Highlights
► SARS-coronavirus is a rather unique example of a correlation between virus affinity to a cellular receptor, entry efficiency and receptor mediated-pathogenicity with involvement of an ectoprotease cleaving both envelope glycoprotein and receptor. ► Henipaviruses is exemplary in mimicking the binding of physiological ligand to their cellular receptors to enter mainly via macropinocytosis. ► Filoviruses use a long and complex entry pathway involving macropinocytosis, a postulated cellular receptor and a multistep activation of the envelope protein.
Abstract
Viruses enter the host cell by binding cellular receptors that allow appropriate delivery of the viral genome. Although the horizontal propagation of viruses feeds the continuous emergence of novel pathogenic viruses, the genetic variation of cellular receptors can represent a challenging barrier. The SARS coronavirus, henipaviruses and filoviruses are zoonotic RNA viruses that use bats as their reservoir. Their lethality for man has fostered extensive research both on the cellular receptors they use and their entry pathways. These studies have allowed new insights into the diversity of the molecular mechanisms underlying both virus entry and pathogenesis.
Mankind is under a permanent threat from novel pathogens qualified as emerging [1, 2••, 3]. Here I review the receptors and mode of entry of three emerging zoonotic viruses, responsible for rare but deadly diseases, whose natural reservoir is the bat: severe acute respiratory syndrome coronavirus (SARS-CoV), Hendra (HeV), Nipah (NiV), Ebola (EboV), and Marburg (MarV) viruses.
SARS-CoV: a dangerous affinity
SARS-CoV has a ∼30 kb positive RNA genome and the crown-like shape typical of the Coronavidae. A regular array of viral spike glycoprotein (S) trimers constitutes the viral envelope. S mediates binding to the cellular receptor Angiotensin Converting Enzyme 2 (ACE2) [4••], and ensures the viral-cell membrane fusion that allows virus entry.
As an ectometalloprotease with monocarboxypeptidase activity, ACE2 cleaves the vasoconstrictor Angiotensin II octapeptide into the vasodilatator Ang1-7 heptapeptide. ACE2 protects the heart, lung and kidney from deleterious vasoconstriction and prevents the onset of an acute respiratory distress syndrome [4••, 5, 6, 7, 8]. The tissue distribution of ACE2 (pneumocytes I and II, lung epithelium progenitor cells, small intestine enterocytes, kidney, heart cardiomyocytes and endothelium) mostly correlates with the known replication sites of SARS-CoV, and could explain the poor lung repair following SARS infection [4••, 9].
The ectopeptidase Type II transmembrane protease serine subfamily member 2 (TMPRSS2) was recently identified as a companion molecule of ACE2 [10•, 11•, 12•]. TMPRSS2 is detected on the epithelium of the small intestine and respiratory tract, that is the major cell targets of SARS-CoV, but not on the endothelium, which is refractory to SARS-CoV infection [13, 14]. TMPRSS2 and ACE2 physically interact [10•]. Only a few S proteins get cleaved by TMPRSS2 to allow a pH- and cathepsin-independent efficient entry of SARS-CoV [10•, 12•]. TMPRSS2 cleaves S protein at sites distinct from those ascribed to trypsin and cathepsin L [12•]. Upon contact of ACE2 with S protein, ACE2 is also cleaved by TMPRSS2 [10•]. When expressed on opposing membranes, SARS-CoV S and the ACE2 + TMPRSS2 complex induce intercellular fusion [11•]. However, newly expressed S proteins escape cleavage by TMPRSS2 allowing the production of virions decorated with uncleaved S [10•, 11•, 12•], possibly because the tripartite association is prevented intracellularly.
The present model of virus entry predicts the following (Figure 1a): SARS-CoV S protein binds to the ACE2 receptor via the concave S424–494 region of the receptor binding site (RBD) that cradles over 17 nm2 of the outer surface of the N-terminal lobe of the ACE peptidase domain, that is outside the enzymatic site [15••]. This activates TMPRSS2 to cleave a few S proteins into fusion-competent S1-S2 homodimers [10•, 11•, 12•, 16], which immediately undergo typical class I fusion protein structural changes [17] which permit the viral envelope to fuse with the plasma membrane. S1-S2 heterodimers are probably too unstable to be incorporated into infectious virus particles [16, 18]. Moreover, activated TMPRSS2, and possibly ADAM17/TACE (TNFα converting enzyme) [4••, 19], concomitantly cleave ACE2. This results in the massive shedding of ACE2 ectodomains [10•], probably due to amplification of the constitutive pathway [5].
Figure 1.
(a) Entry steps of SARS-CoV and pathogenic consequences, with (1) virus attachment to the cell via binding of S to ACE2, (2) activation of TMPRSS2 ectoprotease that leads to the cleavage and activation of virus S envelope protein into the fusion competent S1–S2 heterodimer, (3) fusion of the virus envelope with the plasma membrane to deliver the nucleocapsid into the cytoplasm and allow virus replication, (4) proteolytic cleavage by TMPRSS2 (and/or ADAM17/TACE) ectoprotease with shedding of the ectodomain of the majority of ACE2 molecules independently of their use by SARS-CoV leading to (5) lung failure. (b) Relationship between the affinity of SARS-CoV S protein with the ACE2 receptor, entry efficiency, pathogenicity and inter-human transmission.
ACE2 shedding is not required for SARS-COV entry [5, 20], but is probably responsible for the associated major lung failure. Indeed, soluble S both induces ACE2 shedding and worsens the clinical signs of SARS [21]. In humans and according to virus strain diversity, S/ACE2 affinity correlates with the efficacy of virus entry, the level of ACE2 cleavage and the intensity of the pathology. Furthermore, the highest affinity is associated with inter-human transmission [20, 22••] (Figure 1b). Antibodies targeting the S binding site on ACE2 strongly inhibit viral infection [23]. The correlation between S/ACE2 affinity and SARS-CoV pathogenicity extends to the host range for other mammal species (except bats) in determining whether a particular ACE2 protein can act as a receptor for SARS-CoV or not (including among bats) (Figure 2b) [23, 24, 25, 26, 27]. Consequently, the accuracy of an animal model of human infection critically depends on the capacity to mimic the affinity between the human ACE2 and viral S protein. In mammals, except bats, SARS-CoV induces a ‘toxic-like’ syndrome by triggering a devastating massive cleavage of the cellular receptor used for entry, the molecular basis of which remains to be documented. Why bats do not exhibit clinical signs of infection by SARS-CoV remains puzzling.
Figure 2.
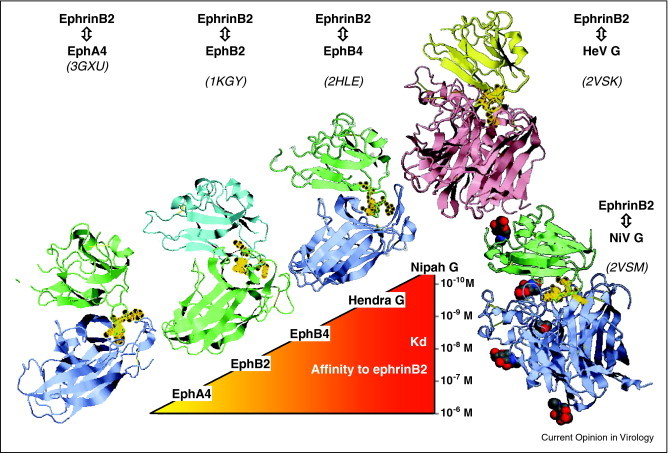
Crystal structure of ephrinB2 in complex with EphA4, EphB2, EphB4, HeV and NiV G showing nearly identical binding modes and affinity ranking of ligands. Structures (PDB identification codes indicated in parenthesis) were drawn using FirstGlance in Jmol (http://molvis.sdsc.edu/fgij/). Phe120, Asn123, Trp125 and Leu129 of the F–G loop of ephrinB2 are decorated with golden circles for easier perception.
Henipaviruses: a universal receptor?
NiV and HeV constitute the Henipavirus genus of the Paramyxoviridae family and are responsible for fatal respiratory and neurological diseases. Their non-segmented negative strand RNA genomes code for two envelope glycoproteins. The fusion protein F is synthesized as a precursor maturated into a functional F1–F2 heterodimer by cathepsin L via a clathrin-mediated recycling endosomal pathway [28, 29]. The attachment protein G is a tetramer consisting of two disulfide bridged dimers. Like the morbillivirus H and parainfluenzae HN proteins, its C-terminal globular head is folded into a six β-sheet blade propeller surmounting a stalk, transmembrane region and cytosolic tail. The sugar-free β1–β6 dimer interface is conserved. The two heads rotate relative to each other by 0°, 63° and 30–40° for henipavirus G, measles virus H and HN, respectively, while the buried area is 9–10 nm2 for G and H against 18 nm2 for HN [30]. The sugars of G are poor in the DC-SIGN ligand oligomannose but rich in the sugar moiety recognized by LESCtin, a lectin specifically expressed on the sinusoidal endothelial cells of lymph node and liver [31].
G protein attaches to the cellular receptors ephrinB2 and ephrinB3 [32]. Ephrins are ligands of Eph receptor tyrosine kinases involved in cell homeostasis [33•]. EphB4 and its ligand ephrinB2 are expressed on the endothelial cells of veinules and small arteries, respectively [34]. Their reciprocal trans-endocytosis acts as a repulsive signal during vasculogenesis [33•]. EphrinB2 is also expressed in the smooth muscle cells of vessels [34]. The tissue distribution of ephrinB3 is restricted to the spinal cord and the corpus callosum. [35]. EphrinB2/B3 expression correlates with the tropism of NiV in infected humans [32].
Ephrin binding sites on NiV and HeV G map to the top of the globular head over the β5 and β6 blades [36••], that is away from the center and side of the propeller, where sialic acid and protein receptors bind parainfluenza HN proteins and measles H, respectively [37]. Astonishingly, both the physiological ligands of ephrinB2, EphB2, EphB4 and EphA4, and the G proteins of NiV and HeV bind the same site (F113–K131) on the G–H loop [30, 31, 36••, 38, 39] (Figure 2). However, the henipavirus G proteins exhibit the highest affinity to ephrinB2 [38, 40•].
EphrinB2 from mammal species, including human, horse, mouse, pig, cat, dog, and bat, can act as efficient cellular receptor for NiV. Indeed, the G–H loop FTIKFQE(F,Y)SPNLWG(L,H)EF sequence is highly conserved between ephrinB2 and ephrinB3 including in those from the distantly related species zebrafish and chicken as supported by successful replication of NiV in chicken embryo [41]. Bronchial epithelium gets infected in the pig and cat [42, 43, 44] but not in human [45] suggesting ectopic expression of ephrinB2/3 in the former species. A LW/YM substitution prevents ephrinB1 from acting as a receptor for NiV and HeV and binding to EphB4 and EphB2 receptor [35, 46, 47]. Correlatively, (i) all ligands compete with each other, (ii) the ephrinB2/3 binding site on NiV-G is exquisitely neutralized by specific antibodies in vitro and in vivo [48, 49•] and (iii) there is a cross-protection between NiV and HeV [50]. The affinity of NiV-G and HeV-G for ephrinB2/B3 correlates with the efficiency of virus entry [48, 51].
Interestingly, whereas NiV G and F induce fusion of cells expressing ephrinB2/B3, NiV preferentially enters after internalization via macropinocytosis [52••], though acidic pH is not required [52••, 53]. Virus entry, but not membrane fusion, is inoperative when the cytosolic tail of ephrinB2 has its PDZ-binding motif deleted or Tyr304 mutated [52••]. These two motifs recruit Grb4 and the P21-activated kinase 1 (Pak1)/CdC42/Rac1 complex that govern macropinocytosis [33•, 54, 55, 56•, 57, 58, 59]. The need for macropinocytosis while the fusion machinery is operative at the cell surface is puzzling. Macropinocytosis occurs very rapidly upon contact [60] and could be faster than the fusion step but then macropinocytosis inhibitors would not be expected to prevent virus entry. Several hypotheses can be proposed: (i) fusion requires a specific Ca++ [61] and/or Na+ ionic environment as documented for Semliki Forest virus [62]. (ii) NiV replication requires a specific conditioning of the cytoplasm induced by contacting ephrinB2/B3. (iii) The nucleocapsid needs to reach a particular cytoplasmic location deeper in the cells, more favorable for viral polymerase activity. The latter would not be unprecedented since forced rerouting of virus normally entering by fusion at the cell surface into the endocytic pathway results in hampered infectivity as shown for pseudotyped measles virus and lentivectors [63, 64, 65].
Filoviruses: an elusive receptor
The Filoviridae EboV and MarV cause severe hemorrhagic fevers and septic-like shock in humans [66]. Their non-segmented negative RNA genomes code for the envelope glycoprotein GP which ensures both attachment to a (still elusive) cellular receptor and membrane fusion. GP is cleaved by a furin-like protease into mature GP1–GP2 heterodimers [67]. Curiously, mutation of the furin-cleavage site does not abolish GP-mediated virus entry due to alternative cleavage [68]. GP is heavily glycosylated with sugar moieties recognized by LESCtin and DC-SIGN/R lectins that can enhance but not mediate infection [69, 70, 71, 72]. This high glycan content shields MHC class I and β-integrin from antibody recognition [73, 74], a finding that explains the previously reported apparent downregulation of the latter [75].
A cellular receptor of glycoprotein nature is predicted on the basis of saturable binding of soluble GP [76] and loss of binding after protease, periodate or tunicamycin treatment [76, 77]. In infected animals, the virus disseminates in many tissues [66]. The EboV receptor is stocked in trans-Golgi network membranes in all cell types including the non-permissive T and B lymphocytes. It is exported to the cell surface upon cell adhesion and internalized via a microtubule-dependent and actin-dependent pathway, respectively [78•, 79•]. EboV and MarV GP cross-compete for binding suggesting the use of a common receptor [76, 80]. However, 3 out of 4 key lysines (at positions 114,115 and 140) defining the receptor binding region (RBR) of EboV GP1 [76] are not conserved in MarV GP1 [81••]. The structure of a soluble trimeric form of GP1–GP2 reveals a GP1-based chalice form, lined by the RBR. The fusion competent GP2 trimers cradle the chalice stem, with the internal fusion peptide flanked by two β-sheets. The RBR is mostly shielded by a glycan cap and a mucin-like domain [82••], the cleavage of which by cathepsins strongly enhances GP1–GP2 binding to the cell surface [76, 83]. However, lowering the pH neither allows EboV entry at the cell surface, nor cell–cell fusion by mucin-deleted GP1–GP2, and the GP/receptor interaction is stable at acidic pH [76, 77, 84••].
In effect, EboV mostly enters by macropinocytosis with a requirement for lipid rafts, the Na+/H+ exchanger, Pak 1, Rac1, Rab5, Rab7, RhoC GTPase and the vacuole closure protein C-terminal binding protein 1 of E1A, CtBP/BARS [59, 85••, 86••, 87, 88, 89]. Constitutive macropinocytosis in dendritic cells and macrophages fits with their permissiveness to EboV infection [90, 91]. Activation of Ax1 enhances both macropinocytosis and EboV entry [92] although the latter may be mediated by serum Gas6, which was recently reported to mediate non-specific entry for several enveloped viruses [93].
The EboV (and MarV) entry process lasts for about 1 h [94, 95] and can be schematized as follows (Figure 3 ): Firstly, (i) EboV attaches to the cells via the GP1/GP2 interaction with DC-SIGN/R and/or LECStin and is (ii) immediately internalized by constitutive and/or virus-contact-induced macropinocytosis. (iii) After ∼30 min of endocytic trafficking, EboV reaches a late endosomal compartment, where (iv) the resident cathepsin B cleaves off the mucin-like domain [83, 84••, 96] to (v) expose GP1's RBR so that the putative receptor can be recruited; then, (vi) a late pH-dependent activation step of the mucin-deleted GP1/GP2 complex triggers the fusion activity of GP2, possibly via the reduction of a disulfide bridge [84••].
Figure 3.
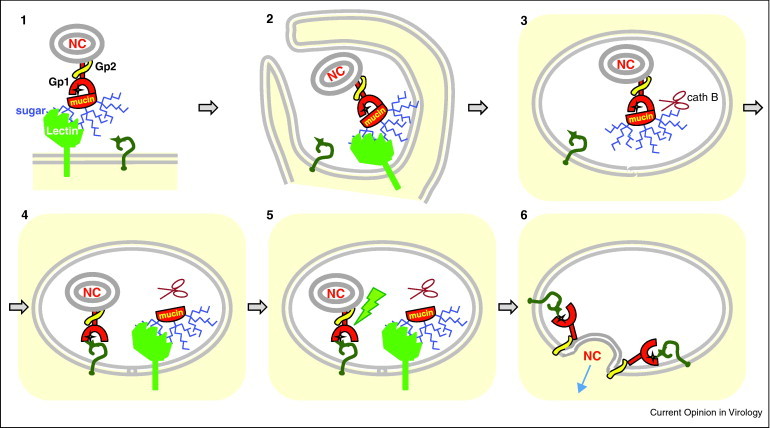
Model of filovirus entry. Virus binds to the cell surface via recognition of sugar moieties (blue branches) of GP1 by DC-SIGN or LECStin (light green) (1) to be immediately internalized by macropinocytosis (2) and migrate through the endocytic pathway (3) until the mucin-like domain (mucin) is cleaved off from GP1 by resident cathepsin B (cath B). This results in RBR (yellow star) being accessible for binding to the postulated receptor (dark green question mark) (4). An additional activation event (disulfide bridge reduction?) occurs (5). This triggers the conformational changes of GP2 (yellow) that mediates fusion of viral and endosomal membranes and ensures the delivery of the nucleocapsid (NC) into the cytosol (pale yellow background) where replication occurs (6). Because the postulated receptor (question mark) is predicted to be expressed at the cell surface, it has been included in every step of virus internalization. Although not yet documented, endosome structures 4 to 6 may successively represent early, maturing, late and possibly endolysosomes, because of the ∼1 h delay between the cell attachment and membrane fusion steps.
In conclusion, several lessons can be taken home. (i) Susceptibility to a disease can be driven by the affinity level between the viral attachment glycoprotein and its cellular receptor. (ii) Evolutionary conserved orthologs of a viral receptor can allow an extended host-range. (iii) A timely proteolytic activation of membrane fusion can occur only upon binding to the receptor. (iv) A viral glycoprotein may follow a complex maturation pathway during endosomal trafficking.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
I apologize for numerous works not cited in this review because of space constraint. I thank R. Buckland, V. Volchkov and B. Horvat for their proof reading of the manuscript and useful comments. This work would not have been possible without the support from the Centre National de la Recherche Scientifique and 08_AFF-IDP grant from the Agence Nationale de la Recherche.
References
- 1.Snowden F.M. Emerging and reemerging diseases: a historical perspective. Immunol Rev. 2008;225:9–26. doi: 10.1111/j.1600-065X.2008.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Morens D.M., Folkers G.K., Fauci A.S. Emerging infections: a perpetual challenge. Lancet Infect Dis. 2008;8:710–719. doi: 10.1016/S1473-3099(08)70256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; An historical overview that allows easy understanding of the “emerging infection” concept.
- 3.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ace2: a peptidase in the renin-angiotensin system, a sars receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review describing known physiological functions of ACE2 that allow the understanding of its key role in the severe lung injury induced by SARS-CoV.
- 5.Jia H.P., Look D.C., Tan P., Shi L., Hickey M., Gakhar L., Chappell M.C., Wohlford-Lenane C., McCray P.B., Jr. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;297:L84–96. doi: 10.1152/ajplung.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oudit G.Y., Herzenberg A.M., Kassiri Z., Wong D., Reich H., Khokha R., Crackower M.A., Backx P.H., Penninger J.M., Scholey J.W. Loss of angiotensin-converting enzyme-2 leads to the late development of angiotensin ii-dependent glomerulosclerosis. Am J Pathol. 2006;168:1808–1820. doi: 10.2353/ajpath.2006.051091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Chan V.S., Zheng B., Chan K.Y., Xu X., To L.Y., Huang F.P., Khoo U.S., Lin C.L. A novel subset of putative stem/progenitor cd34+oct-4+ cells is the major target for sars coronavirus in human lung. J Exp Med. 2007;204:2529–2536. doi: 10.1084/jem.20070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; These three convergent and complementary papers describe the TMPRSS2 ectoprotease as a partner of ACE2 receptor responsible for both activation of SARS-CoV S protein allowing virus entry and shedding of ACE2 ectodomain.
- 11•.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; see ref. [10•]
- 12•.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; see ref. [10•]
- 13.Vaarala M.H., Porvari K.S., Kellokumpu S., Kyllonen A.P., Vihko P.T. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ace2 protein, the functional receptor for sars coronavirus. A first step in understanding sars pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Li F., Li W., Farzan M., Harrison S.C. Structure of sars coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]; The solved structure illuminates the viral spike binding site on ACE2 receptor.
- 16.Belouzard S., Chu V.C., Whittaker G.R. Activation of the sars coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche S., Albertini A.A., Lepault J., Bressanelli S., Gaudin Y. Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell Mol Life Sci. 2008;65:1716–1728. doi: 10.1007/s00018-008-7534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kam Y.W., Okumura Y., Kido H., Ng L.F., Bruzzone R., Altmeyer R. Cleavage of the sars coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS ONE. 2009;4:e7870. doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Sasazuki T., Ishizaka Y. Modulation of tnf-alpha-converting enzyme by the spike protein of sars-cov and ace2 induces tnf-alpha production and facilitates viral entry. Proc Natl Acad Sci USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J. Differential downregulation of ace2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus nl63. J Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L. A crucial role of angiotensin converting enzyme 2 (ace2) in sars coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22••.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A. Receptor and viral determinants of sars-coronavirus adaptation to human ace2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]; The solved structure illuminates the viral spike binding site on ACE2 receptor.
- 23.Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ace2-s-protein interactions. J Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y., Peng C., Yu M., Li Y., Han Z., Li F., Wang L.F., Shi Z. Angiotensin-converting enzyme 2 (ace2) proteins of different bat species confer variable susceptibility to sars-cov entry. Arch Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu M., Tachedjian M., Crameri G., Shi Z., Wang L.F. Identification of key amino acid residues required for horseshoe bat angiotensin-i converting enzyme 2 to function as a receptor for severe acute respiratory syndrome coronavirus. J Gen Virol. 2010;91:1708–1712. doi: 10.1099/vir.0.020172-0. [DOI] [PubMed] [Google Scholar]
- 28.Diederich S., Moll M., Klenk H.D., Maisner A. The nipah virus fusion protein is cleaved within the endosomal compartment. J Biol Chem. 2005;280:29899–29903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 29.Pager C.T., Craft W.W., Jr., Patch J., Dutch R.E. A mature and fusogenic form of the nipah virus fusion protein requires proteolytic processing by cathepsin l. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowden T.A., Crispin M., Harvey D.J., Jones E.Y., Stuart D.I. Dimeric architecture of the hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84:6208–6217. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden T.A., Crispin M., Harvey D.J., Aricescu A.R., Grimes J.M., Jones E.Y., Stuart D.I. Crystal structure and carbohydrate analysis of nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee B. Envelope-receptor interactions in nipah virus pathobiology. Ann N Y Acad Sci. 2007;1102:51–65. doi: 10.1196/annals.1408.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Pitulescu M.E., Adams R.H. Eph/ephrin molecules—a hub for signaling and endocytosis. Genes Dev. 2010;24:2480–2492. doi: 10.1101/gad.1973910. [DOI] [PMC free article] [PubMed] [Google Scholar]; Complete review to understand the role of Eph/ephrin molecules in cell–cell communication and homeostasis.
- 34.Shin D., Garcia-Cardena G., Hayashi S., Gerety S., Asahara T., Stavrakis G., Isner J., Folkman J., Gimbrone M.A., Jr., Anderson D.J. Expression of ephrinb2 identifies a stable genetic difference between arterial and venous vascular smooth muscle as well as endothelial cells, and marks subsets of microvessels at sites of adult neovascularization. Dev Biol. 2001;230:139–150. doi: 10.1006/dbio.2000.9957. [DOI] [PubMed] [Google Scholar]
- 35.Negrete O.A., Chu D., Aguilar H.C., Lee B. Single amino acid changes in the nipah and hendra virus attachment glycoproteins distinguish ephrinb2 from ephrinb3 usage. J Virol. 2007;81:10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Bowden T.A., Aricescu A.R., Gilbert R.J., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of nipah and hendra virus attachment to their cell-surface receptor ephrin-b2. Nat Struct Mol Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]; Crystal structure of Henipavirus G proteins with ephrinB2 receptor showing the central role of the G–H loop of ephrinB2 in the interface and location of the binding site on the edge of the top side of the globular head of G proteins.
- 37.Hashiguchi T., Ose T., Kubota M., Maita N., Kamishikiryo J., Maenaka K., Yanagi Y. Structure of the measles virus hemagglutinin bound to its cellular receptor slam. Nat Struct Mol Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 38.Qin H., Noberini R., Huan X., Shi J., Pasquale E.B., Song J. Structural characterization of the epha4-ephrin-b2 complex reveals new features enabling eph-ephrin binding promiscuity. J Biol Chem. 2010;285:644–654. doi: 10.1074/jbc.M109.064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himanen J.P., Saha N., Nikolov D.B. Cell–cell signaling via eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Negrete O.A., Wolf M.C., Aguilar H.C., Enterlein S., Wang W., Muhlberger E., Su S.V., Bertolotti-Ciarlet A., Flick R., Lee B. Two key residues in ephrinb3 are critical for its use as an alternative receptor for nipah virus. PLoS Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]; By mutagenesis of two amino acid that differ between ephrinB1 and ephrinB3 within the G–H loop and using binding and virus entry assays, the molecular basis of ephrinB2/B3 exclusive usage as henipavirus receptor was established.
- 41.Tanimura N., Imada T., Kashiwazaki Y., Sharifah S.H. Distribution of viral antigens and development of lesions in chicken embryos inoculated with nipah virus. J Comp Pathol. 2006;135:74–82. doi: 10.1016/j.jcpa.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., Goldsmith C.S. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 43.Middleton D.J., Westbury H.A., Morrissy C.J., van der Heide B.M., Russell G.M., Braun M.A., Hyatt A.D. Experimental nipah virus infection in pigs and cats. J Comp Pathol. 2002;126:124–136. doi: 10.1053/jcpa.2001.0532. [DOI] [PubMed] [Google Scholar]
- 44.Mungall B.A., Middleton D., Crameri G., Halpin K., Bingham J., Eaton B.T., Broder C.C. Vertical transmission and fetal replication of nipah virus in an experimentally infected cat. J Infect Dis. 2007;196:812–816. doi: 10.1086/520818. [DOI] [PubMed] [Google Scholar]
- 45.Wong K.T., Shieh W.J., Kumar S., Norain K., Abdullah W., Guarner J., Goldsmith C.S., Chua K.B., Lam S.K., Tan C.T., Goh K.J. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161:2153–2167. doi: 10.1016/S0002-9440(10)64493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chrencik J.E., Brooun A., Kraus M.L., Recht M.I., Kolatkar A.R., Han G.W., Seifert J.M., Widmer H., Auer M., Kuhn P. Structural and biophysical characterization of the ephb4*ephrinb2 protein–protein interaction and receptor specificity. J Biol Chem. 2006;281:28185–28192. doi: 10.1074/jbc.M605766200. [DOI] [PubMed] [Google Scholar]
- 47.Himanen J.P., Rajashankar K.R., Lackmann M., Cowan C.A., Henkemeyer M., Nikolov D.B. Crystal structure of an eph receptor–ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- 48.Bossart K.N., Tachedjian M., McEachern J.A., Crameri G., Zhu Z., Dimitrov D.S., Broder C.C., Wang L.F. Functional studies of host-specific ephrin-b ligands as henipavirus receptors. Virology. 2008;372:357–371. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 49•.Bossart K.N., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J., McEachern J.A., Green D., Hancock T.J., Chan Y.P., Hickey A.C. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5:e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of receptor binding site on NiV G protein as potent target for neutralizing antibody.
- 50.Mungall B.A., Middleton D., Crameri G., Bingham J., Halpin K., Russell G., Green D., McEachern J., Pritchard L.I., Eaton B.T., Wang L.F. Feline model of acute nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J Virol. 2006;80:12293–12302. doi: 10.1128/JVI.01619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aguilar H.C., Aspericueta V., Robinson L.R., Aanensen K.E., Lee B. A quantitative and kinetic fusion protein-triggering assay can discern distinct steps in the nipah virus membrane fusion cascade. J Virol. 2010;84:8033–8041. doi: 10.1128/JVI.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Pernet O., Pohl C., Ainouze M., Kweder H., Buckland R. Nipah virus entry can occur by macropinocytosis. Virology. 2009;395:298–311. doi: 10.1016/j.virol.2009.09.016. [DOI] [PubMed] [Google Scholar]; The use of chemical inhibitors and molecular tools strongly supports macropinocytosis as a major pH independent entry pathway for NiV with a key role for ephrinB2 cytosolic tail although G/F complex can mediate fusion at the cell surface.
- 53.Porotto M., Orefice G., Yokoyama C.C., Mungall B.A., Realubit R., Sganga M.L., Aljofan M., Whitt M., Glickman F., Moscona A. Simulating henipavirus multicycle replication in a screening assay leads to identification of a promising candidate for therapy. J Virol. 2009;83:5148–5155. doi: 10.1128/JVI.00164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cowan C.A., Henkemeyer M. The sh2/sh3 adaptor grb4 transduces b-ephrin reverse signals. Nature. 2001;413:174–179. doi: 10.1038/35093123. [DOI] [PubMed] [Google Scholar]
- 55.Bruckner K., Pasquale E.B., Klein R. Tyrosine phosphorylation of transmembrane ligands for eph receptors. Science. 1997;275:1640–1643. doi: 10.1126/science.275.5306.1640. [DOI] [PubMed] [Google Scholar]
- 56•.Mercer J., Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]; A useful review describing the molecular machinery of macropinocytosis and defining molecular criteria required for validating the use of this entry pathway by viruses.
- 57.Mercer J., Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 58.Meier O., Boucke K., Hammer S.V., Keller S., Stidwill R.P., Hemmi S., Greber U.F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amstutz B., Gastaldelli M., Kalin S., Imelli N., Boucke K., Wandeler E., Mercer J., Hemmi S., Greber U.F. Subversion of ctbp1-controlled macropinocytosis by human adenovirus serotype 3. EMBO J. 2008;27:956–969. doi: 10.1038/emboj.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammarfjord O., Wallin R.P. Dendritic cell function at low physiological temperature. J Leukoc Biol. 2010;88:747–756. doi: 10.1189/jlb.0310155. [DOI] [PubMed] [Google Scholar]
- 61.Zhu M.X., Ma J., Parrington J., Galione A., Evans A.M. Tpcs: endolysosomal channels for ca2+ mobilization from acidic organelles triggered by naadp. FEBS Lett. 2010;584:1966–1974. doi: 10.1016/j.febslet.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helenius A., Kielian M., Wellsteed J., Mellman I., Rudnick G. Effects of monovalent cations on semliki forest virus entry into bhk-21 cells. J Biol Chem. 1985;260:5691–5697. [PubMed] [Google Scholar]
- 63.Spielhofer P., Bachi T., Fehr T., Christiansen G., Cattaneo R., Kaelin K., Billeter M.A., Naim H.Y. Chimeric measles viruses with a foreign envelope. J Virol. 1998;72:2150–2159. doi: 10.1128/jvi.72.3.2150-2159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frecha C., Costa C., Negre D., Gauthier E., Russell S.J., Cosset F.L., Verhoeyen E. Stable transduction of quiescent t cells without induction of cycle progression by a novel lentiviral vector pseudotyped with measles virus glycoproteins. Blood. 2008;112:4843–4852. doi: 10.1182/blood-2008-05-155945. [DOI] [PubMed] [Google Scholar]
- 65.Funke S., Schneider I.C., Glaser S., Muhlebach M.D., Moritz T., Cattaneo R., Cichutek K., Buchholz C.J. Pseudotyping lentiviral vectors with the wild-type measles virus glycoproteins improves titer and selectivity. Gene Ther. 2009;16:700–705. doi: 10.1038/gt.2009.11. [DOI] [PubMed] [Google Scholar]
- 66.Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Volchkov V.E., Feldmann H., Volchkova V.A., Klenk H.D. Processing of the ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wool-Lewis R.J., Bates P. Endoproteolytic processing of the ebola virus envelope glycoprotein: cleavage is not required for function. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powlesland A.S., Fisch T., Taylor M.E., Smith D.F., Tissot B., Dell A., Pohlmann S., Drickamer K. A novel mechanism for lsectin binding to ebola virus surface glycoprotein through truncated glycans. J Biol Chem. 2008;283:593–602. doi: 10.1074/jbc.M706292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuno K., Kishida N., Usami K., Igarashi M., Yoshida R., Nakayama E., Shimojima M., Feldmann H., Irimura T., Kawaoka Y., Takada A. Different potential of c-type lectin-mediated entry between marburg virus strains. J Virol. 2010;84:5140–5147. doi: 10.1128/JVI.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gramberg T., Hofmann H., Moller P., Lalor P.F., Marzi A., Geier M., Krumbiegel M., Winkler T., Kirchhoff F., Adams D.H., Becker S. Lsectin interacts with filovirus glycoproteins and the spike protein of sars coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gramberg T., Soilleux E., Fisch T., Lalor P.F., Hofmann H., Wheeldon S., Cotterill A., Wegele A., Winkler T., Adams D.H., Pohlmann S. Interactions of lsectin and dc-sign/dc-signr with viral ligands: differential ph dependence, internalization and virion binding. Virology. 2008;373:189–201. doi: 10.1016/j.virol.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Francica J.R., Varela-Rohena A., Medvec A., Plesa G., Riley J.L., Bates P. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reynard O., Borowiak M., Volchkova V.A., Delpeut S., Mateo M., Volchkov V.E. Ebolavirus glycoprotein gp masks both its own epitopes and the presence of cellular surface proteins. J Virol. 2009;83:9596–9601. doi: 10.1128/JVI.00784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takada A., Watanabe S., Ito H., Okazaki K., Kida H., Kawaoka Y. Downregulation of beta1 integrins by ebola virus glycoprotein: implication for virus entry. 2000;278:20–26. doi: 10.1006/viro.2000.0601. [DOI] [PubMed] [Google Scholar]
- 76.Dube D., Brecher M.B., Delos S.E., Rose S.C., Park E.W., Schornberg K.L., Kuhn J.H., White J.M. The primed ebolavirus glycoprotein (19-kilodalton gp1,2): sequence and residues critical for host cell binding. J Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; Soluble GP and mutagenesis rationally designed from prefusion GP 3D structure allowed the detailed delineation of the RBR of EboV GP1.
- 77.Takada A., Robison C., Goto H., Sanchez A., Murti K.G., Whitt M.A., Kawaoka Y. A system for functional analysis of ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Dube D., Schornberg K.L., Shoemaker C.J., Delos S.E., Stantchev T.S., Clouse K.A., Broder C.C., White J.M. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci USA. 2010;107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two papers demonstrate the intracellular pool of RBR binding protein of EboV that can be exported to the cell surface upon cell adhesion including in lymphocyte, which are refractory to EboV infection probably because they poorly able to prime GP.
- 79•.Dube D., Schornberg K.L., Stantchev T.S., Bonaparte M.I., Delos S.E., Bouton A.H., Broder C.C., White J.M. Cell adhesion promotes ebola virus envelope glycoprotein-mediated binding and infection. J Virol. 2008;82:7238–7242. doi: 10.1128/JVI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; see ref. [78•]
- 80.Kuhn J.H., Radoshitzky S.R., Guth A.C., Warfield K.L., Li W., Vincent M.J., Towner J.S., Nichol S.T., Bavari S., Choe H., Aman M.J. Conserved receptor-binding domains of lake victoria marburgvirus and zaire ebolavirus bind a common receptor. J Biol Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 81••.Brindley M.A., Hughes L., Ruiz A., McCray P.B., Jr., Sanchez A., Sanders D.A., Maury W. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J Virol. 2007;81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systematic mutagenesis study that has allowed the delineation of key residues of EboV RBR within GP1.
- 82••.Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]; A first structure of prefusion EboV GP1-GP2 heterodimer showing the mucin-like domain covering the RBR site and location of GP2 at the basis of the GP1 chalice fold.
- 83.Kaletsky R.L., Simmons G., Bates P. Proteolysis of the ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84••.Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; First demonstration for a two-step activation of EboV GP occurring in the endosomal pathway using chemical inhibitors, RNA silencing, and biochemical cleavage by cathepsins.
- 85••.Nanbo A., Imai M., Watanabe S., Noda T., Takahashi K., Neumann G., Halfmann P., Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using Ebolavirus like particles made of GP and VP40, a close mimic of the filamentous EboV, infectious EboV and dedicated molecular tools and chemical inhibitors, entry by macropinocytosis was demonstrated by these two papers.
- 86••.Saeed M.F., Kolokoltsov A.A., Albrecht T., Davey R.A. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]; see ref. [85••]
- 87.Quinn K., Brindley M.A., Weller M.L., Kaludov N., Kondratowicz A., Hunt C.L., Sinn P.L., McCray P.B., Jr., Stein C.S., Davidson B.L., Flick R. Rho gtpases modulate entry of ebola virus and vesicular stomatitis virus pseudotyped vectors. J Virol. 2009;83:10176–10186. doi: 10.1128/JVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liberali P., Kakkonen E., Turacchio G., Valente C., Spaar A., Perinetti G., Bockmann R.A., Corda D., Colanzi A., Marjomaki V., Luini A. The closure of pak1-dependent macropinosomes requires the phosphorylation of ctbp1/bars. EMBO J. 2008;27:970–981. doi: 10.1038/emboj.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kalin S., Amstutz B., Gastaldelli M., Wolfrum N., Boucke K., Havenga M., DiGennaro F., Liska N., Hemmi S., Greber U.F. Macropinocytotic uptake and infection of human epithelial cells with species b2 adenovirus type 35. J Virol. 2010;84:5336–5350. doi: 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Norbury C.C., Chambers B.J., Prescott A.R., Ljunggren H.G., Watts C. Constitutive macropinocytosis allows tap-dependent major histocompatibility complex class i presentation of exogenous soluble antigen by bone marrow-derived dendritic cells. Eur J Immunol. 1997;27:280–288. doi: 10.1002/eji.1830270141. [DOI] [PubMed] [Google Scholar]
- 91.Norbury C.C., Hewlett L.J., Prescott A.R., Shastri N., Watts C. Class i mhc presentation of exogenous soluble antigen via macropinocytosis in bone marrow macrophages. Immunity. 1995;3:783–791. doi: 10.1016/1074-7613(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 92.Hunt C.L., Kolokoltsov A.A., Davey R.A., Maury W. The tyro3 receptor kinase axl enhances macropinocytosis of zaire ebolavirus. J Virol. 2011;85:334–347. doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morizono K., Xie Y., Olafsen T., Lee B., Dasgupta A., Wu A.M., Chen I.S. The soluble serum protein gas6 bridges virion envelope phosphatidylserine to the tam receptor tyrosine kinase axl to mediate viral entry. Cell Host Microbe. 2011;9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saeed M.F., Kolokoltsov A.A., Freiberg A.N., Holbrook M.R., Davey R.A. Phosphoinositide-3 kinase-akt pathway controls cellular entry of ebola virus. PLoS Pathog. 2008;4:e1000141. doi: 10.1371/journal.ppat.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schelhaas M. Come in and take your coat off—how host cells provide endocytosis for virus entry. Cell Microbiol. 2010;12:1378–1388. doi: 10.1111/j.1462-5822.2010.01510.x. [DOI] [PubMed] [Google Scholar]
- 96.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]


