Aedes species [6]. The two known mosquito vectors, Aedes aegypti and Aedes albopictus, are ubiquitously present in tropics and have spread to the subtropics including the US and European countries [7]. Since 2005, there have been several reports of CHIKV outbreaks in several countries with millions of people suffering from high fever, myalgia, headache, rash and characteristic polyarthralgia that may persist for several months.
CHIKV has a large number of cellular targets including epithelial cells, fibroblastic cells and primary immune cells which are susceptible to infection. Upon infection with the virus, these cells undergo significant modulation in some of their basic functions like innate immune response, apoptosis, autophagy and ER stress response [8]. A number of earlier studies have been carried out to understand the host–virus interaction and pathogenesis of CHIKV by proteomic approaches. These include studies using cell lines like CHME-5, a microglial cell [9], WRL-68, a human hepatic fetal epithelial cell line [10] and SJCRH30, a human rhabdomyosarcoma cell line [11], and those in patient serum [12], in white blood cells isolated from Chikungunya fever patients [13] and in experimentally infected mice tissues such as the liver, brain and muscle [14], [15], [16]. A recent comparison of these studies [17] indicated that even though the results identified common pathways that are being modulated upon infection, there was less consistency in identifying individual proteins.
HEK293 (Human Embryo Kidney) cells are one of the most widely used cellular models for CHIKV infection studies as per the available literature [8]. In the present study, we performed a high throughput proteomic analysis in CHIKV infected HEK293 cells by multidimensional Liquid Chromatography–tandem mass spectrometry. Further, we compared our results with that of earlier studies and identified a common set of proteins. The modulation of a few selected proteins from this list was re-confirmed by transcript level analysis of the genes at different time-points post-infection by quantitative real-time PCR and by double immunofluorescence.
2. Materials and methods
2.1. Virus and cell line
HEK293 cells, a human embryonic kidney epithelial cell line (American Type Culture Collection, ATCC), were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1 × antibiotic–antimycotic mixture (all from Sigma) at 37 °C in a humidified atmosphere containing 5% CO2. A CHIKV strain, RGCB355/KL08 (Genbank accession no: GQ428214), which was isolated from a classical Chikungunya patient and characterized by whole-genome sequencing [18] was used for the study. The virus was isolated by three serial passages in Vero cells prior to use for infecting HEK293 cells, and the viral titer was quantified by plaque assay as previously described [8].
2.2. Infection in HEK293 and immunofluorescence staining
Cells grown on glass cover slips were infected at 70–80% confluency with RGCB355/KL08 in DMEM with 2% FBS. All infections were carried out at a multiplicity of infection of one (MOI 1) for 2 h at 37 °C. After giving a Phosphate Buffered Saline (PBS, pH 7.4) wash to remove the unadsorbed virus, the cells were maintained in DMEM with 2% FBS at 37 °C. At 48 h post-infection, cells were fixed by 4% paraformaldehyde in PBS for 15 min at 4 °C. The cells were permeabilized in 0.2% TritonX-100 for 10 min at room temperature and blocked by incubating in 5% normal goat serum in PBS for 1 h at 37 °C. Cells were incubated in 1:50 dilution of primary antibody (in-house rabbit anti-CHIKV polyclonal serum against recombinant E2 protein), and then with 1:2000 dilution of mouse anti-rabbit IgG Alexa fluor 488 secondary antibody (Invitrogen), each for an hour. DAPI (final concentration: 1 μg/ml) staining for 15 min was done to reveal the nucleus morphology. Three PBS washes were included between each step to remove excess antibodies and stain. After the final wash the cells were imaged using an inverted fluorescent microscope (Nikon TiS) equipped with EMCCD camera (LucaR; Andor). The images were captured using NIS elements software (Nikon) under identical exposure and gain settings for the infected cells as well as the control cells.
For double immunofluorescence detection of Nucleophosmin (NPMI), the fixed monolayers were first stained with anti-CHIKV E2 antibody and anti-rabbit IgG Alexa fluor 488 secondary antibody (Invitrogen) as described above, and followed with overnight incubation with anti-NPMI antibody (Santacruz; B23-sc32256; 1:50 dilution) at 4 °C and anti-mouse Cy3 conjugate (1:200 dilution) as secondary antibody. This was followed by DAPI staining and extensive washes using PBS. The images was captured using confocal microscopy (Nikon A1Rsi) and analyzed using NIS elements software (Nikon) under identical exposure and gain settings for the infected cells as well as the control cells.
2.3. Nuclear condensation by Hoechst staining
100 μl of 5 μg/ml Hoechst 33342 Trihydrochloride Trihydrate (Invitrogen) in PBS was added to infected or uninfected HEK293 cells at the required time points post-infection. Cells were incubated for 20 min at 37 °C in the dark before visualization. The images were captured as above, and the condensed nuclei were quantified using the NIS Elements software.
2.4. Flow cytometry analysis for apoptosis/necrosis detection
Apoptosis/necrosis detection in infected cells was carried out using GFP-Certified Apoptosis/Necrosis detection kit (ENZ-51002; Enzo Life sciences, USA). HEK 293 cells were infected or mock infected at MOI 1. The cells were collected by trypsinization at respective time points, washed twice with cold PBS and re-suspended in 500 μl of Dual Detection Reagent containing 5 μl of Annexin V-EnzoGold (enhanced Cyanine-3) conjugate and 5 μl of Necrosis Detection Reagent (Red) and incubated in dark for 15 min at room temperature, as per the instructions from the manufacturer. The positive control sample was pretreated with the Apoptosis Inducer (Staurosporine) at a final concentration of 2 mM for 4 h and was treated in the same manner. After incubation, the samples were immediately analyzed by flow cytometry (FACS AriaII, BD, USA) at 585/42 and 695/40 band filters.
2.5. Infection in HEK293 cells and sample preparation and tryptic peptides
HEK293 cells, 48 h post-CHIKV infection, and uninfected control cells were collected by trypsinization and washed four times with PBS. Total proteins were extracted from these samples by cell homogenization and cell lysis using Rapigest (Waters) as detergent in 50 mM ammonium bicarbonate. The samples were incubated at 80 °C for 15 min to increase protein solubilization. Protein concentration was estimated by Bradford assay, and 100 μg of total protein from each sample was subjected to in-solution trypsin digestion to generate peptides. Initially, the protein disulfide bonds were reduced by treating the sample with 5 μl of 100 mM dithiothreitol in 50 mM ammonium bicarbonate for 30 min at 60 °C and alkylation with 200 mM iodoacetamide in 50 mM ammonium bicarbonate at room temperature for 30 min. Proteins were then digested with 4 μg of sequencing grade modified trypsin (Sigma) in 50 mM ammonium bicarbonate by incubating overnight at 37 °C. The trypsin digestion reaction was stopped by 1 μl of 100% formic acid. The digested peptide solutions were centrifuged at 14,000 rpm for 12 min, and the collected supernatant was stored at − 20 °C until the LC/MS/MS analysis.
Before the LC/MS/MS analysis of the peptide samples, appropriate peptide standards of various known proteins were spiked in different ratios between the samples for the eventual protein expression analysis. We used Mass Prep Digestion Standards (MPDS), tryptic digested peptides from a set of four proteins, viz. bovine serum albumin (ALBU_BOVIN), Rabbit Glycogen phosphorylase (PYGM_RABIT), Alcohol dehydrogenase from Saccharomyces cerevisiae (ADH1_YEAST), and Enolase from S. cerevisiae (ENO1_YEAST). These MPDS were available as MPDS 1 and MPDS 2 (Waters) having 1:2:8:0.5 fold abundance of peptides for ADH1_YEAST, ENO1_YEAST, ALBU_BOVIN, and PYGM_RABIT, respectively in MPDS 2. MPDS 1 was spiked in the control sample, and MPDS 2 was spiked in the test sample.
2.6. Liquid-chromatography
The peptide samples were analyzed by nanoLC–MSE (MS at elevated energy) using a nanoAcquity UPLC system with 2D technology (Waters Corporation) coupled to a Quadrupole-Time of Flight (Q-TOF) mass spectrometer (SYNAPT-G2-HDMS, Waters Corporation). Both the systems were operated and controlled by MassLynx software. In the 2D nanoLC, the peptides were separated by reverse phase (RP) chromatography at high pH in the first dimension, followed by an orthogonal separation at low pH in the second dimension. An online dilution of the effluent was performed after the first dimension while trapping prior to the second dimension. The chromatography configuration and conditions are given in the Supplementary Table 1 and the method is as follows.
Briefly, 5 μl of sample was injected in partial loop mode and was loaded into the first dimension column (RP column-1) with 20 mM ammonium formate at pH 8 (aqueous mobile phase A) while unwanted solutes were washed to waste. The peptides were eluted from the RP column-1 by the binary solvent manager-1 (BSM-1) into 4 different fractions sequentially based on % B (11.8, 15.3, 19.3, 50% respectively) and are then trapped into the trap column. The organic mobile phase B was 100% ACN. To maximize sample recovery on the second dimension trap column from the organic-containing fractions, an aqueous flow of 0.1% formic acid in water at a flow rate of 20 μl/min was delivered with the second dimension pump (BSM-2). Each fraction was resolved further in the second dimension RP column by the BSM-2. The second dimension separation was performed on a 75 μm × 100 mm BEH C18 1.7 μm particle size column (Waters), with 1–40% B, for a 30 minute gradient at 0.3 μl/min flow rate. After separation, the column was washed with 85% mobile phase B for 4 min and re-equilibrated with 1% mobile phase B for 14 min. The column temperature was maintained at 40 °C.
2.7. Mass spectrometry
Mass spectrometric analysis of eluting peptides from the LC was performed on SYNAPT G2 High Definition Mass Spectrometer — HDMS (Waters). It is a hybrid, quadrupole, ion mobility, orthogonal acceleration, time-of-flight mass spectrometer controlled by MassLynx software. The system combines exact-mass, high-resolution mass spectrometry with high-efficiency ion-mobility-based measurements and separations (IMS-MS).
The parameters used were the following: nanoESI capillary voltage — 3.5 kV, sample cone — 30 V, extraction cone — 4 V; transfer CE — 2 V, trap gas flow — 2 ml/min, IMS gas flow — 90 ml/min (N2), and trap direct current bias — 45 V. To minimize unintended ion activation each of the preceding voltages was chosen to run as low as possible while facilitating ion transmission. The traveling wave height was ramped over 100% of the IMS cycle between 8 and 20 V. The time of flight analyzer (TOF) was calibrated with a solution of glu-fibrinopeptide B (500 fmol/μl) and the lock mass acquisition was done every 30 s by the same peptide delivered through the reference sprayer of the NanoLockSpray source at a flow rate of 500 nl/min. This calibration set the analyzer to detect ions up to 2000 m/z. The radio frequency (RF) generator supplying the quadrupole allowed isolation of ions up to 2400 m/z. Full mass spectra were acquired with a trap CE of 4 V. For mass-to-charge selected ions (in resolving quad-mode) LM resolution was set to 4.7, and HM resolution to 15.0 with the quadrupole set to isolate the m/z of interest. Experiment was run in Resolution mode, and the data format was Continuum. Fragmentation was done in HDMSE mode. The parameters were: Trap MS Collision Energy Low — (eV) 4, Trap MS Collision Energy High — (eV) 4, Transfer MS Collision Energy Low — (eV) 20, and Transfer MS Collision Energy High — (eV) 45.0. For the fragmentation in HDMSE mode, the collision energy was linearly ramped in the Transfer region of TriWave from 20 to 45 V across the duration of high energy scan. Generally, each spectrum was acquired for 0.9 s.
2.8. Relative protein quantitation and identification
The LC–MSE data was analyzed by ProteinLynx Global Server version 2.5.3 (PLGS, Waters) for protein identification as well as for the relative protein quantification. Raw data of all the fractions of control and infected samples were processed and then merged as single control file and test file. Data processing includes lock mass correction post-acquisition. Noise reduction thresholds for low energy scan ion, high energy scan ion and peptide intensity combined across charge states and isotopes were fixed at 150, 50 and 500 counts respectively. Human reviewed database downloaded from UniProt was used as a database for the database search. During database search, the protein false positive rate was set at 4%. The ion accounting parameters were precursor ion tolerance 6 ppm and product ion tolerance 15 ppm. A peptide was required to have at least a single assigned fragment, and protein was required to have at least one assigned peptide and three assigned fragments for identification. Oxidation of methionine was selected as variable modification and cysteine carbamidomethylation was selected as a fixed modification. Trypsin was chosen as the enzyme used with specificity of 1 missed cleavage. Data sets were normalized using the ‘internal standard-normalization’ function of Protein Lynx Global Server software, and quantitative analyses were performed by comparing the normalized peak area/intensity of identified peptides between the samples. ADH was chosen as the internal standard for normalization during expression analysis, and top three intense peptides of ADH were used for that purpose.
2.9. Bioinformatics and bio-statistical analysis
Functional classification and sub-cellular localization were carried out based on Swiss-Prot/TrEMBL database (http://www.uniprot.org/uniprot). In addition to this, enrichment analysis of differentially expressed genes based on Gene Ontology terms for its molecular functions, molecular processes and sub-cellular localizations was performed using the web-based tool GOrilla (Gene Ontology enRIchment anaLysis and visuaLizAtion tool) (http://cbl-gorilla.cs.technion.ac.il/) (updated March 08 2013) [19]. In GORILLA, the running mode had two lists of genes (target and background sets) with a p-value threshold set at 10− 5 and it searches for GO terms that are enriched in the target set compared to the background set using the standard hypergeometric statistics. Functional association studies were performed using the manually curated STRING database (the Search Tool for the Retrieval of Interacting Genes/Proteins) (http://string-db.org) [20]. Protein–protein interactions at high level of confidence (score of > 0.7) was considered. The enrichment was further done in KEGG pathways. The p-values were corrected for multiple testing using the Bonferroni test.
2.10. Quantitative real time PCR
HEK293 cells were infected with CHIKV or mock infected at MOI 1 and incubated for different time points 0 h, 12 h, 24 h, 36 h and 48 h. The cells were collected in Trizol (Invitrogen) reagent and stored in − 80 °C. Total RNA was isolated by phenol–chloroform extraction as per manufacturer's protocols and quantified using NANODROP. 2 μg of RNA samples was treated with RQ1 RNase Free DNase (Promega) for 30 min at 37 °C as per manufacturer's protocol. DNase treated RNA was reverse-transcribed using AMV-RT for 1 h at 42 °C as per manufacturer's protocol. Quantitative real-time PCR was done in ABI7500 Real-time PCR (Applied Biosystems) using gene-specific primers (Supplementary Table 2). Comparison of mRNA expression between samples (control vs. infected) was performed with 1 μl 1:15 diluted cDNA in a 5 μl PCR reaction with SYBR GREEN Jump Start Ready Mix S4438 (Sigma). The thermal cycling profile was set as follows — an initial denaturation of 94 °C for 2 min; followed by 40 cycles of denaturation at 94 °C for 30 s, primer annealing at 65 °C for 30 s, and extension at 72 °C for 30 s. Following amplification, a melt curve analysis was done to confirm the amplified product. Relative changes in gene expression were calculated using − ΔΔCt method by comparing with the expression in mock infected cells, and using β-actin gene expression as the internal control. All experiments were done in three biological and three technical replicates, and the results were analyzed statistically using GraphPad software.
3. Results
3.1. Infection of CHIKV in HEK293 cells and optimization of time point for proteomics
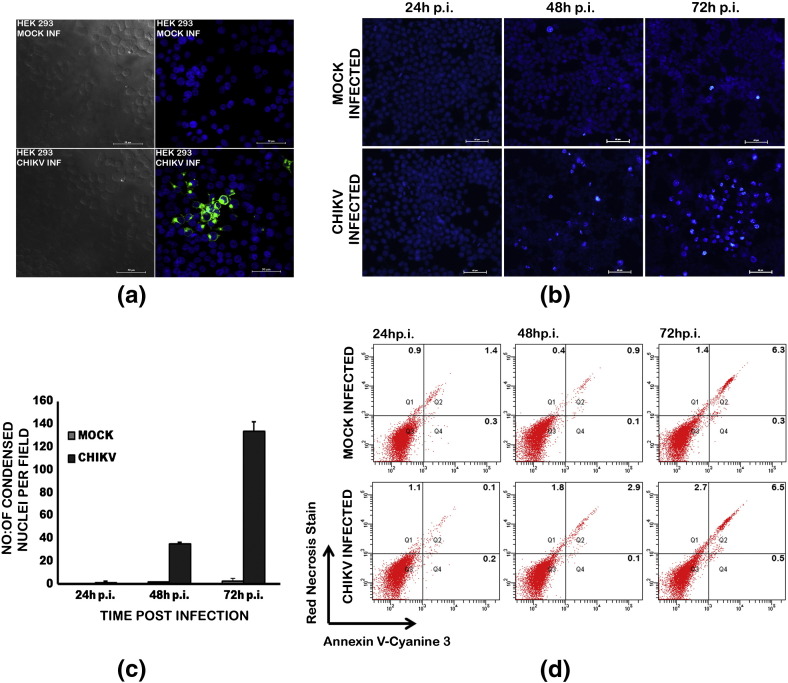
We confirmed CHIKV infection in HEK293 cells by immunofluorescence. Immunostaining for viral envelope protein E2 expression detected the virus replication as distinct cytoplasmic fluorescence in infected cells (Fig. 1a). The infection at MOI 1 resulted in significantly high percentage of infection (about 85%) at 48 h p.i. without causing much cytopathic effect. The Hoechst staining showed only very few cells with condensed nuclei at 48 h p.i. indicating a very low number of apoptotic cells (Fig. 1b & c). When cells were double stained with Annexin V-Cy3 and Necrosis Detection Reagent and analyzed through flow cytometry, we could find that the 95.2% of infected cells (Q3) at 48 h were viable and about 2.9% of cells in Q2 and 1.8% of cells (Q1) were non-viable and in late apoptosis and necrosis (Fig. 1d). Hence we selected this time point for sample collection for the proteomics analysis.
Fig. 1.
(a) Immunofluorescence images of CHIKV infection in HEK293 cells. Monolayer cultures of the cells were infected at MOI 1. At 48 h p.i., CHIKV infected and mock-infected cells were fixed using 4% paraformaldehyde and subjected to immunofluorescence analysis. The virus infection was detected using anti-CHIKV E2 envelope protein rabbit polyclonal serum at 1:50 dilution. Alexafluor 488 anti-rabbit IgG was used as the secondary antibody. A focus of infection in the infected cells indicated by the green cytoplasmic fluorescence is shown. (b) Hoechst staining of CHIKV infected HEK293 cells. Monolayer cultures of the cells were infected at MOI 1. Infected and mock-infected cells were stained at 24 h, 48 h, and 72 h p.i. with the nuclear staining dye Hoechst 33342 for 20 min at 37 °C in the dark before visualization. Images were captured with a LucaR (Andor) EMCCD camera using NIS elements software under identical exposure and gain settings for the infected cells as well as the controls. (c) Quantitative estimation of the condensed nuclei in infected HEK293 cells. Each value is an average of four fields from three independent experiments. (d) Quantification of apoptosis and necrosis in infected HEK 293 cells. Cells infected at MOI 1 and mock infected cells were collected at respective time points and stained with Annexin V-EnzoGold and Necrosis Detection Reagent as per manufacturer's directions and analyzed by flow cytometry (FACS Aria Special order system) at 585/42 and 695/40 band filters. Q1 represents the non-viable cells undergone necrosis, Q2 has cells from both late apoptosis and necrosis stages, Q3 the most viable healthy cells and Q4, cells from early apoptosis stage. Representative images of two independent experiments are shown.
3.2. Identification of differentially expressed proteins
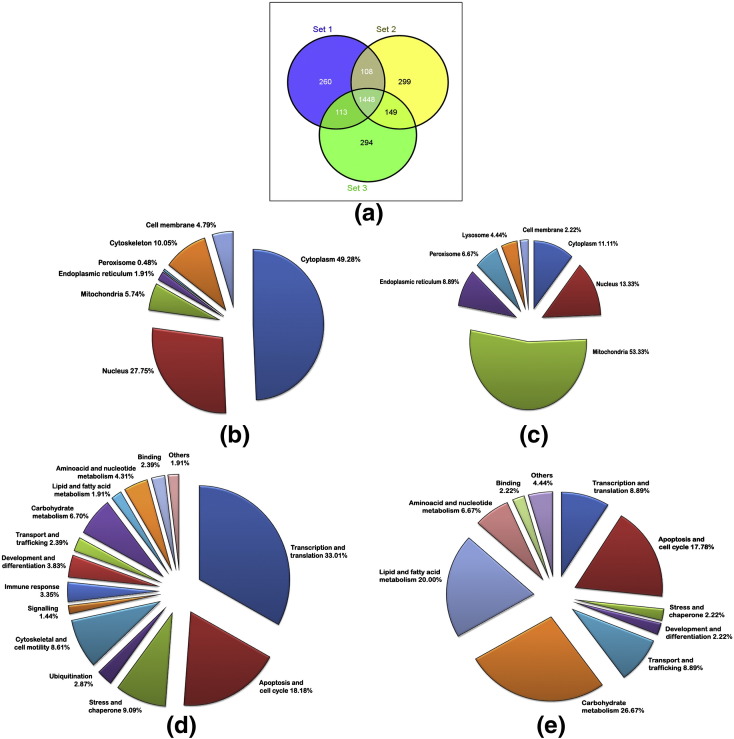
The raw data sets of differentially expressed proteins obtained by the analysis of infected as well as control samples were first filtered by selecting only those proteins that were consistently identified in all the three biological replicates using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) [21]. A total of 1448 proteins were identified by this criterion (Fig. 2a). By setting a false positive cut off and data normalization steps mentioned in Section 2.7, 1047 proteins were short-listed. Subsequent filtering by applying statistical criteria of a 95% up- or down-regulation likelihood ([p > 1] < 0.05 or [p > 1] > 0.95) and a fold-change higher than 30% (ratio of either < 0.70 or > 1.3) as significantly altered levels of expression, we could short-list 209 proteins as up-regulated and 45 proteins as down-regulated compared to uninfected control samples. There were 11 proteins that were unique to the infected samples and five to the control samples. The details of the short-listed proteins from the proteomics analysis are given in the Supplementary Table 3.
Fig. 2.
Number of proteins identified in proteomic analysis, sub-cellular localization and biological functions. (a) Number of proteins identified in proteomic analysis. The data from the three biological replicates were analyzed in Venny software and the results are shown. (b) Sub-cellular localization of up-regulated proteins (11 unique proteins in infected + 198 up-regulated = 209); (c) sub-cellular localization of down-regulated proteins (5 unique in control + 40 down-regulated = 45); (d) biological functions of up-regulated proteins; (e) biological functions of down-regulated proteins. Analysis was done using existing information from Swiss-Prot/TrEMBL database.
3.3. Sub-cellular localization and biological classification of differentially expressed proteins
Functional classification and sub-cellular localization of differentially expressed proteins were done in two sets: as up-regulated (11 unique in infected + 198 up-regulated = 209) group and down-regulated (5 unique in control + 40 down-regulated = 45) based on existing information from Swiss-Prot/TrEMBL database. It was seen that about half of the up-regulated proteins were located in the cytoplasm (49%), while the down-regulated ones were seen mostly in mitochondria (53%) (Fig. 2b & c). When the differentially expressed proteins were categorized based on biological functions, up-regulated proteins were mainly associated with transcription and translation (33%) whereas the down-regulated proteins were mainly associated with energy metabolism (54%). Both groups had proteins which are involved in regulating apoptosis and cell cycle to an equal extent (18%) (Fig. 2d & e).
The identified sub-cellular localization patterns, molecular functions and biological processes of the proteins were cross-checked in GORILLA software. The analysis was done for enriched GO terms in the target list of genes compared to a background list of genes. The target list of genes was our final, refined data set of 209 up-regulated and 45 down-regulated proteins, whereas the background list included all the differentially expressed proteins short-listed from the three sets of experiments (1047). The analysis showed that in the sub-cellular localization patterns of the modulated proteins, the highest p-value was for the cytoplasmic component; in the molecular functions, the highest p-values were for the protein binding and RNA binding; and in the biological process, these were for the gene expression and mRNA metabolism. The details are given in Supplementary Fig. 1 and Supplementary Table 4. The results largely supported the observations in the analysis using Swiss-Prot/TrEMBL database.
3.4. Network analysis of cellular pathways associated with infection
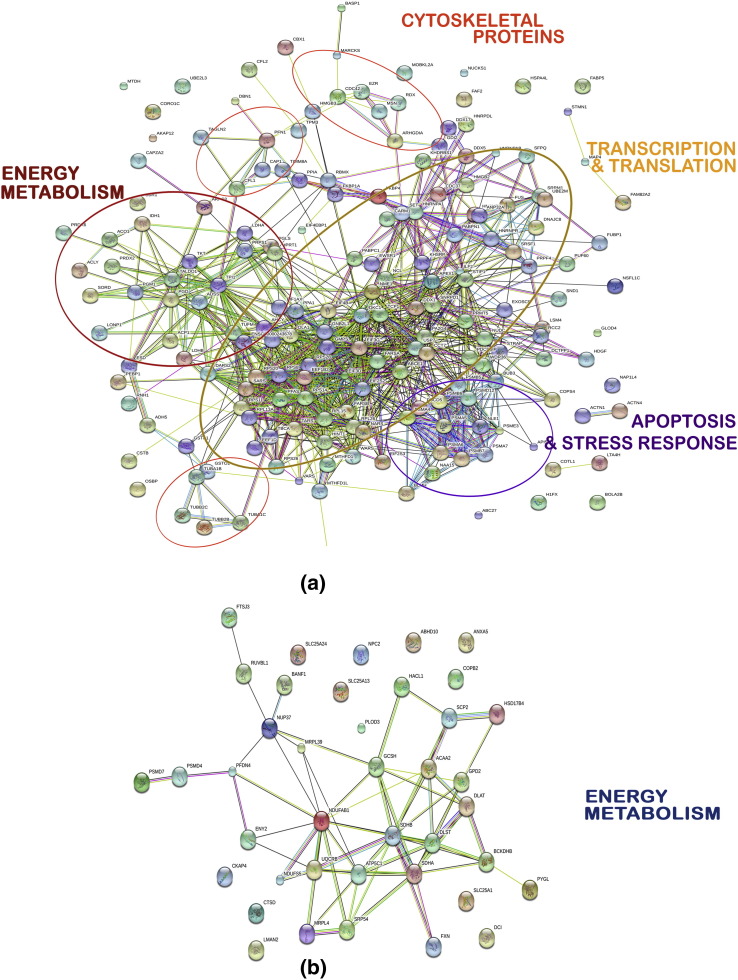
The list of the differentially expressed proteins were submitted individually to STRING 9.0.5 to elucidate the direct (physical) and indirect (functional) associations of these proteins. Analysis was carried out at three confidence levels in the increasing order — 0.4, 0.7 and 0.9. At 0.4 confidence level, out of the 209 proteins up-regulated, 184 proteins were used by the STRING program for protein–protein interactions and 941 interactions involving major cellular pathways were found (Fig. 3 ). Further analysis of these interactions by KEGG pathway enrichment identified ten major biological processes which were significantly involved, with the proteasome pathway having the highest p-value (Supplementary Table 5). Out of 45 proteins down-regulated, 40 were analyzed in STRING and 64 interactions were found at 0.4 confidence level (Supplementary Table 5).
Fig. 3.
Network interactions of differentially expressed proteins by STRING analysis at confidence level 0.4. A subset of the total proteins, which were identified by the software, were used in the analysis: (a) up-regulated proteins (184 out of 209 proteins) and (b) down-regulated proteins(40 out of 45 proteins).
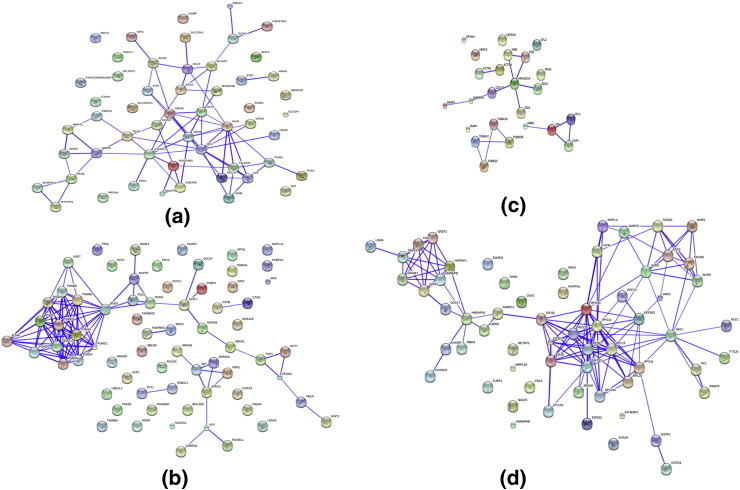
At a confidence level setting of 0.7 in the STRING analysis using the whole data set of up-regulated and down-regulated proteins, the network was refined further with the identification of the interactions by 53 proteins classified under energy metabolism, 68 proteins in apoptosis, cell cycle and stress response, 61 proteins in transcription and translation and 23 proteins in cytoskeleton and cell motility. A few high interacting clusters were identified in the group of proteins involved in apoptosis and stress response, as well as in cytoskeleton and cell motility (Fig. 4 ). To get an insight into interactions at a very high confidence level, the analysis was carried out at a confidence level of 0.9 and a number of proteins were short-listed for a detailed review of their role in viral infections (Table 1 ).
Fig. 4.
Network interactions of differentially expressed proteins under various biological functions by STRING analysis at confidence level 0.7. Proteins functionally categorized under each group based on existing information from Swiss-Prot/TrEMBL database were analyzed: (a) energy metabolism (53 proteins); (b) apoptosis, cell cycle and stress response (68 proteins); (c) cytoskeleton (23 proteins); (d) transcription and translation (61 proteins).
Table 1.
Major pathways identified in STRING analysis under high confidence level (0.9) settings.
| Pathways associated | Gene name | ||
|---|---|---|---|
| 1 | Energy metabolism | Glycolysis and gluconeogenesis | PGM1, TPI1, LDHB, ADH5 |
| Pentose phosphate pathway | PGLS, PRPS1, TKT, TALDO1, PGD, PGM1 | ||
| Glyoxylic acid synthesis pathway | MTHFD1, MTHFD1L, ACO1 | ||
| One carbon folate metabolism | MTHFD1, MTHFD1L | ||
| Glutathione metabolism | IDH1, PGD, GSTP1, GSTO1 | ||
| Fructose and mannose metabolism | SORD, TPI1 | ||
| Fatty acid metabolism | ADH5, ACAA2, DCI, IDH1, SCP2, HSD17B4, HACL1 | ||
| Transmamination and deamination | CBR1, LTA4H, LDHB, AHCY, WARS, ADH5 | ||
| Purine and pyrimidine metabolism | PRPS1, HPRT1, PGM1, ADSS, HMPS, PFAS, NME1, ENSG00000243678 | ||
| Citric acid cycle | ACO1, IDH1, ACLY, DLAT, DLST | ||
| Oxidative phosphorylation | PPA1, SDHA, SDHB, NDUFAB1, NDUFS5, ATP5C1, UQCRB | ||
| 2 | Cellular stress response | Proteasome machinery | PSMA4, PSMA5, PSMA6, PSMA7, PSMB5, PSMB6, PSMB7, PSMD12, PSMD4, PSMD7, PSME3, UBE2M, UBE2L3 |
| 3 | Transcription & translation machinery | Splicing machinery | HNRNPA1, SRSF1, PRPF4, PUF60, SNRPD1, LSM4, DDX5 |
| RNA degradation process | EXOSC9 and LSM4 | ||
| tRNA synthetases | DARS2, SARS, TARS, VARS, WARS, NARS, FARSA, FARSB | ||
| Ribosomal proteins | RPS8, RPS11, RPS20, RPS26, RPS28, RPL13A, RPL15, RPL26 | ||
| mTOR signaling pathway | EIF4B, EIF4EBP1 | ||
| 4 | Cytoskeletal proteins | Regulation of actin cytoskeleton | EZR, CDC42, RDX, MSN, CFL1, CFL2, PFN1, ARPC3, ACTN1, ACTN4 |
| Fc gamma receptor mediated phagocytosis | CFL1, CFL2, ARPC3, CDC42, MARCKS | ||
| Leukocyte transendothelial migration | CDC42, EZR, MSN, ACTN1, ACTN4 | ||
| MAPK signaling pathway | CDC42, STMN1 | ||
| Regulation of gap junction | TUBA1B, TUBA1C, TUBB2B, TUBB2C |
3.5. Comparison with other proteomic studies in CHIKV infection and further validation with quantitative RT-PCR
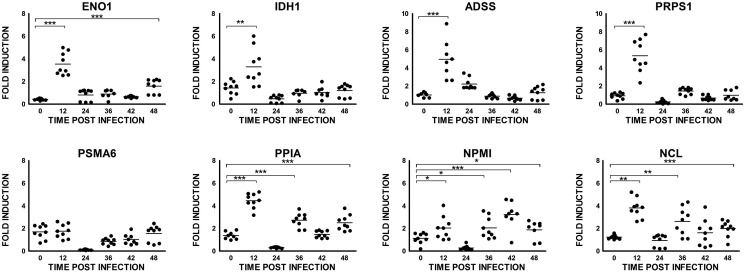
To generate a list of a/the common set of proteins modulated in CHIKV infection, we compared our data with all other proteomic studies published so far on CHIKV infection. We could identify 30 proteins that were seen modulated in at least one of the eight earlier studies (Table 2 ). Based on our STRING interaction analysis and based on this common list of proteins, we selected eight proteins to check the kinetics of transcript level variations during CHIKV infection. In the quantitative real-time RT-PCR, except for one of the proteins selected, all the remaining showed the expected variation in HEK293 cells, albeit at different time points post-infection, reconfirming our observations on proteomic analysis (Fig. 5 ).
Table 2.
List of common proteins identified in the present study and earlier studies which are modulated in Chikungunya virus infection.
| Sl. no | Name of the protein, abbreviation and UniProt ID | Function | Present study HEK293 |
Dhanwani et al. [14], [15] Mice |
Abere et al. [9] CHME |
Thio et al. [10] WRL-68 |
Puttamallesh et al. [12] Human serum |
Wikan et al. [13] WBCs |
Fraisier et al. [16] Mice |
Isaac et al., 2014 SJCRH30 [11] |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | α-Enolase ENO1 — P06733; Eno1 — P17182 |
Energy production and metabolism | ↑ | ↑ | – | ↓ | – | – | – | – |
| 2 | Adenylsuccinate synthetase isozyme 2 ADSS — P30520 |
Purine nucleotide biosynthesis | ↑ | – | – | ↓ | – | – | – | – |
| 3 | Annexin V ANXA5 — P08758 |
Anticoagulant protein | ↓ | – | – | – | – | – | – | ↓↑ |
| 4 | ATP citrate synthase ACLY — P53396, Acly — Q91V92 |
Energy production and metabolism (lipid synthesis) | ↑ | – | – | – | – | – | ↑ | – |
| 5 | DEAD box polypeptide 17 DDX17 — Q92841 |
Transcription and translation | ↑ | – | – | – | – | ↓ | – | – |
| 6 | Dihydrolipoyllysine residue succinyltransferase component of 2 oxoglutarate dehydrogenase complex DLST — P36957, Dlst — Q9D2G2 |
Amino acid and nucleotide metabolism | ↓ | – | – | – | – | – | ↓ | – |
| 7 | Elongation factor Tu mitochondrial TUFM — P49411 |
Transcription and translation | ↑ | – | – | – | – | – | – | ↓↑ |
| 8 | Fatty-acid binding protein, epidermal FABP5 — Q01469 |
High specificity for fatty acids | ↑ | – | – | ↓ | – | – | – | – |
| 9 | Guanine nucleotide-binding protein subunit beta-2-like 1 GNB2L1 — P63244 |
Apoptosis, cell cycle and translational regulation | ↑ | – | – | ↓ | – | – | – | – |
| 10 | Heterogeneous nuclear ribonucleoprotein A/B isoform a HNRNPAB — Q99729 | mRNA processing and translation | ↑ | – | ↑ | – | – | – | – | – |
| 11 | Heterogeneous nuclear ribonucleoprotein C1/C2 HNRNPC — P07910 |
mRNA processing and translation | ↑ | – | – | ↑ | – | – | – | – |
| 12 | High mobility group protein B2 HMGB2 — P26583, Hmgb2 — P30681 |
DNA binding protein | ↑ | ↑ | – | – | – | – | – | |
| 13 | High mobility group protein B3 HMGB3 — O15347, Hmgb3 — A2AP78 |
Apoptosis and cell cycle DNA binding protein | ↑ | – | – | – | – | – | ↓ | – |
| 14 | Isocitrate dehydrogenase, cytoplasmic IDH1 — O75874 |
Glyoxylate bypass and TCA cycle | ↑ | – | – | ↓ | – | – | – | – |
| 15 | Metadherin (protein LYRIC) MTDH — Q86UE4 |
Activates NF-kB transcription factor | ↑ | – | ↑ | – | – | – | – | – |
| 16 | Mitotic checkpoint protein BUB3 BUB3 — O43684, Bub3 — Q9WVA3 |
Apoptosis and cell cycle | ↑ | – | – | – | – | – | ↑ | – |
| 17 | Nucleolin NCL — P19338 |
Transcriptional machinery Induces chromatin decondensation |
↑ | – | ↑ | – | – | – | – | – |
| 18 | Nucleophosmin (B23) NPM1 — P06748 |
Host virus interaction | ↑ | – | ↑ | – | – | – | ↓↑ | |
| 19 | Peptidyl-prolyl cis-trans isomerase A (cyclophilin A) PPIA — P62937 |
Protein folding | ↑ | – | – | ↓ | – | – | – | – |
| 20 | Peroxiredoxin 2 PRDX2 — P32119 |
Stress response and antioxidant property | ↑ | – | – | – | ↓ | – | – | – |
| 21 | Peroxiredoxin 6 PRDX6 — P30041, Prdx6 — O08709 |
Stress response and antioxidant property | ↑ | ↑ | – | – | – | – | – | – |
| 22 | Proteasome subunit alpha type-6 PSMA6 — P60900 |
Ubiquitin proteasome pathway | ↑ | – | – | ↓ | – | – | – | – |
| 23 | Protein SET SET — Q01105 |
Apoptosis, and cell cycle regulation | ↑ | – | – | ↑ | – | – | – | – |
| 24 | Rab GDP dissociation inhibitor beta GDI2 — P50395 |
Regulates the GDP/GTP exchange reaction | ↑ | – | – | ↓ | – | – | – | – |
| 25 | Ribose phosphate pyrophosphokinase 1 PRPS1 — P60891 |
Nucleotide synthesis | ↑ | – | – | ↓ | – | – | – | – |
| 26 | 40S ribosomal protein SA RPSA — P08865 |
Laminin receptor 1 | ↑ | – | – | – | – | – | – | ↓↑ |
| 27 | S-formylglutathione hydrolase ESD — P10768 |
Serine hydrolase involved in the detoxification of formaldehyde | ↑ | – | – | ↓ | – | – | – | – |
| 28 | Stress induced phosphoprotein 1 STIP1 — P31948, Stip1 — Q60864 |
Stress response | ↑ | ↑ | – | – | – | – | – | – |
| 29 | Triosephosphate isomerase TPI1 — P60174 |
Glycolysis and gluconeogenesis | ↑ | – | – | ↓ | – | – | – | – |
| 30 | Vesicular integral membrane protein VIP36 LMAN2 — Q12907, Lman2 — Q9DBH5 |
Lectin, mannose-binding 2 | ↓ | – | – | – | – | – | ↓ | – |
HEK293 — Human Embryo Kidney cells; CHME — human microglial cells; WRL-68 — human embryonic liver epithelial cells; WBCs — white blood cells; SJCRHR30 — human rhabdomyosarcoma cell line. ‘–’ indicates either absence of modulation or no reporting. ‘↑↓’ indicates both up-regulation or down-regulation, but at different time points.
Fig. 5.
Differential expression analyses of the transcripts of selected proteins by quantitative real-time RT-PCR. The fold-change of expression was calculated with respect to the expression levels in mock-infected controls at the respective time points, after normalizing the values with that of the house-keeping gene β-actin. Values from three independent experiments each done in triplicates are indicated for the different time points.
3.6. Nucleophosmin expression in CHIKV infected cells
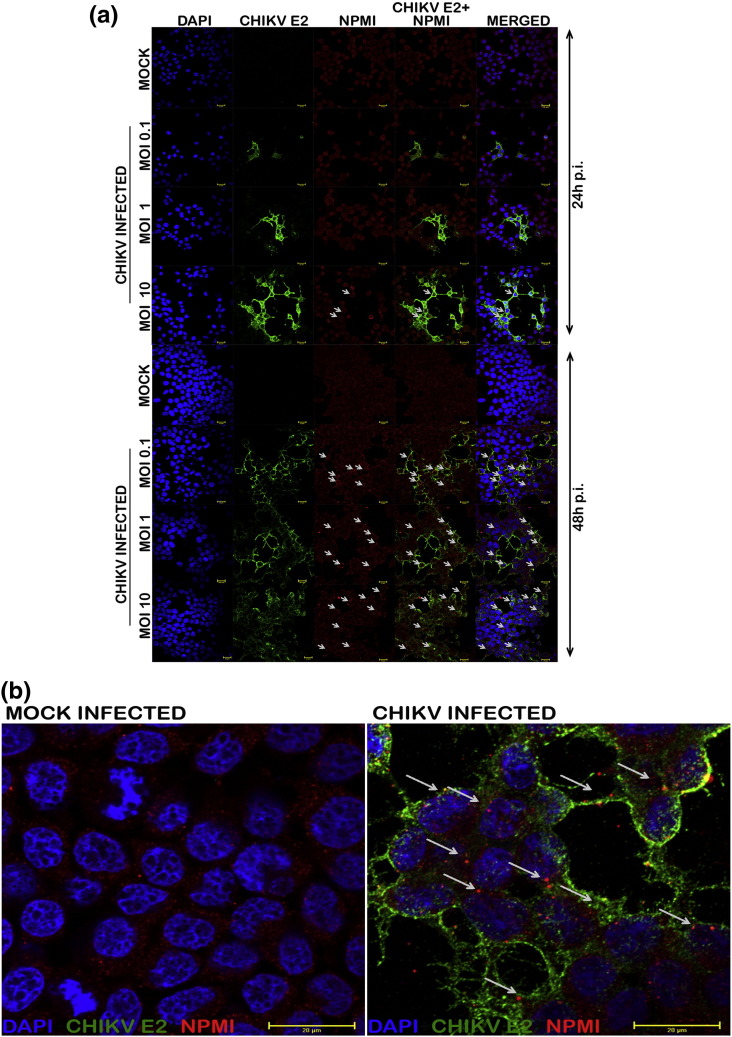
Double immunofluorescence analysis for detecting the CHIKV protein and the cellular protein NPMI was done in both CHIKV and mock infected cells at 24 and 48 h p.i. at multiplicity of infection 0.1, 1 and 10. The results indicated expression of NPMI in both control and infected cells. However, the infected cells had an enhanced expression of NPMI where it had a speckled appearance in the cytoplasm. The NPMI expression was altered only in infected cells that co-expressed the viral proteins, and varied as per time and MOI. Maximum aggregation was found at 48 h p.i. in cells infected at an MOI of 10 (Fig. 6a, b).
Fig. 6.
NPMI expression in CHIKV infected cells. (a) Monolayer cultures of the cells were infected at different MOIs. At 24 and 48 h p.i., CHIKV infected and mock-infected cells were fixed using 4% paraformaldehyde and subjected to immunofluorescence analysis. The virus infection was detected using anti-CHIKV E2 envelope protein rabbit polyclonal serum at 1:50 dilution. Alexafluor 488 anti-rabbit IgG was used as the secondary antibody. For NPMI detection, the cells were again stained using 1:50 dilution of NPMI antibody and anti mouse Cy3 was used as the secondary antibody. Nuclei was counterstained with DAPI staining and visualized at 60 × objective through confocal microscopy.(b) Zoomed image of mock and CHIKV infected cells (zoom factor 2.5) showing the speckle formation of NPMI in infected cells.
4. Discussion
The interphase of protein network interactions occurring during viral infection is complex and regulates two antagonistic processes — one favoring efficient viral replication resulting in the subversion of host survival and the other, enhancing the host antiviral responses to counteract the viral replication. These reactions lead to changes in basic cellular physiology like cellular metabolism, mRNA biogenesis, translation machinery, stress response, and apoptosis. We evaluated the cellular responses to CHIKV infection in HEK293 cells choosing an MOI that causes infection in majority of the cells [8], and an optimal time point (48 h) that would reflect most of the cellular changes before the cells undergo death due to infection. As revealed in the high confidence level (0.9) STRING analysis (Table 1), the infection broadly affected four major pathways, which are discussed below.
4.1. Changes in cellular energy metabolism
Cellular energy metabolism is a key factor that is expected to be modulated in infection with actively replicating viruses. The increase in macromolecular synthesis for the generation of viral components also trigger synthesis of host factors that promote cell sustenance and survival resulting in an overall increase in the host cell biosynthetic activity [22]. Earlier studies in Sindbis virus infection, the prototype Alphavirus, have identified modulation of mitochondrial bioenergetics to favor ATP synthesis that is required to support active viral replication [23]. Similarly, in our study we see that the mitochondrial proteins are regulated during CHIKV infection in the HEK293 cell line. Most of the enzymes involved in carbohydrate metabolism like gluconeogenesis and glycolysis leading to the citric acid cycle are modulated upon infection. All the products of nutrient catabolism are fed to the central pathway of citric acid cycle by the formation of acetylCoA. Modulation of several key enzymes in this pathway indicates that the CHIKV infection regulates the oxidative phosphorylation leading to ATP synthesis by the electron transport chain.
CHIKV, being an enveloped virus, seems to have a significant effect on host fatty-acid metabolism. This observation is evident from the modulation of various key enzymes in associated pathways. The enveloped viruses in general stimulate increased fatty-acid metabolism for generation of membranes from which the envelopes can be derived [24]. Apart from this, CHIKV infection also regulates the expression of several enzymes in the amino-acid metabolism which have a role in both transamination and deamination processes.
4.2. Changes in cellular stress response
The cellular ubiquitin–proteasome pathway (UPP) is an important protein degradation pathway that plays a crucial role in the regulation of many fundamental cellular functions. Many positive-sense RNA viruses use this pathway to eliminate excess viral proteins that prevent viral replication [25]. The proteasome is a multi-catalytic proteinase complex that has an ATP-dependent proteolytic activity and is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu. The cytosolic 26S proteasome has one 20S protein subunit and two 19S regulatory cap subunits. We found the modulation of several proteasomal proteins as a key component in CHIKV infected cells. Also, there was modulation of ubiquitin-conjugating enzymes that catalyze the covalent attachment of ubiquitin to other proteins. These observations strongly suggest that the CHIKV infection activates the ubiquitin-mediated proteasomal pathway (UPP) leading to protein degradation. The increase in the UPP thus helps in selectively eliminating the abnormally folded or damaged proteins produced during infection. Similar observations on the requirement of the components of the ubiquitin–proteasome system have been shown for the maturation and release of a number of viruses [26]. Proteomics studies have also shown the functional role of UPP in infection by many viruses such as Dengue virus [27], Hepatitis B virus [28] and Japanese encephalitis virus [29].
4.3. Changes in cytoskeletal proteins
Several viruses, including CHIKV, are thought to employ the mechanism of clathrin-mediated endocytosis (CME) for cellular entry [30]. Right from the entry to egress, the virus strategically reorganizes the cytoskeleton to facilitate intracellular trafficking and localized assembly of viral components [31]. During active replication of the viruses, the virion proteins and other viral products are transported intracellularly either by the cytoplasmic membrane traffic or direct interaction with the cytoskeletal transport machinery [32], [33]. The resulting cytoskeletal re-organization was observed in many viral infections like Rubella virus, Rabies virus, vesicular stomatitis virus, and Respiratory Syncytial Virus [34], [35].
The cytoskeleton of mammalian cells consists of three main components: microfilament, microtubulin, and intermediate filaments. CHIKV was found to modulate several proteins associated with these components indicating cytoskeletal reconfiguration or reorganization during infection. Some of the proteins in this category, such as CDC42 and stathmin 1 (STMN1) are also involved in signaling pathways such as MAPK signaling and chemokine signaling. In many viral infections, the alterations of cytoskeletal networks are identified by proteomics and such cytoskeletal reorganization might be essential for virus assembly [36], [37], [38].
4.4. Changes in transcription and translation machinery
In most of the viral infections, especially RNA viruses that replicate in the host cytoplasm, the host transcriptional and translational shut-off is induced mainly through the interaction of viral proteins [39]. In CHIKV infection, activation of the cellular protein, protein kinase R (PKR), plays a role in the inhibition of host protein synthesis [40]. Also, similar to other Alphaviruses, nsP2 induces a cellular shut-off that blunts the host antiviral response and finally kills infected cells [41]. The cellular shut-off is a multi-step process in which several components are regulated. One of the key steps is post-transcriptional modification, like splicing. This splicing mechanism is done by spliceosome which is assembled from snRNPs and protein complexes. Several proteins that play a role in splicing machinery are up-regulated in CHIKV infected cells. Modulation of the spliceosome assembly and regulation were also detected in other viral infections like Herpes simplex virus in proteomic analysis [42]. Also, several tRNA synthetases are up-regulated during CHIKV infected HEK293 cells as shown in other viral infections [36,43]. CHIKV infection also led to the up-regulation of some of the ribosomal proteins that are involved in the translational machinery. Eukaryotic translation initiation factor 4B (EIF4B) and its binding protein EIF4EBP1 are important in cellular mRNA translation and play a role in mTOR signaling pathway in the apoptotic processes. Hyperphosphorylation of 4E-BP1 in other viral infections such as in African Swine Fever Virus infection has been reported [44]. Both these proteins are up-regulated in CHIKV infected cells.
4.5. Common set of pathways and proteins modulated in CHIKV infection
A number of proteomics studies have been carried out in CHIKV infection previously (Table 2), and the results were compared in a recent review by Smith [17]. All the four major pathways that we have identified in our study have been found to be modulated during CHIKV infection in one or more of these earlier studies. Among the four, the energy metabolism and stress response pathways were found as the major affected pathways in common. Our analysis to identify common proteins from the present study and earlier reports identified 30 proteins (Table 2), which are discussed below. Our results are more comparable to the proteomic study done by Thio et al. [10] using WRL-68 cells, which is another cell line of epithelial origin like HEK293. Irrespective of an inverse pattern of modulation (up-regulation vs down-regulation) of many of them, a number of proteins were in common in these two studies that were modulated during infection.
α-Enolase (ENO1), a glycolytic enzyme which catalyzes the dehydration of 2-phosphoglycerate to phosphoenolpyruvate, plays an important role in many pathophysiological conditions [45]. The protein is seen up-regulated in HEK293 cells and the liver and brain of infected mice [14] and down-regulated in WRL-68 cell line [10] upon CHIKV infection. A few of the proteins involved in carbohydrate metabolism (IDH, TPI1) and in nucleotide metabolism (ADSS, PRPS1) are seen modulated in both studies using epithelial cell lines-HEK293 and WRL-68. The same is true for FABP5, which is involved in metabolism and intracellular transport of lipids, and ACLY involved in lipid synthesis. Another protein, DLST that is involved in the amino acid and nucleotide pathway was seen down-regulated in our study and also in mice [14] during CHIKV infection. In our study, we could reconfirm these observations by real-time RT-PCR detection of the transcript level up-regulation of a few of the above-listed proteins (ENO1, ADSS, IDH1 and PRPS1) (Fig. 5).
A major group of proteins that showed a very strong interaction in STRING analysis and could be seen modulated in multiple studies are those in the ubiquitin–proteasome pathway, prompting us to believe that it is a component playing a vital role in the infection process. About ten proteasomal subunits are seen modulated in WRL-68 cell line, and three of them (PSMA4, PSMA6, and PSMB5) are the same as in our study. In real-time PCR, PSMA6 mRNA level showed suppression at 24 h post-infection, regained to its original level by 48 h (Fig. 5).
Viral infections lead to significant stress response in host cells. Peroxiredoxins (PRDX) are a family of proteins involved in managing reactive oxygen species. The cellular level of PRDX2 is high in infected HEK293 cells while, in patient serum [12], the level is down-regulated. PRDX6 and STIP1 are also seen modulated in mouse tissues as well as in HEK cells during infection. Our proteomic study shows the modulation of 8 heterogeneous nuclear ribonucleoproteins. Of these, human hnRNP C1/C2, which is involved in mRNA biogenesis, protein–protein interaction, and nuclear localization, was up-regulated both in HEK293 cells and WRL-68 cells. In previous studies, hnRNP C1/C2 was found to interact with NS1 protein in Dengue virus infection, and also play a role in poliovirus infection [46], [47], [48]. hnRNPA/B isoform A, another heterogeneous nuclear ribonucleoprotein, is seen up-regulated in our study and also in CHME-5 cells of human microglial origin [9]. High mobility group proteins are DNA-binding proteins involved in the transcriptional machinery and are seen modulated in many previous CHIKV proteomics study. HMGB1 and HMGB2 are seen up-regulated in CHIKV infected mouse brain, whereas HMGB2 and HMGB3 are seen up-regulated in HEK293.
Nucleolin, another protein involved in the rRNA and ribosomes biogenesis, is seen up-regulated in HEK cells and also in CHME cells [9]. The transcript level of nucleolin also showed up-regulation at 12 h of post-infection (Fig. 5). It is a protein widely seen in nucleolus and is seen to be involved in virus replication in many viral infections [54]. It is also thought to be important in the nuclear egress of viral particles like HSV type I [55] and entry of human parainfluenza virus in A549 cells [56]. Nucleolin interacts with the viral proteins like NS5B of Hepatitis B virus [57] and capsid protein of Dengue [58]. It acts as a cellular receptor for human Respiratory Syncytial Virus [59]. Cyclophilin A (PPIA) is a protein with peptidyl-prolyl cis-trans isomerase activity and acts as a chaperone in protein folding mechanism. PPIA is important in viral infections like HIV, influenza A virus, SARS coronavirus, vaccinia virus, rotavirus, vesicular stomatitis virus, and HCMV and HCV infection, and may serve to play a critical role in the life cycle of viruses [49], [50]. In our proteomics study, we could find that PPIA is up-regulated while it is down-regulated in WRL-68 cells. Similarly, we could also find a significant transcript level modulation of PPIA mRNA 12 h post-infection in HEK293 cells (Fig. 5).
From among the short listed proteins, an important protein that we analyzed specifically for its expression at protein level was Nucleophosmin (NPMI). It was originally identified as a human histone chaperone which activates chromatin transcription in an acetylation dependent manner, and was found to be important in host–virus interaction, especially in HIV infection and HBV infection [51], [52]. It is a multifunctional protein involved in ribosome biogenesis, nucleo-cytoplasmic trafficking and regulation of cell proliferation and apoptosis [53]. Interestingly in earlier studies on CHIKV proteomics, this protein was found to be up-regulated at cellular level in both WRL-68 cells and SJCRH30 cells [10], [11]. In our study, the modulation of NPMI in HEK293 cells was prominently visible at 48 h p.i. as a speckle formation in the cell cytoplasm (Fig. 6a & b). The aggregation of these proteins could be because of its interaction with viral proteins or with other cellular proteins, which is an interesting lead for further investigation.
4.6. Conclusion
Our study in CHIKV infected HEK293 cells reveals involvement of four major pathways by high throughput proteomics analysis — the ones regulating cellular metabolism, cytoskeletal networks, spliceosome and proteasomal machinery. Interestingly, all these pathways were documented in earlier proteomics studies done in CHIKV infection. Further, a comparative analysis of the results from our study and earlier studies identified 30 proteins that were consistently modulated in all the studies. Preliminary analysis indicates that the nuclear chaperone, Nucleophosmin, might be a player in CHIKV infection also, as seen in certain other viral infections in earlier studies [51], [52]. These short-listed proteins will serve as a starting point for targeted studies using single-molecule approach to decipher CHIKV–host interaction.
Transparency Document
Transparency document.
Acknowledgments
The authors are grateful to the Director, RGCB, for providing the infrastructure facilities and Mr. Pradip Fulmali for providing reagents for FACS studies. RA is supported by INSPIRE fellowship from Department of Science and Technology, Government of India. The authors acknowledge the financial support from the Department of Biotechnology, Ministry of Science and Technology, Government of India (grant no. BT/PR5315/MED/29/476/2012).
Footnotes
The Transparency document associated with this article can be found, in the online version.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2015.03.007.
Appendix A. Supplementary data
Supplementary Table 1: The chromatography configuration and conditions of LC/MS/MS.
Supplementary Table 2. List of primers used for quantitative real-time PCR analysis.
Supplementary Table 3: List of differentially altered proteins in HEK 293 cells in response to CHIKV infection.
Supplementary Table 4 : The molecular functions, molecular processes and sub-cellular localizations analysis in GORILLA (Gene Ontology enRIchment anaLysis and visuaLization tool).
Supplementary Table 5: Results of STRING analysis at 0.4 level confidence settings indicating p-values.
Supplementary Fig.1 GO Enrichment of differentially expressed proteins after CHIKV infection
References
- 1.Maxwell K.L., Frappier L. Viral proteomics. Microbiol Mol Biol Rev. 2007;71:398–411. doi: 10.1128/MMBR.00042-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J., Tan B.H., Sugrue R., Tang K. Current approaches on viral infection: proteomics and functional validations. Front Microbiol. 2012;3:393. doi: 10.3389/fmicb.2012.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langley S.R., Dwyer J., Drozdov I., Yin X., Mayr M. Proteomics: from single molecules to biological pathways. Cardiovasc Res. 2012;97:612–622. doi: 10.1093/cvr/cvs346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.List E.O., Berryman D.E., Bower B., Sackmann-Sala L., Gosney E., Ding J. The use of proteomics to study infectious diseases. Infect Disord Drug Targets. 2008;8:31–45. doi: 10.2174/187152608784139640. [DOI] [PubMed] [Google Scholar]
- 5.Cauchemez S., Ledrans M., Poletto C., Quenel P., de Valk H., Colizza V. Local and regional spread of chikungunya fever in the Americas. Euro Surveill. 2014;19:20854. doi: 10.2807/1560-7917.es2014.19.28.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss E.G., Strauss J.H. The Togaviridae and Flaviviridae. Springer; New York: 1986. Structure and replication of the alphavirus genome; pp. 35–90. [Google Scholar]
- 7.Nasci Roger S. Movement of chikungunya virus into the Western hemisphere. Emerg Infect Dis. 2014;20(8):1394. doi: 10.3201/eid2008.140333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham R., Mudaliar P., Padmanabhan A., Sreekumar E. Induction of cytopathogenicity in human glioblastoma cells by chikungunya virus. PLoS One. 2013;8:e75854. doi: 10.1371/journal.pone.0075854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abere B., Wikan N., Ubol S., Auewarakul P., Paemanee A., Kittisenachai S. Proteomic analysis of chikungunya virus infected microgial cells. PLoS One. 2012;7:e34800. doi: 10.1371/journal.pone.0034800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thio C.L., Yusof R., Abdul-Rahman P.S., Karsani S.A. Differential proteome analysis of chikungunya virus infection on host cells. PLoS One. 2013;8:e61444. doi: 10.1371/journal.pone.0061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issac T.H., Tan E.L., Chu J.J. Proteomic profiling of chikungunya virus-infected human muscle cells: reveal the role of cytoskeleton network in CHIKV replication. J Proteomics. 2014;108:445–464. doi: 10.1016/j.jprot.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Puttamallesh V.N., Sreenivasamurthy S.K., Singh P.K., Harsha H.C., Ganjiwale A., Broor S. Proteomic profiling of serum samples from chikungunya-infected patients provides insights into host response. Clin Proteomics. 2013;10:14. doi: 10.1186/1559-0275-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wikan N., Khongwichit S., Phuklia W., Ubol S., Thonsakulprasert T., Thannagith M. Comprehensive proteomic analysis of white blood cells from chikungunya fever patients of different severities. J Transl Med. 2014;12:96. doi: 10.1186/1479-5876-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhanwani R., Khan M., Alam S.I., Rao P.V., Parida M. Differential proteome analysis of Chikungunya virus-infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics. 2011;11:1936–1951. doi: 10.1002/pmic.201000500. [DOI] [PubMed] [Google Scholar]
- 15.Dhanwani R., Khan M., Lomash V., Rao P.V., Ly H., Parida M. Characterization of chikungunya virus induced host response in a mouse model of viral myositis. PLoS One. 2014;9:e92813. doi: 10.1371/journal.pone.0092813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraisier C., Koraka P., Belghazi M., Bakli M., Granjeaud S., Pophillat M. Kinetic analysis of mouse brain proteome alterations following Chikungunya virus infection before and after appearance of clinical symptoms. PLoS One. 2014;9:e91397. doi: 10.1371/journal.pone.0091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D.R. Global protein profiling studies of chikungunya virus infection identify different proteins but common biological processes. Rev Med Virol. 2014 doi: 10.1002/rmv.1802. [DOI] [PubMed] [Google Scholar]
- 18.Sreekumar E., Issac A., Nair S., Hariharan R., Janki M.B., Arathy D.S. Genetic characterization of 2006–2008 isolates of Chikungunya virus from Kerala, South India, by whole genome sequence analysis. Virus Genes. 2009;40:14–27. doi: 10.1007/s11262-009-0411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinforma. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliveros J.C. BioinfoGP, CNB-CSIC. 2007. VENNY. An interactive tool for comparing lists with Venn Diagrams. [Google Scholar]
- 22.Munger J., Bajad S.U., Coller H.A., Shenk T., Rabinowitz J.D. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva da Costa L., da Silva AP Pereira, Da Poian A.T., El-Bacha T. Mitochondrial bioenergetic alterations in mouse neuroblastoma cells infected with Sindbis virus: implications to viral replication and neuronal death. PLoS One. 2012;7:e33871. doi: 10.1371/journal.pone.0033871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y., Clippinger A.J., Alwine J.C. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends Microbiol. 2011;19:360–367. doi: 10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi A.G., Wong J., Marchant D., Luo H. The ubiquitin–proteasome system in positive-strand RNA virus infection. Rev Med Virol. 2012;23:85–96. doi: 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao G., Luo H. The ubiquitin–proteasome pathway in viral infections. Can J Physiol Pharmacol. 2006;84:5–14. doi: 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- 27.Kanlaya R., Pattanakitsakul S.N., Sinchaikul S., Chen S.T., Thongboonkerd V. The ubiquitin–proteasome pathway is important for dengue virus infection in primary human endothelial cells. J Proteome Res. 2010;9:4960–4971. doi: 10.1021/pr100219y. [DOI] [PubMed] [Google Scholar]
- 28.Tong A., Wu L., Lin Q., Lau Q.C., Zhao X., Li J. Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics. 2008;8:2012–2023. doi: 10.1002/pmic.200700849. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L.K., Chai F., Li H.Y., Xiao G., Guo L. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J Proteome Res. 2013;12:2666–2678. doi: 10.1021/pr400011k. [DOI] [PubMed] [Google Scholar]
- 30.Solignat M., Gay B., Higgs S., Briant L., Devaux C. Replication cycle of chikungunya: a re-emerging arbovirus. Virology. 2009;393:183–197. doi: 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor M.P., Koyuncu O.O., Enquist L.W. Subversion of the actin cytoskeleton during viral infection. Nat Rev Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dohner K., Sodeik B. The role of the cytoskeleton during viral infection. Curr Top Microbiol Immunol. 2005;285:67–108. doi: 10.1007/3-540-26764-6_3. [DOI] [PubMed] [Google Scholar]
- 33.Radtke K., Dohner K., Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 34.Cudmore S., Reckmann I., Way M. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 1997;5:142–148. doi: 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 35.Ulloa L., Serra R., Asenjo A., Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53:13–25. doi: 10.1016/s0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- 36.Pastorino B., Boucomont-Chapeaublanc E., Peyrefitte C.N., Belghazi M., Fusai T., Rogier C. Identification of cellular proteome modifications in response to West Nile virus infection. Mol Cell Proteomics. 2009;8:1623–1637. doi: 10.1074/mcp.M800565-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zandi F., Eslami N., Torkashvand F., Fayaz A., Khalaj V., Vaziri B. Expression changes of cytoskeletal associated proteins in proteomic profiling of neuroblastoma cells infected with different strains of rabies virus. J Med Virol. 2009;85:336–347. doi: 10.1002/jmv.23458. [DOI] [PubMed] [Google Scholar]
- 38.Vester D., Rapp E., Gade D., Genzel Y., Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics. 2009;9:3316–3327. doi: 10.1002/pmic.200800893. [DOI] [PubMed] [Google Scholar]
- 39.Lyles D.S. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev. 2000;64:709–724. doi: 10.1128/mmbr.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White L.K., Sali T., Alvarado D., Gatti E., Pierre P., Streblow D. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J Virol. 2011;85:606–620. doi: 10.1128/JVI.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourai M., Lucas-Hourani M., Gad H.H., Drosten C., Jacob Y., Tafforeau L. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol. 2012;86:3121–3134. doi: 10.1128/JVI.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antrobus R., Grant K., Gangadharan B., Chittenden D., Everett R.D., Zitzmann N. Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics. 2009;9:3913–3927. doi: 10.1002/pmic.200900207. [DOI] [PubMed] [Google Scholar]
- 43.Clarke P., Leser J.S., Bowen R.A., Tyler K.L. Virus-induced transcriptional changes in the brain include the differential expression of genes associated with interferon,apoptosis, interleukin 17 receptor A, and glutamate signaling as well as the flavivirus-specific upregulation of tRNA synthetases. mBio. 2014;5:e00902–e00914. doi: 10.1128/mBio.00902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castello A., Quintas A., Sanchez E.G., Sabina P., Nogal M., Carrasco L. Regulation of host translational machinery by African swine fever virus. PLoS Pathog. 2009;5:e1000562. doi: 10.1371/journal.ppat.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noisakran S., Sengsai S., Thongboonkerd V., Kanlaya R., Sinchaikul S., Chen S.T. Identification of human hnRNP C1/C2 as a dengue virus NS1-interacting protein. Biochem Biophys Res Commun. 2008;372:67–72. doi: 10.1016/j.bbrc.2008.04.165. [DOI] [PubMed] [Google Scholar]
- 47.Kanlaya R., Pattanakitsakul S.N., Sinchaikul S., Chen S.T., Thongboonkerd V. Vimentin interacts with heterogeneous nuclear ribonucleoproteins and dengue nonstructural protein 1 and is important for viral replication and release. Mol Biosyst. 2010;6:795–806. doi: 10.1039/b923864f. [DOI] [PubMed] [Google Scholar]
- 48.Brunner J.E., Nguyen J.H., Roehl H.H., Ho T.V., Swiderek K.M., Semler B.L. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watashi K., Shimotohno K. Cyclophilin and viruses: cyclophilin as a cofactor for viral infection and possible anti-viral target. Drug Target Insights. 2007;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- 50.Nigro P., Pompilio G., Capogrossi M.C. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gadad S.S., Rajan R.E., Senapati P., Chatterjee S., Shandilya J., Dash P.K. HIV-1 infection induces acetylation of NPM1 that facilitates Tat localization and enhances viral transactivation. J Mol Biol. 2011;410:997–1007. doi: 10.1016/j.jmb.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 52.Lee S.J., Shim H.Y., Hsieh A., Min J.Y., Jung G. Hepatitis B virus core interacts with the host cell nucleolar protein, nucleophosmin 1. J Microbiol. 2009;47:746–752. doi: 10.1007/s12275-009-2720-z. [DOI] [PubMed] [Google Scholar]
- 53.Grisendi S., Mecucci C., Falini B., Pandolfi P.P. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 54.Hiscox J.A. The nucleolus—a gateway to viral infection? Arch Virol. 2002;147:1077–1089. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sagou K., Uema M., Kawaguchi Y. Nucleolin is required for efficient nuclear egress of herpes simplex virus type 1 nucleocapsids. J Virol. 2010;84:2110–2121. doi: 10.1128/JVI.02007-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bose S., Basu M., Banerjee A.K. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. J Virol. 2004;78:8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirano M., Kaneko S., Yamashita T., Luo H., Qin W., Shirota Y. Direct interaction between nucleolin and hepatitis C virus NS5B. J Biol Chem. 2003;278:5109–5115. doi: 10.1074/jbc.M207629200. [DOI] [PubMed] [Google Scholar]
- 58.Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tayyari F., Marchant D., Moraes T.J., Duan W., Mastrangelo P., Hegele R.G. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med. 2011;17:1132–1135. doi: 10.1038/nm.2444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.
Supplementary Table 1: The chromatography configuration and conditions of LC/MS/MS.
Supplementary Table 2. List of primers used for quantitative real-time PCR analysis.
Supplementary Table 3: List of differentially altered proteins in HEK 293 cells in response to CHIKV infection.
Supplementary Table 4 : The molecular functions, molecular processes and sub-cellular localizations analysis in GORILLA (Gene Ontology enRIchment anaLysis and visuaLization tool).
Supplementary Table 5: Results of STRING analysis at 0.4 level confidence settings indicating p-values.
Supplementary Fig.1 GO Enrichment of differentially expressed proteins after CHIKV infection