Abstract
Background
The Middle East respiratory syndrome (MERS) has been reported for the first time infecting a human being since 2012. The WHO was notified of 27 countries have reported cases of MERS, the majority of these cases occur in the Arabian Peninsula, particularly in Saudi Arabia. Dromedary camels are likely to be the main source of Middle East respiratory syndrome virus (MERS-CoV) infection in humans.
Methods
MERS-CoV infection rates among camels in livestock markets and slaughterhouses were investigated in Saudi Arabia. A total of 698 nasal swabs were collected and examined with Rapid assay and rtRT-PCR. Ten MERS-CoV positive samples were subjected to full genomic sequencing. In addition, the sensitivity and specificity of the Rapid immunochromatographic assay (BioNote, South Korea) was evaluated as a diagnostic tool for MERS-CoV compared to rtRT-PCR.
Results
The results showed a high percentage of dromedaries (56.4%) had evidence for nasal MERS-CoV infection. Phylogenetic analysis of the ten MERS-CoV isolates showed that the sequences were closely related to the other MERS-CoV strains recovered from camels and human cases. Moreover, the results showed that 195 samples were positive for MERS-CoV by rapid assay compared to 394 positive samples of rtRT-PCR, which showed low rapid assay sensitivity (49.49%) while, the specificity were found to be 100%.
Conclusion
These findings indicate that these sites are a highly-hazardous to zoonotic diseases.
Keywords: MERS, Slaughterhouses, Livestock markets, Saudi Arabia
Introduction
Middle East respiratory syndrome (MERS) was first reported from Saudi Arabia in 2012, in the patient’s respiratory samples with severe pneumonia leading to acute respiratory distress syndrome and death [1]. Since September 2012, 27 countries have reported cases of MERS and WHO has been notified of 2079 laboratory-confirmed cases with at least 722 deaths by the end of August 2017 [2]. MERS coronavirus (MERS-CoV) is a novel virus that belongs to the family Coronaviridae and the genus Betacoronavirus causes the disease [3]. Human-to-human transmission appears to be limited to family and health care settings [4]. In general, a significant proportion of the cases are suspected to be a result of zoonotic transmission. Serological evidence of MERS-CoV infection of dromedaries was reported from Saudi Arabia, United Arab Emirates (UAE), Oman, Qatar, Jordan, Pakistan and Africa [2], [5], [6]. In addition, MERS-CoV RNA has been detected in nasal swabs of dromedaries in Qatar, Oman, Saudi Arabia, Egypt and UAE [2], [6], [7], [8]. Moreover, widespread circulation of different genetic variants of MERS-CoV in camels, with geographic clustering of human and camel MERS-CoV sequences [6], [9]. Few studies have provided evidence for zoonotic transmission of MERS-CoV from camels [6], [10], [11], but the mechanisms of direct or indirect zoonotic transmission have yet to be known.
Recently, OIE has certified BIONOTE® Rapid MERS-CoV Ag assay for the qualitative detection of MERS Coronavirus antigens from nasal swabs in dromedary camels with high virus titer (herd test) and as a complementary test, to estimate spread of infection to facilitate risk analysis, e.g. surveys, herd health schemes and disease control programs [12].
In this study, MERS prevalence rates in relation to locations and seasons were investigated in livestock markets and slaughterhouses in Saudi Arabia. Moreover, the sensitivity and specificity of the BIONOTE® Rapid MERS-CoV Ag test was assessed compared with the real-time reverse transcriptase PCR (rtRT-PCR).
Materials and methods
Sampling
This study was conducted from December, 2015 to August, 2017 at the Ministry of Environment, Water and Agriculture (MEWA), Riyadh, Saudi Arabia. A total of 698 dromedary camels were examined including 435 animals in the livestock markets (Jeddah, Al-Gandria, Al-Toki, Arar and Najran) and 263 animals in South Riyadh slaughterhouse (Table 1 ). Two swabs were collected from each camel, one on viral transport media (COPAN Italia, Italy), examined for rtRT-PCR and the second swab on the buffer of Rapid MERS-CoV Ag test.
Table 1.
Prevalence and seasonality rates of MERS-CoV infections in livestock markets and slaughterhouses in Saudi Arabia.
| No of samples | Time of samples collection | No of positive samples by BIONOTE® Rapid | Prevalence rate (%) | No of positive samples by RT-qPCR | Prevalence rate (%) | χ2 test | P-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Region | Jeddah livestock market (Jeddah) | 112 | December 2015 | 43 | 38.1% | 95 | 85% | 369.23 | 0.0000* |
| Al-Ganderia livestock market (Riyadh) | 100 | April 2016 | 24 | 24% | 33 | 33.7% | |||
| Arar livestock market (Northern boundaries) | 20 | June 2016 | 0 | 0% | 2 | 10% | |||
| Najran livestock market (Najran) | 14 | May 2016 | 2 | 14.3% | 4 | 28.6% | |||
| Al-Toki livestock market (Riyadh) | 50 | January 2017 | 19 | 38% | 38 | 76% | |||
| South Riyadh slaughterhouse (Riyadh) | 226 | January–February 2017 | 102 | 45.1% | 207 | 91.5% | |||
| Al-Toki livestock market (Riyadh) | 35 | May 2017 | 4 | 11.4% | 7 | 20% | |||
| Al-Toki livestock market (Riyadh) | 104 | August 2017 | 0 | 0% | 5 | 4.8% | |||
| South Riyadh slaughterhouse (Riyadh) | 37 | August 2017 | 1 | 2.7% | 3 | 8.1% | |||
| Season | |||||||||
| Winter | 537 | December–May | 189 | 35.2% | 384 | 71.5% | 214.82 | 0.0000* | |
| Summer | 161 | June–August | 6 | 3.7% | 10 | 6.2% | |||
| Age | |||||||||
| <2 years | 423 | 156 | 36.9% | 303 | 71.6 | 100.69 | 0.0001* | ||
| >2 years | 275 | 39 | 14.2% | 91 | 33% |
Non significant difference at (P > 0.05).
Significant difference at (P ≤ 0.05).
All collected nasal swabs from dromedaries on viral transport media, were transferred to the Riyadh veterinary laboratory within 24–72 h after collection to be investigated for MERS-CoV RNA by rtRT-PCR.
Camels less than two years of age were considered young, while those over two years old were considered adult. The majority camels in slaughterhouses were young (less than 2 year age). Sampling procedures were approved by the Ethics Committee of the MEWA, Saudi Arabia.
Detection of MERS-CoV by Rapid MERS-CoV Ag assay
BIONOTE® Rapid MERS-CoV Ag Test Kit is a qualitative test. It was performed for all samples according to the manufacturing protocol. Briefly, the camel nasal swabs were transferred directly into a tube containing the assay diluents. The test strip was then placed into the test tube, with the arrows on the strip pointing down, and the results were read after 15 min. The test was considered negative when only the control (C) line appeared, whereas it was considered positive when both the test line (T) and the control line (C) appeared. In the absence of the control line (C), the test was considered invalid.
Detection of MERS-CoV by rtRT-PCR and genome sequencing
MERS-CoV RNAs were extracted from nasal samples by Qiagen viral RNA extraction kit, according to the manufacturer’s protocol (Qiagen GmbH, Hilden, Germany). The rtRT-PCR targeting upstream gene (UpE) of MERS-CoV was used for screening [13]. Confirmation was done using the open reading frame (ORF) 1a. 5 μL of extracted RNA was subjected to rtRT-PCR using UpE primers using a LightMix Molecular Dx MERS-CoV upE kits (Roche) according to the manufacturer’s protocol. All positive samples by the UpE assay were then confirmed by ORF1a, previously described [14].
To investigate the genetic relationship between the ten MERS-CoV isolates and other strains whose genomes are available in GenBank, the whole-genome for ten positive rtRT-PCR samples {Riyadh (5) and Jeddah (5)} were Ion Torrent sequenced according to the primer/amplicon combinations (123 sequencing reactions) as described earlier [15]. The evolution analysis based on full genome sequencing of MERS-CoV was carried out with MEGA7. The evolutionary distances were estimated by means of the Neighbor-Joining method, based on Kimura 2-Parameter method. Bootstrap analyses were performed with 1000 repeat samples of the data sets [16].
Data management and statistical analysis
Data collected from the study animals were entered in a Microsoft excel sheet, then imported into the statistical Package for Social Sciences (SPSS) for windows® Version 22.0 (SPSS Inc., Chicago, Illinois) for statistical analyses appropriate for each variable. The association between MERS-CoV prevalence in camels and the study variables (location, Season and age) were analyzed using the 2-tailed chi-square test and logistic regression model. The statistical significance was considered when P ≤ 0.05.
Results
Detection of MERS-CoV by Rapid MERS-CoV Ag assay
The results of Rapid MERS-CoV Ag assay showed that positive samples of MERS-CoV were 23 in Jeddah livestock market, 14 in Al-Gandria, 13 in Al-Toki, 2 in Najran, 58 in South Riyadh slaughterhouse and no positive samples were recorded in Northern boundaries (Table 1). In general, the prevalence of MERS infection in camels in animal markets and slaughterhouses by rapid screening assay showed 15.8%.
Detection of MERS-CoV by rtRT-PCR and genome sequencing
MERS-CoV RNA detection rate by rtRT-PCR from nasal swabs was showed that 95 positive samples for MERS-CoV RNA in Jeddah market, 33 samples in Al-Ganderia market, 50 in Al-Toki market, 2 in Northern boundaries market, 4 in Najran market and 210 in South Riyadh slaughterhouse (Table 1). In general, the prevalence of MERS infection in camels in animal markets and slaughterhouses by rtRT-PCR was 56.4%.
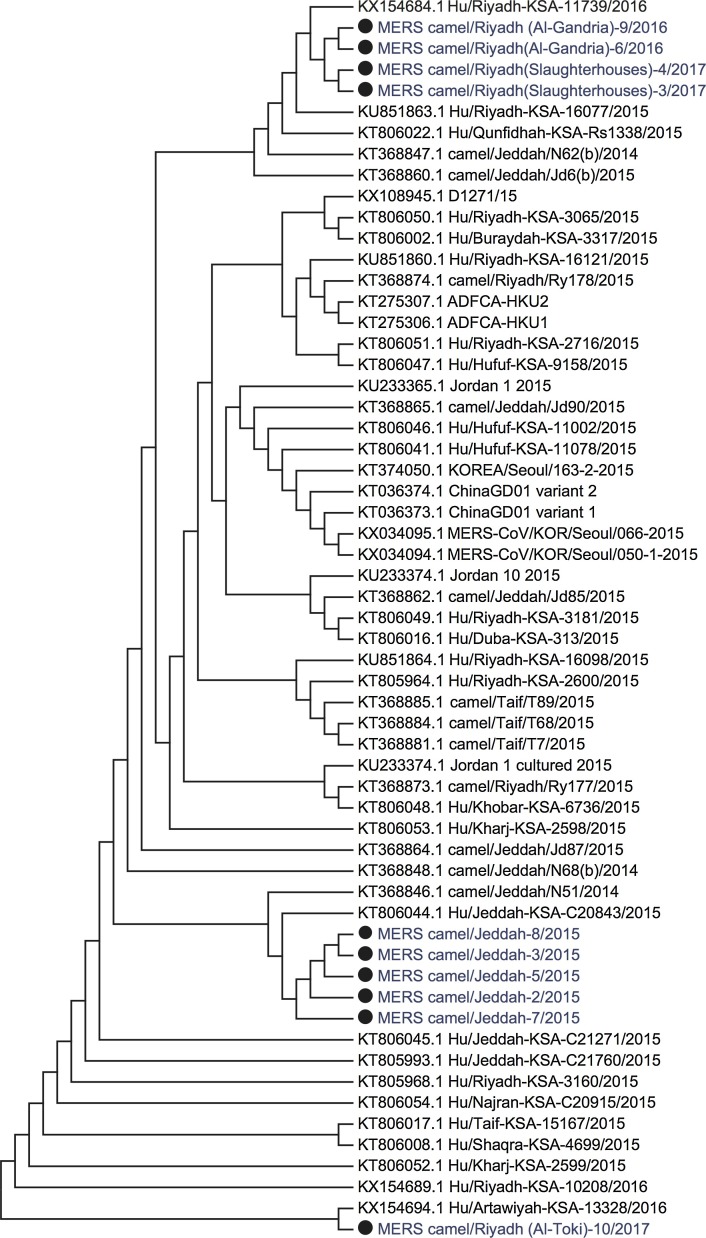
The genomic sequences obtained from ten camel’s nasal samples, were assembled with MERS-CoV genomic sequences obtained from the GenBank. The analysis showed complete similarity between MERS-CoV camel isolates and MERS-CoV sequences recovered from human cases (Fig. 1 ).
Fig. 1.
The evolutionary analysis of full genome sequences of ten MERS-CoV isolates recovered from camels using MEGA7. The ten MERS-CoV camel samples were aligned with MERS-CoV reference strains retrieved from the GenBank. The analysis was inferred using the Neighbor-Joining method and distance calculations were computed using Kimura 2-Parameter method. Sequences from the current study are indicated by solid circle.
Evaluation of Rapid MERS-CoV Ag assay with rtRT-PCR
The Rapid MERS-CoV Ag assay was used to examine 698 camel nasal swabs compared to rtRT-PCR. 195 nasal swabs were positive in the Rapid screening assay showing Rapid specificity of 100%. In addition, 199 nasal samples that were negative in the Rapid assay compared to rtRT-PCR, showed Rapid assay sensitivity of 49.49% (Table 2 ).
Table 2.
Evaluation of Rapid immunochromatographic assay with real-time PCR.
| Real-time PCR | ||||
|---|---|---|---|---|
| +ve samples | −ve samples | Total samples | ||
| Rapid IC assay | +ve samples | 195 | 0 | 195 |
| −ve samples | 199 | 304 | 503 | |
| Total samples | 394 | 304 | 698 | |
| Sensitivity% | 49.49% | |||
| Specificity% | 100% | |||
Effect of seasonality and age on the prevalence of MERS-CoV infection
The result revealed that a high prevalence rate of MERS infection in winter months, ranging from 20% in Al-Toki livestock market (May, 2017), 28.6% in Najran market, 33.7% in Al-Ganderia market, 76% in Al-Toki market (January, 2017), 85% in Jeddah market to 91.5% in South Riyadh slaughterhouse.
The prevalence of MERS infection showed a decreased pattern in summer months ranging from 4.8% in Al-Toki livestock market (August, 2017), 8.1% in South Riyadh slaughterhouse (August, 2017) to 10% in Arar livestock market.
The overall prevalence of MERS-CIV RNA in dromedaries in summer mothers (June–August) were lower either by Rapid assay (0.62%) or by rtRT-PCR (6.2%) in comparison to higher prevalence rates in winter months (December–May) by Rapid assay (20.3%) or by rtRT-PCR (71.5%) (Table 1).
The prevalence rates of MERS-CoV RNA were significantly higher in young animals (<2 years of age) indicating increased infection of young camels and high rates of positive samples recorded either by Rapid assay (22.3%) or by rtRT-PCR (71.6%). While MERS-CoV RNA prevalence rates were lower in dromedaries >2 years of age either by Rapid assay (5.8%) or by rtRT-PCR (33%) (Table 1).
Discussion
Dromedary camels are likely to be the primary source of Middle East respiratory syndrome virus (MERS-CoV) infection in humans. The routes of direct or indirect zoonotic transmission are yet unknown. In this study, the prevalence rates of MERS-CoV were investigated in dromedaries at the livestock markets and slaughterhouse in Saudi Arabia. This study was based on the screening of nasal swab samples by BIONOTE® Rapid MERS-CoV Ag Test Kit and by real-time PCR assay targeting the UpE and ORF1a genes and characterizing genetic diversity of the MERS-CoV genome sequences.
The presented study showed that high rates of MERS-CoV infection among dromedary camels in livestock markets and slaughterhouses in Saudi Arabia. The overall prevalence of MERS infection in camels in livestock markets and slaughterhouses by rtRT-PCR was 56.4%. Previous studies reported a prevalence of 29.2% of MERS-CoV infection in Al-Ahsa Province slaughterhouse [17]. Also a high proportion of 59% of dromedaries (62/105) shed MERS-CoV RNA in the central Doha animal market and adjoining slaughterhouse in Qatar [18]. In Egypt, the MERS-CoV RNA prevalence in nasal swabs at slaughterhouses was 14.7% % among 584 camels [7].
The analysis of the obtained data based on the season demonstrated that high MERS infection rates in winter months, varied from 20% in Al-Toki livestock market to 91.5% in South Riyadh slaughterhouse. While, the prevalence of MERS infection showed a decreased pattern in summer months varied from 4.8% in Al-Toki livestock market to 10% in Arar livestock market. In general, prevalence rate of MERS infection in camels in winter months by rtRT-PCR was 71.5% and summer months was 6.2%. In addition, the analysis of the data based on age showed that young camels (<2 years of age) had higher prevalence of MERS-CoV RNA (71.6%) compared with adult camels (>2 years of age) (33%), indicating increased rates of MERS-CoV infection of young camels. These results were in agreement with the previous reports, showed that the MERS-CoV infection in camels was recorded in winter season and young age camels appear to be the highest risk of MERS-CoV infection [5], [6], [7].
The sequences of MERS-CoV genome were aligned with reference sequences obtained from the GenBank. The phylogenetic analysis was performed using MEGA7. Genetic analysis revealed a complete identity between camel MERS-CoV isolates from Riyadh and Jeddah regions and MERS isolates recovered from human patients. The similarity of MERS-CoV sequences recovered from camels and human patients indicating the increased risk of potential zoonosis and camels act as intermediate hosts transmitting the virus to humans. These results are consistent with many previous studies [6], [9], [10].
The performance of commercial Rapid MERS-CoV Ag assay to detect MERS-CoV antigen was compared with the MERS UpE and open reading frame 1a (Orf1a) real-time reverse transcriptase PCR (rtRT-PCR) assay in camel nasal swabs. The data showed that Out of the 698 camel nasal swabs tested, 195 were MERS-CoV positive and 503 were negative by the ICA, while 394 were positive and 304 were negative by the UpE and Orf1a real-time RTPCR, therefore, the sensitivity and specificity of the ICA compared to those of the real-time RT-PCR were 49.49% and 100%, respectively. Previous reports recorded sensitivity and screening quality of the ICA assay found to be 93.9% and 100%, respectively, compared with that of rtRT-PCR. Also they reported that the ICA was less sensitive for the detection of MERS-CoV antigen (105 TCID50) than was the UpE real-time PCR (104 TCID50). The difference between the assay sensitivities might be regarded to the release of subgenomic RNA after the onset of cytopathogenic effect (CPE) in cell culture, including the UpE target fragment, as previously reported [13], [14], [19]. Also the Rapid MERS-CoV Ag assay cannot detect MERS-CoV loads < 104 or Ct value >30. Previous report pointed out that the MERS-CoV rtRT-PCR assay is the specific assay for molecular studies in a variety of animal species and human [12], [13]. Overall, rapid screening tests are less sensitive than are confirmatory tests; however, the advantages of using rapid screening tests are the high throughput and rapid turnaround time, without requirements of sample preparation and the use of special equipment. Therefore, the ICA is considered satisfactory to be used for screening camel herds against MERS-CoV antigen across animal markets, and slaughterhouses, followed by a confirmatory test for positive samples.
In conclusion, the high prevalence of MERS-CoV infection among dromedary camels in livestock markets and slaughterhouses, especially in winter months and young age animals, should be considered as high-risk areas for zoonosis, especially for peoples with professional contact with live camels and their bodies. In addition to these locations are the drivers of MERS-CoV circulation.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
Acknowledgements
This study was done by Ministry of Environment, Water and Agriculture (MEWA), Saudi Arabia. We thank the King Abdulaziz City for Science and Technology (KACST) for their help in conducting MERS-CoV sequencing.
References
- 1.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . WHO; Geneva, Switzerland: 2017. Middle East respiratory syndrome coronavirus (MERS-CoV)http://www.who.int/emergencies/merscov/ Available at. [Google Scholar]
- 3.Bermingham A., Chand M.A., Brown C.S. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East. EuroSurveill. 2012;17:20290. [PubMed] [Google Scholar]
- 4.ECDC . ECDC; Stockholm: 2015. Severe respiratory disease associated with Middle East respiratory syndrome coronavirus (MERS-CoV) _ seventeenth update, 11 June 2015. [Google Scholar]
- 5.Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Said M.Y. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992–2013. Emerg Infect Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasem S., Qasim I., Al-Hufofi A., Hashim O., Alkarar A., Abu-Obeida A. Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia. J Infect Public Health. 2017 doi: 10.1016/j.jiph.2017.09.022. pii: S1876-0341(17)30257-5 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali M.A., Shehata M.M., Gomaa M.R. Systematic, active surveillance for Middle East respiratory syndrome coronavirus in camels in Egypt. Emerg Microbes Infect. 2017;6(1):e1-. doi: 10.1038/emi.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saqib M., Sieberg A., Hussain M., Mansoor M., Zohaib A., Lattwein E. Serologic evidence for MERS-CoV infection in dromedary camels, Punjab, Pakistan, 2012–2015. Emerg Infect Dis. 2017;23(3):550–551. doi: 10.3201/eid2303.161285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alagaili A.N., Briese T., Mishra N., Kapoor V., Sameroff S.C., deWit E. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5 doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azhar E.I., El-Kafrawy S.A., Farraj S.A., Hassan A.M., Al-Saeed M.S., Hashem A.M.M.T. Evidence for camel-to-human transmission of MERS coronavirus. New Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. pmid:24896817. [DOI] [PubMed] [Google Scholar]
- 11.Briese T., Mishra N., Jain K., Zalmout I.S., Jabado O.J., Karesh W.B. Middle East respiratory syndrome coronavirus quasi species that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio. 2014;5(3) doi: 10.1128/mBio.01146-14. e01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.OIE . 2016. Register of diagnostic kits certified by the OIE as validated as fit for purpose.http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/OIEGS2016_Resolution15_eng.pdf Available at. [Google Scholar]
- 13.Corman V.M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 14.Corman V.M., Muller M.A., Costabel U., Timm J., Binger T., Meyer B. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17 doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 15.Graham R. 10 July 2014. MERS-CoV PCR/sequencing primers. Protocol Exchange. 10.1038/protex.2014.022. [DOI]
- 16.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalafalla A.I., Lu X., Al-Mubarak A., Dalab A.S., Al-Busadah K., Erdman D.D. MERS-CoV in upper respiratory tract and lungs of dromedary camels, Saudi Arabia, 2013–2014. Emerg Infect Dis. 2015;21(7):1153–1158. doi: 10.3201/eid2107.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farag E.A., Reusken C.B., Haagmans B.H., Khaled Mohran A.K.A., Raj V.S., Pas S.D. High proportion of MERS-CoV shedding dromedaries at slaughterhouse with a potential epidemiological link to human cases, Qatar 2014. Infect Ecol Epidemiol. 2015;5 doi: 10.3402/iee.v5.28305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song D., Ha G., Serhan W., Eltahir Y., Yusof M., Hashem F. Development and validation of a rapid immunochromatographic assay for detection of Middle East respiratory syndrome coronavirus antigen in dromedary camels. J Clin Microbiol. 2015;53:1178–1182. doi: 10.1128/JCM.03096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]