Highlights
-
•
New plant viruses diseases emerge periodically in crops.
-
•
These viruses jump species from other crops, or from wild plants.
-
•
The role of biodiversity loss, or ecosystem simplification, in this process is not well understood.
-
•
A few clear examples are summarized, but generalization of these principles will require better understanding of plant virus biodiversity.
Abstract
Plant viruses can emerge into crops from wild plant hosts, or conversely from domestic (crop) plants into wild hosts. Changes in ecosystems, including loss of biodiversity and increases in managed croplands, can impact the emergence of plant virus disease. Although data are limited, in general the loss of biodiversity is thought to contribute to disease emergence. More in-depth studies have been done for human viruses, but studies with plant viruses suggest similar patterns, and indicate that simplification of ecosystems through increased human management may increase the emergence of viral diseases in crops.
Current Opinion in Virology 2015, 10:56–62
This review comes from a themed issue on Emerging viruses: interspecies transmission
Edited by Antoine Gessain and Fernando Garcia-Arenal
For a complete overview see the Issue and the Editorial
Available online 29th January 2015
http://dx.doi.org/10.1016/j.coviro.2015.01.005
1879-6257/© 2015 Elsevier B.V. All rights reserved.
Introduction
Plant virus emergence is a complex, incompletely understood, process involving multiple ecological and genetic factors that may be considered as acting sequentially: first, the encounter between the virus and the new host must occur; then the virus must adapt to the new host so that infection is productive enough to ensure between-host transmission; and, last, epidemiological dynamics must change to optimize between-host transmission in the new host population [1•]. Here we are concerned with the first and third steps of emergence. It is widely accepted, although empirical and experimental evidence is scarce, that ecosystem simplification including biodiversity loss will favor both new virus–host encounters, and new epidemiological dynamics resulting in emergence. Linked to this idea is the concept that emergent viruses generally are multi-host pathogens that spillover into a new host population from the original host population in which the pathogen is permanently maintained, that is, from a reservoir host [2], usually a wild host. The focus has been on virus emergence in crops; however, understanding plant virus emergence in crops is limited by a lack of knowledge about the occurrence and dynamics of virus species and strains in wild plant communities [3]. As this gap is being filled it is becoming increasingly clear that crops may also be reservoirs for emergence into wild plants, with consequences potentially as socioeconomically relevant as emergence in crops. Although plant virus-like sequences have also been found in the feces of a number of animals, including humans [4•], these are almost certainly acquired in the diet, and there is no evidence that any of these can replicate, disseminate or cause disease in humans.
Virus infections in crops and wild plants
In general viruses in crop plants have been discovered because they caused symptoms, and surveys for asymptomatic viruses are extremely limited. Viruses that cause disease in some crops can be asymptomatic in others. For example Peanut stunt virus causes severe disease in peanuts and beans, but establishes a completely asymptomatic infection in clover [5, 6]. Cucumber mosaic virus (CMV) can asymptomatically infect some cultivars of rice (Xu P, Roossinck MJ, unpublished data), and even though both the crop and the virus are extremely well-studied, this has not been reported. The result is that the literature on plant viruses in crops is inherently biased, and the real situation in crops is still unknown. Similar scenarios exist for human and other animal hosts and their viruses.
Until recently very few studies looked at viruses in wild plants, with the exception of a few surveys conducted on weeds in areas surrounding crops, and these only tested for known viruses [3]. With the advent of next generation sequencing however, methods have been developed to look for virus-like sequence elements without the bias of only looking for known viruses, and plant viruses are being found in wild plants and a number of other environments [4•, 7]. These studies indicate that what is found in wild plants is vastly different from what is known in crop plants [4•]. This does not mean that the viruses are truly vastly different however, because in depth surveys for all viruses in crop plants have not yet been done. Perhaps most interesting in the wild plants is the preponderance of persistent viruses [8] that make up more than half of the sequences that have relatives in GenBank. Another interesting observation is that although some known acute plant viruses are found in wild plants they are most often asymptomatic, and there is no correlation between the presence of a virus and the presence of symptoms in wild plants [8] (but see [9•]). In addition, many wild plants appear to have multiple infections, including both acute and persistent viruses [10•].
At least 60% of the sequences obtained from virus-enriched samples in plants have no relatives in GenBank, and are currently classified as unknowns. This is common in virus surveys, and in marine virus surveys these numbers are close to 90%. New methods are being developed to try to understand these entities [11], but we are still along way from definitive answers to the question: what viruses are infecting crops and wild plants?
Another question relevant to understanding emergence is if virus infection patterns are the same in crops and in wild plants. Again, data are limited, and consider only acute viruses infecting both crops and wild plants. In one study, the incidence of two begomoviruses with narrow host ranges, Pepper golden mosaic virus and Pepper huasteco yellow vein virus in wild pepper (Capsicum annuum var. glabriusculum) populations in Mexico increased with ecosystem simplification due to habitat anthropization [9•]. The main predictor of begomovirus incidence was the species diversity of the habitat (Figure 1 ). However, diversity was unrelated to the incidence of the generalist virus CMV [9•]. Patterns of begomovirus and CMV incidence may be related to the dilution effect and amplification effect hypotheses linking host diversity and emergence risk (see below). The yearly variation in begomovirus incidence was higher in cultivated than in wild populations of the host plant [9•, 12••], suggesting that ecosystem simplification favors epidemic (vs. endemic) dynamics of virus infection. Large yearly incidence variation in crops, but not in wild hosts, was also reported for CMV, with the extent of temporal variation in wild hosts being positively correlated with ecosystem simplification [13]. Further work is required to determine if these patterns can be generalized to other acute viruses, or to persistent viruses.
Figure 1.
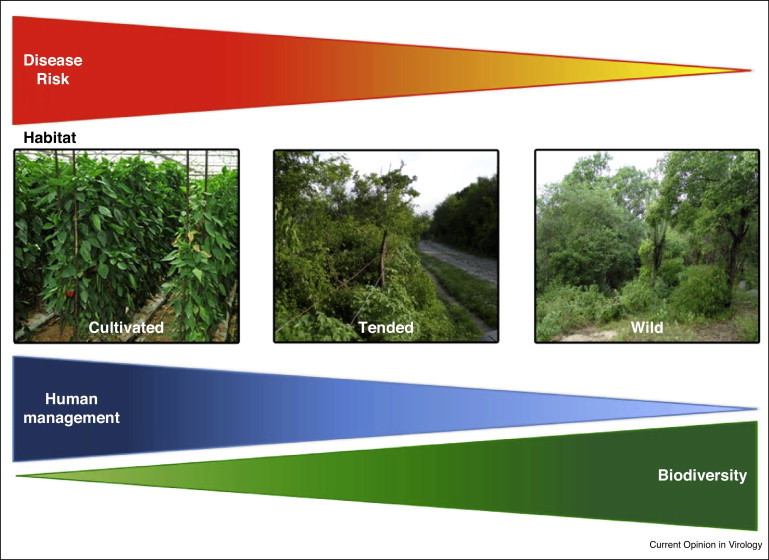
Schematic of the relationship between habitat heterogenenity and disease risk. Disease is greatest in fully cultivated plants, intermediate in wild plants that are tended by humans in anthropic habitats, and lowest in fully wild plants. On the other hand, biodiversity is highest in the habitats of wild plants, and lowest in those of cultivated plants.
The relationships between biodiversity and infection risks
Loss of biodiversity can increase incidence of emerging diseases [14••], and change disease dynamics [15] although with very limited data on the incidence of infectious agents in wild hosts it is difficult to know if this assumption is general [16]. Diversity of available hosts could drive virus divergence by providing expanded niches for viruses when competition is strong [17]. Critically, we have a very limited inventory of the global pathome, the worldwide diversity and distribution of potentially pathogenic microbes in wild and domesticated plants and the environment. Some recent large-scale statistical studies have set the stage for identifying hot spots for potential emergence events for human pathogens [18, 19, 20, 21, 22]. These consider many potential drivers of pathogen biodiversity and emergence including climate factors, human demographics, population changes, land use changes, prior host species and pathogen control. While they disagree on some fundamental points (e.g. the proportion of emergence events due to viruses, the relationship between latitude and propensity for emergence), they highlight the importance of pathogen host range, local host species diversity, land use changes and agriculture. Although similar analyses have not been reported for plant viruses, plant pathologists have postulated for a long time that ecological simplification associated with agriculture favors the appearance of new diseases in crops, as well as their incidence and severity. Specifically, it is considered that reduced biodiversity in agroecosystems, in terms of both plant species richness and the genetic diversity of the focal host, will favor emergence [23, 24]. Support for these hypotheses derives mostly from circumstantial or historical evidence. For plant viruses, empirical evidence has been reported from the analysis of Cereal/Barley yellow dwarf luteoviruses which have been the object of detailed experimental studies on their effects in wild grassland ecosystems in western North America [25•, 26, 27, 28, 29•], and for the begomovirus infecting wild pepper in Mexico, as described above [9•]. Current declines in biodiversity are at the root of a renewed interest on the relationship between biodiversity and disease emergence [14••]. For multi-host pathogens, heterogeneity in the ability of the pathogen to infect and multiply in different hosts has been shown to either reduce (‘dilution effect’ [30]) or increase (‘amplification effect’ [31]) the overall prevalence of infection in diverse host communities. The ‘dilution effect’ and ‘amplification effect’ represent extremes of a continuum based on the degree of host specialization by the pathogen. Again, available knowledge is scant here, as information on the host ranges and the degree of host preference of plant viruses derives mostly from assays on collections of experimental hosts, rather than from host range analyses in the field. Theoretical studies have speculated that host heterogeneity should drive selection for pathogen virulence [32] and may drive evolutionary branching in the parasite population [33]. These predictions have been tested for CMV though model analyses with biologically realistic, experimentally determined, model parameters [34].
The role of parasite biodiversity in disease emergence is equivocal, though many studies point to the critical role played by parasite intraspecific diversity in parasite adaptability [35]. Most studies of disease emergence do not directly address plant-virus systems but many of the questions and hypotheses they raise are relevant. For example, pathogens with broad host ranges (generalists) are hypothesized to be more likely to emerge in new species than those with narrow host ranges (specialists), and only a small fraction of parasites capable of moving from animals into humans are likely to spread from person to person [22]. Underlying this hypothesis is the concept of across-host fitness trade-offs (see Elena and Bedhomme in this volume), which should drive evolution toward specialists.
Phylogenetic approaches to study virus emergence, evolution and spread
Little is known about the selective forces that drive viral evolution in natural ecosystems, which contrasts with the more detailed population genetics studies in crop plants that have revealed the importance of mutation rates [36, 37], recombination [38, 39, 40], genetic drift [1•], and, to a lesser extent, migration in virus evolution. In recent years, the methodology of phylogenetic inference, relying on maximum likelihood, Bayesian and other quantitative frameworks, has made great strides and started to unravel life histories of viruses with a sensitivity not attainable previously [36, 41, 42, 43, 44, 45, 46]. Approaches based on the coalescent theory and other statistical models have been developed [47, 48, 49], allowing investigators to quantify the effects of the individual evolutionary factors, including particularly revealing results on positive vs. purifying selection, interhost vs. intrahost evolution patterns, etc. (see Ref. [50] for a recent monographic treatment).
Phylogenetics can be used to model patterns of evolution and migration [51]. Various mathematical and statistical approaches to model emergence events have been proposed, providing retrospective understanding of parasites that have previously moved into novel host species [52, 53, 54] and tools for rapidly identifying hotspots of molecular evolution [37, 55, 56]. These methods recently have been used to assess whether environmental heterogeneity determines the evolutionary dynamics of plants viruses, and to reconstruct the spatio-temporal pattern of the colonization of new host populations [12••]. However, we still lack the data and data-driven models that would allow us to anticipate future movement of parasites among different host species/populations [57].
Spillover and spillback
Emergence requires that a virus infects a new host from a reservoir, that is, a natural host that may be either wild or domestic. This first encounter between virus and host is termed spillover. Virus adaptation to the new host results in infection dynamics becoming independent from further spillover [1•], and eventually the virus may spillback from the new host to the natural host (Figure 2 ).
Figure 2.
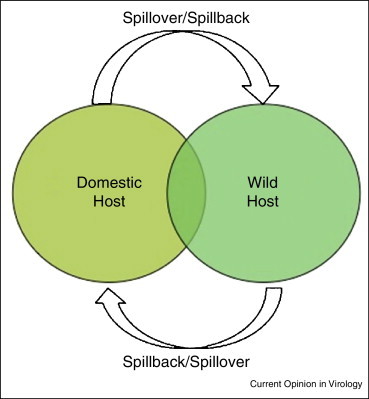
Spillover and spillback between wild and domestic hosts. Spillover occurs when a virus moves from its natural (either a domestic or a wild host) into a new host (either wild or domestic). Spillback occurs when a virus moves from the new host back into the natural host. Natural host here means the source of the virus in this ecosystem, but does not necessarily mean the host where the virus originally evolved (this is often unknown).
There have been very few studies on the role of wildlands in plant virus diseases in crops, although considerably more work has been done in human virology. Human pathogenic viruses can emerge from unapparent infections in wildlife via a range of routes, including air-born, vector-born and body fluid-born transmission, as documented for Human immunodeficiency virus that apparently emerged numerous times from the closely related Simian immunodeficiency virus, endemic in chimpanzees [58], and only rarely pathogenic [59], and more recently for Severe acute respiratory syndrome virus that apparently emerged from bats [60, 61]. West Nile virus has been a spillover human pathogen for some time, but only recently appeared in North America. It is carried by wild birds, and transmitted by mosquitoes [62]. Influenza virus has been a human pathogen for a long time, but new strains emerge periodically from wild waterfowl populations, usually via a secondary domesticated host like swine [63, 64, 65, 66], and are a frequent source of concern. Disease dynamics between domestic and wild hosts have been studied in Dengue virus (mosquito-born), mostly in the context of movement from sylvatic hosts to humans. The movement of Dengue from humans to simian hosts may have occurred but has not been studied beyond phylogenetics of extant viruses [67] and even extensive treatments of the sylvatic cycle of Dengue and other zoonotic viruses do not consider spillback [68], although understanding the sylvatic cycle is recognized as critical for understanding human Dengue dynamics [67]. Often assumptions are made that humans are dead-end hosts for zoonotic viruses [69], but this has not been well tested, and the role of wild host diversity in disease dynamics has been studied for only a few cases [14••, 16]. Although the studies relating plant virus emergence to wild host reservoirs are fewer than for human or animal viruses, the classical work on the emergence of Cacao swollen shoot virus as an important pathogen of cacao (Malvaceae family, native to the Americas) in West Africa from lowland forest and native savanna trees belonging to the Malvaceae, and the more recent studies on the emergence of Maize streak virus on maize in Africa after recombination of strains infecting local grasses [70•, 71] demonstrate the role of spillover in plant virus disease emergence (Table 1 ). In other instances, evidence strongly suggests emergence from a wild reservoir, as for Sweet potato feathery mottle virus [72] or Pepino mosaic virus [73] or a wild reservoir is suspected, but has not been identified, as for African cassava mosaic virus and East African cassava mosaic virus [70•]. As with humans, crops may be epidemiological dead ends. With Mal de Rio Cuarto virus that causes the major maize viral disease in Argentina, maize infection is strictly dependent on the migration of viruliferous delphacid vectors from wild grasses or winter grain crops to the young maize plants [74]. In western Australia Hardenbergia mosaic virus that naturally infects a native legume can spillover into introduced legumes and cause severe crop losses [75].
Table 1.
Examples of plant viruses that have emerged in crops grown in non-native environments
| Virus | Geographical emergence | Date first reported | Origin of crop plant | Source of virus | Reference |
|---|---|---|---|---|---|
| Cassava mosaic geminiviruses | East Africa | 1894 | South America | Unknown | [70•] |
| Cacao swollen shoot virus | West Africa | 1900 | Central America | Local trees from the Malvaceae | [80] |
| Maize streak virus | Africa | 1928 | Central America | Wild native grasses | [81] |
| Tomato yellow leaf curl virus | Israel | 1930 | South America | Infects many wild hosts, origin not clear | [82] |
| Sugarcane yellow leaf virus | Southern United States, Central and South America | 1994 | Southern Asia | Host unknown, but originated in Columbia | [83] |
| Pepino mosaic virus | Peru, but has emerged in tomato around the world | 1980 | South America | Wild native Solanum species | [73] |
| Tomato torroado virus | Spain | 1996 | South America | Unknown, infects many Solanaceae | [84] |
| Iris yellow spot virus | Brazil | 1981 | Worldwide | Unknown, but common in weeds | [85] |
| Plum pox virus | United Statesa | 1999 | China | Unknown, may have arrived from Europe on nursery stock | [86] |
| Wheat mosaic virusb | United States | 1993 | Turkey | Unknown but also found in maize (a native crop) | [87] |
Widespread in Europe, emerged in eastern US recently.
Also called High plains virus.
The role of domestic hosts as reservoirs for virus emergence in wild hosts has been documented for viruses affecting wildlife, due to the high impact of emerging viruses in conservation programs, particularly for endangered species. Thus, the introduction of Rinderpest virus into Africa with infected cattle from India had a catastrophic impact on native wild African ruminants in the late 19th century [76]. More recent examples from the late 20th century include the outbreaks of rabies that decimated the populations of African wild dogs and Ethiopian wolves, probably from domestic dogs [77] or the outbreaks of Porcine distemper virus that decimated North Sea and Caspian seal populations, the virus evolving from Canine distemper virus after jumping species [76, 78]. No similarly documented cases have been reported for plants, but broad-range strains of Bean yellow mosaic virus have spilled over from ornamental plants to the native flora of Australia, possibly with negative effects [79]. In Costa Rica, surveys have found acute viruses from local crops in wild plants, but without any associated negative effects. There is also evidence that some of these viruses spillback into domestic crops with attenuation of their pathology (Roossinck, unpublished data). Other data on spillback and its potential affects on disease dynamics are lacking.
Conclusions
As human populations increase, the need for food continuously increases, with concomitant increases in conversion of wild lands for agricultural use. Increased use of wild lands for crops has resulted in significant losses in biodiversity, or ecosystem simplification. Virus disease emergence in crops is a complex process that involves interactions among wild and domestic hosts, insect and other virus vectors, and changes in ecosystems. Although the data is quite limited, evidence to date indicates that loss of biodiversity will lead to increased incidence of virus disease. The need for more studies on the presence of viruses in wild and domestic plants, particularly without regard to symptoms, is apparent, and a thorough understanding of these dynamic processes will not be possible until more data are available. In addition, the role of crop viruses in diseases of wild plants is largely unknown. Spillover from domestic plants to wild plants could contribute to increased simplification of wild ecosystems, further increasing the potential for newly emerging diseases in crops.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported in part by the Pennsylvania State University College of Agricultural Science, by the National Science Foundation grant numbers EF-0627108, EPS-0447262, IOS-0950579, and IOS-1157148, the United States Department of Agriculture grant number OKLR-2007-01012; and Universidad Politécnica de Madrid and by Spain's Plan Nacional de I+D+I grant AGL2008-02458.
References
- 1•.Elena S.F., Fraile A., García-Arenal F. Evolution and emergence of plant viruses. Adv Virus Res. 2014;88:161–191. doi: 10.1016/B978-0-12-800098-4.00003-9. [DOI] [PubMed] [Google Scholar]; A comprehensive review of the evolution and ecology of plant virus emergence.
- 2.Haydon D.T., Cleaveland S., Taylor L.H., Laurenson M.K. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Inf Dis. 2002;8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper I., Jones R.A.C. Wild plants and viruses: underinvestigated ecosystems. Adv Virus Res. 2006;67:1–47. doi: 10.1016/S0065-3527(06)67001-2. [DOI] [PubMed] [Google Scholar]
- 4•.Roossinck M.J. Plant virus metagenomics: biodiversity and ecology. Annu Rev Genet. 2012;46:357–367. doi: 10.1146/annurev-genet-110711-155600. [DOI] [PubMed] [Google Scholar]; A recent review on what is known about plant virus biodiversity.
- 5.Sherwood R.T. Viruses of white clover in pastures of Pennsylvania, New York, and Vermont. Plant Dis. 1997;81:817–820. doi: 10.1094/PDIS.1997.81.7.817. [DOI] [PubMed] [Google Scholar]
- 6.Anderson J.A., Ghabrial S.A., Taylor N.L. Natural incidence of peanut stunt virus infection in hybrid populations of Trifolium ambiguum X T. repens. Plant Dis. 1991;75:156–159. [Google Scholar]
- 7.Roossinck M.J. The big unknown: plant virus biodiversity. Curr Opin Virol. 2011;1:63–67. doi: 10.1016/j.coviro.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Roossinck M.J. Persistent plant viruses: molecular hitchhikers or epigenetic elements? In: Witzany G., editor. Viruses: Essential Agents of Life. Springer; 2012. pp. 177–186. [Google Scholar]
- 9•.Pagán I., González-Jara P., Moreno-Letelier A., Rodelo-Urrego M., Fraile A., Piñero D., García-Arenal F. Effect of biodiversity changes in disease risk: exploring disease emergence in a plant–virus system. PLoS Path. 2012;8:e1002796. doi: 10.1371/journal.ppat.1002796. [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of infection risk along a continuum of ecosystem simplification in a plant undergoing incipient domestication.
- 10•.Roossinck M.J., Saha P., Wiley G.B., Quan J., White J.D., Lai H., Chavarría F., Shen G., Roe B.A. Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol Ecol. 2010;19:81–88. doi: 10.1111/j.1365-294X.2009.04470.x. [DOI] [PubMed] [Google Scholar]; A pioneering work on virus infection patterns in wild plants in wild ecosystems.
- 11.Stobbe A.H., Roossinck M.J. Plant virus metagenomics: what we know and why we need to know more. Front Plant Sci. 2014;5:150. doi: 10.3389/fpls.2014.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Rodelo-Urrego M., Pagán I., González-Jara P., Betancourt M., Moreno-Letelier A., Ayllón M.A., Fraile A., Piñero D., García-Arenal F. Landscape heterogeneity shapes host–parasite interactions and results in apparent plant–virus co-divergence. Mol Ecol. 2013;22:2325–2340. doi: 10.1111/mec.12232. [DOI] [PubMed] [Google Scholar]; A detailed study of the role of environment variation in virus–host interactions.
- 13.Sacristán S., Fraile A., García-Arenal F. Population dynamics of Cucumber mosaic virus in melon crops and in weeds in central Spain. Phytopathology. 2004;94:992–998. doi: 10.1094/PHYTO.2004.94.9.992. [DOI] [PubMed] [Google Scholar]
- 14••.Keesing F., Belden L.K., Daszak P., Dobson A., Harvell C.D., Holt R.D., Hudson P., Jolles A., Jones K.E., Mitchell C.E. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]; Modeling of the role of changes in ecosystem complexity in disease dynamics.
- 15.Pongsiri M.J., Roman J., Ezenwa V.O., Goldbert T.L., Koren H.S., Newbold S.C., Ostfeld R.S., Pattanayak S.K., Salkeld D.J. Biodiversity loss affects global disease ecology. BioScience. 2009;59:945–954. [Google Scholar]
- 16.Mills J.N. Biodiversity loss and emerging infectious disease: an example from the rodent-borne hemorrhagic fevers. Biodiversity. 2006;7:9–17. [Google Scholar]
- 17.Bono L.M., Gensel C.L., Pfennig D.W., Burch C.L. Competition and the origins of novelty: experimental evolution of niche-width expansion in a virus. Biol Lett. 2013:9. doi: 10.1098/rsbl.2012.0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guernier V., Hochberg M.E., Guégan J.-F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers D.J., Randolph S.E. Studying the global distribution of infectious diseases using GIS and RS. Nat Rev Microbiol. 2003;1:231–237. doi: 10.1038/nrmicro776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolhouse M.E.J., Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Inf Dis. 2005;11:1843–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stukenbrock E.H., McDonald B.A. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- 24.Burdon J.J., Chilvers G.A. Host density as a factor in plant disease ecology. Annu Rev Phytopathol. 1982;20:143–166. [Google Scholar]
- 25•.Power A.G., Borer E.T., Hosseini P., Mitchell C.E., Seabloom E.W. The community ecology of barley/cereal yellow dwarf viruses in Western US grasslands. Vir Res. 2011;159:95–100. doi: 10.1016/j.virusres.2011.05.016. [DOI] [PubMed] [Google Scholar]; A good synthesis of long-term work on the ecology one of the best analysed systems.
- 26.Borer E.T., Seabloom E.W., Mitchell C.E., Power A.G. Local context drives infection of grasses by vector-borne generalist viruses. Ecol Lett. 2010;13:810–818. doi: 10.1111/j.1461-0248.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- 27.Malmstrom C.M., Hughes C.C., Newton L.A., Stoner C.J. Virus infection in remnant native bunchgrasses from invaded California grasslands. New Phytol. 2005;168:217–230. doi: 10.1111/j.1469-8137.2005.01479.x. [DOI] [PubMed] [Google Scholar]
- 28.Power A.G., Mitchell C.E. Pathogen spillover in disease epidemics. Am Nat. 2004;164:S79–S89. doi: 10.1086/424610. [DOI] [PubMed] [Google Scholar]
- 29•.Lacroix C., Jolles A., Seabloom E.W., Power A.G., Mitchell C.E., Borer E.T. Non-random biodiversity loss underlies predictable increases in viral disease prevalence. J R Soc Interf. 2014;11:20130947. doi: 10.1098/rsif.2013.0947. [DOI] [PMC free article] [PubMed] [Google Scholar]; A theoretical treatment of the reasons for increased disease prevalence in cereal virus diseases in California grasslands.
- 30.Schmidt K.A., Ostfeld R.S. Biodiversity and the dilution effect in disease ecology. Ecology. 2001;82:609–619. [Google Scholar]
- 31.Ostfeld R.S., Keesing F. Effects of host diversity on infectious disease. Annu Rev Ecol Evol Syst. 2012;43:157–182. [Google Scholar]
- 32.Osnas E.E., Dobson A.P. Evolution of virulence in heterogeneous host communitites under multiple trade-offs. Evolution. 2011;66:391–401. doi: 10.1111/j.1558-5646.2011.01461.x. [DOI] [PubMed] [Google Scholar]
- 33.Regoes R.R., Nowak M.A., Bonhoeffer S. Evolution of virulence in a heterogeneous host population. Evolution. 2000;54:64–71. doi: 10.1111/j.0014-3820.2000.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 34.Betancourt M., Escriu F., Fraile A., García-Arenal F. Virulence evolution of a generalist plant virus in a heterogeneous host system. Evol Appl. 2013;13:875–890. doi: 10.1111/eva.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vourc’h G., Plantard O., Morand S. How does biodiversity influence the ecology of infectious disease. In: Morand S., Beaudeau F., Cabaret J., editors. New Frontiers of Molecular Epidemiology of Infectious Diseases. Springer; 2012. pp. 291–309. [Google Scholar]
- 36.Duffy S., Holmes E.C. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus Tomato yellow leaf curl virus. J Virol. 2008;82:957–965. doi: 10.1128/JVI.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genetics. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 38.Bruyere A., Wantroba M., Flasinski S., Dzianott A., Bujarski J.J. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J Virol. 2000;74:4214–4219. doi: 10.1128/jvi.74.9.4214-4219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Froissart R., Roze D., Uzest M., Galibert L., Blanc S., Michalakis Y. Recombination every day: abundant recombination in a virus during a single multi-cellular host infection. PLoS Biol. 2005;3:389–395. doi: 10.1371/journal.pbio.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin H.-X., Rubio L., Smythe A.B., Falk B.W. Molecular population genetics of Cucumber mosaic virus in California: evidence for founder effects and reassortment. J Virol. 2004;78:6666–6675. doi: 10.1128/JVI.78.12.6666-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards C.T.T., Homles E.C., Pybus O.G., Wilson D.J., Viscidi R.P., Abrams E.J., Phillips R.E., Drummond A.J. Evolution of the human immunodeficiency virus envelope gene is dominated by purifying selection. Genetics. 2006;174:1441–1453. doi: 10.1534/genetics.105.052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minin V.N., S. Dorman K., Fang F., Suchard M.A. Phylogenetic mapping of recombination hotspots in human immunodeficiency virus via spatially smoothed change-point processes. Genetics. 2007;175:1773–1785. doi: 10.1534/genetics.106.066258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Losada M., Porter M., Crandall K.A. Methods for analyzing virus evolution. In: Roossinck M.J., editor. Plant Virus Evolution. Springer-Verlag; 2008. pp. 165–204. [Google Scholar]
- 45.Redelings B.D., Suchard M.A. Incorporating indel information into phylogeny estimation for rapidly emerging pathogens. BMC Evol Biol. 2007;7:40. doi: 10.1186/1471-2148-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romano C.M., Zanotto P.M.d.A., Holmes E.C. Bayesian coalescent analysis reveals a high rate of molecular evolution in GB virus C. J Mol Evol. 2008;66:292–297. doi: 10.1007/s00239-008-9087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhner M.K. Coalescent genealogy samplers: windows into population history. Trends Ecol Evol. 2008;24:86–93. doi: 10.1016/j.tree.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minin V.N., Bloomquist E., Suchard M.A. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of populaiton dynamics. Mol Biol Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmes E.C. In: The Evolution and Emergence of RNA Viruses. Harvey P.H., May R.M., editors. Oxford University Press; Oxford: 2009. [Google Scholar]
- 51.Grenfell T.B., Pybus O.G., Gog J.R., Wood J.L.N., Daly J.M., Mumford J.A., Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 52.Antia R., Regoes R., Koella J., Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaves L.F., Cohen J.M., Pascual M., Wilson M.L. Social exclusion modifies climate and deforestation impacts on a vector-borne disease. PLoS Neglected Trop Dis. 2008;2:e176. doi: 10.1371/journal.pntd.0000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lloyd-Smith J.O., Schreiber S.J., Kopp P.E., Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005:438. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plotkin J.B., Dushoff J., Fraser H.B. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature. 2004;428:942–945. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- 56.Rambaut A., Pybus O.G., Nelson M.I., VIboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parrish C.R., Holmes E.C., Morens D.M., Park E., Burke D.S., Calisher C.H., Laughlin C.A., Sair L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Micro Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao G., Bailes E., Robertson D.L., Chen Y., Rodenburg C.M., Michael S.F., Cummins L.B., Arthur L.O., Peeters M., Shaw G.M. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 59.Keele B.F., Jones J.H., Terio K.A., Estes J.D., Rudicell R.S., Wilson M.L., Li Y., Learn G.H., Beasley T.M., Schumacher-Stankey J. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.-W., Wong B.H.L., Wong S.S.Y., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory virus syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W., Shi Z., Yu M., Ren W., SMith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 62.Huhn G.D., Sejvar J.J., Montgomery S.P., Dworkin M.S. West Nile Virus in the United States: an update on an emerging infectious disease. Am Fam Phys. 2003;68:653–660. [PubMed] [Google Scholar]
- 63.Gorman O.T., Bean W.J., Webster R.G. Evolutionary processes in influenza viruses: divergence, rapid evolution, and stasis. In: Holland J.J., editor. Genetic Diversity of RNA Viruses. Springer-Verlag; 1992. pp. 75–97. [DOI] [PubMed] [Google Scholar]
- 64.Webster R.G., Bean W.J., Gorman O.T. Evolution of influenza viruses. In: Gibbs A.J., Calishe C.H., Garcia-Arenal F., editors. Molecular Basis of Viral Evolution. Cambridge University Press; 1995. pp. 531–544. [Google Scholar]
- 65.Cohen J., Enserink M. As swine flu circles globe, scientists grapple with basic questions. Science. 2009;324:572–573. doi: 10.1126/science.324_572. [DOI] [PubMed] [Google Scholar]
- 66.Guan Y., Farooqui A., Zhu H., Dong W., Wang J., Kelvin D.J. H7N9 incident, immune status, the elderly and a warning of an influenza pandemic. J Infect Dev Ctr. 2013;7:302–307. doi: 10.3855/jidc.3675. [DOI] [PubMed] [Google Scholar]
- 67.Vasilakis N., Cardosa J., Hanley K.A., Holmes E.C., Weaver S.C. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. 2011;9 doi: 10.1038/nrmicro2595. 543-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanley K.A., Monath T.P., Weaver S.C., Rossi S.L., Richman R.L., Vasilakis N. Fever versus fever: the role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infect Genet Evol. 2013;19:292–311. doi: 10.1016/j.meegid.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeffer M., Dobler G. Emergence of zoonotic arbovirus by animal trade and migration. Parasite Vector. 2010;3:35. doi: 10.1186/1756-3305-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Fargette D., Konaté G., Fauquet C., Muller E., Peterschmitt M., Thresh J.M. Molecular ecology and emergence of tropical plant viruses. Annu Rev Phytopathol. 2006;44:235–260. doi: 10.1146/annurev.phyto.44.120705.104644. [DOI] [PubMed] [Google Scholar]; Detailed information on the mergence of three important viruses in African crops.
- 71.Martin D.P., Willment J.A., Billharz R., Velders R., Odhiambo B., Njuguna J., James D., Rybicki E.P. Sequence diversity and virulence in Zea mays of Maize streak virus isolates. Virology. 2001;288:247–255. doi: 10.1006/viro.2001.1075. [DOI] [PubMed] [Google Scholar]
- 72.Tugume A.K., Cuéllar W.J., Mukasa S.B., Valkonen J.P.T. Molecular genetic analysis of virus isolates form wild and cultivated plants demonstrates that East Africa is a hotspot for the evolution and diversification of Sweet potato feathery mottle virus. Mol Ecol. 2010;19:3139–3155. doi: 10.1111/j.1365-294X.2010.04682.x. [DOI] [PubMed] [Google Scholar]
- 73.Moreno-Pérez M.G., Pagán I., Aragón-Caballero L., Cáceres F., Fraile A., García-Arenal F. Ecological and genetic determinants of Pepino mosaic virus emergence. J Virol. 2014;88:3359–3368. doi: 10.1128/JVI.02980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pardina P.E.R., Giménez M.P., Laguna I.G., Dagoberto E., Truol G. Wheat: a new natural host for the Mal de Río Cuarto virus in the endemic disease area, Río Cuarto, Córdoba Province, Argentina. Plant Dis. 1998;82:149–152. doi: 10.1094/PDIS.1998.82.2.149. [DOI] [PubMed] [Google Scholar]
- 75.Luo H., Wylie S.J., Coutts B., Jones R.A.C., Jones M.G.K. A virus of an isolated indigenous flora spreads naturally in an introduced crop species. Ann Appl Biol. 2011;159:339–347. [Google Scholar]
- 76.Woolhouse M.E.J., Haydon D.T., Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cleaveland S., Laurenson M.K., Taylor L.H. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Phil Trans R Soc B. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Daszak P., Cunningham A.A., Hyatt A.D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001;78:103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- 79.Jones R.A.C. Plant virus emergence and evolution: origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus Res. 2009;141:113–130. doi: 10.1016/j.virusres.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 80.Thresh J.M. Cropping practices and virus spread. Ann Rev Phytopathol. 1982;20:193–218. [Google Scholar]
- 81.Bosque-Pérez N.A. Eight decades of maize streak virus research. Virus Res. 2000;71:107–121. doi: 10.1016/s0168-1702(00)00192-1. [DOI] [PubMed] [Google Scholar]
- 82.Moriones E., Navas-Castillo J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000;71:123–134. doi: 10.1016/s0168-1702(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 83.Moonan F., Molina J., Mirkov T.E. Sugarcane yellow leaf virus: an emerging virus that has evolved by recombination between luteoviral and poleroviral ancestors. Virology. 2000;269:156–171. doi: 10.1006/viro.1999.0162. [DOI] [PubMed] [Google Scholar]
- 84.Verbeek M., Dullemans A.M., vandenHeuvel J.F.J.M., Maris P.C., vanderVlugt A.R.A.A. Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato. Arch Virol. 2007;152:881–890. doi: 10.1007/s00705-006-0917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gent D.H., duToit L.J., Fichtner S.F., Mohan S.K., Pappu H.R., Schwartz H.F. Iris yellow spot virus: an emergning threat to onion bulb and seed production. Plant Dis. 2006;90:1468–1480. doi: 10.1094/PD-90-1468. [DOI] [PubMed] [Google Scholar]
- 86.Levy L., Damsteegt V., Welliver R. First report of Plum pox virus (Sharka disease) in Prunus persica in the United States. Plant Dis. 2000;84:202. doi: 10.1094/PDIS.2000.84.2.202B. [DOI] [PubMed] [Google Scholar]
- 87.Jensen S.G., Lane L.C., Seifers D.L. A new disease of maize and wheat in the high plains. Plant Dis. 1996;80:1387–1390. [Google Scholar]


