Highlights
-
•
Morbilliviruses are important human and animal pathogens.
-
•
Measles virus is the prototype and is the most infectious human pathogen on earth.
-
•
Live attenuated vaccines have been used to control the infections.
-
•
Rinderpest virus is the second virus to be eradicated from earth.
-
•
New morbilliviruses have been identified in cats and vampire bats.
Abstract
Morbilliviruses are pathogens of humans and other animals. Live attenuated morbillivirus vaccines have been used to end endemic transmission of measles virus (MV) in many parts of the developed world and to eradicate rinderpest virus. Entry is mediated by two different receptors which govern virus lymphotropism and epitheliotropism. Morbillivirus transmissibility is unparalleled and MV represents the most infectious human pathogen on earth. Their evolutionary origins remain obscure and their potential for adaption to new hosts is poorly understood. It has been suggested that MV could be eradicated. Therefore it is imperative to dissect barriers which restrict cross species infections. This is important as ecological studies identify novel morbilliviruses in a vast number of small mammals and carnivorous predators.
Current Opinion in Virology 2016, 16:95–105
This review comes from a themed issue on Emerging viruses: interspecies transmission
Edited by Christian Drosten and Yi Guan
For a complete overview see the Issue and the Editorial
Available online 26th February 2016
http://dx.doi.org/10.1016/j.coviro.2016.01.019
1879-6257/© 2016 Elsevier B.V. All rights reserved.
What makes a morbillivirus?
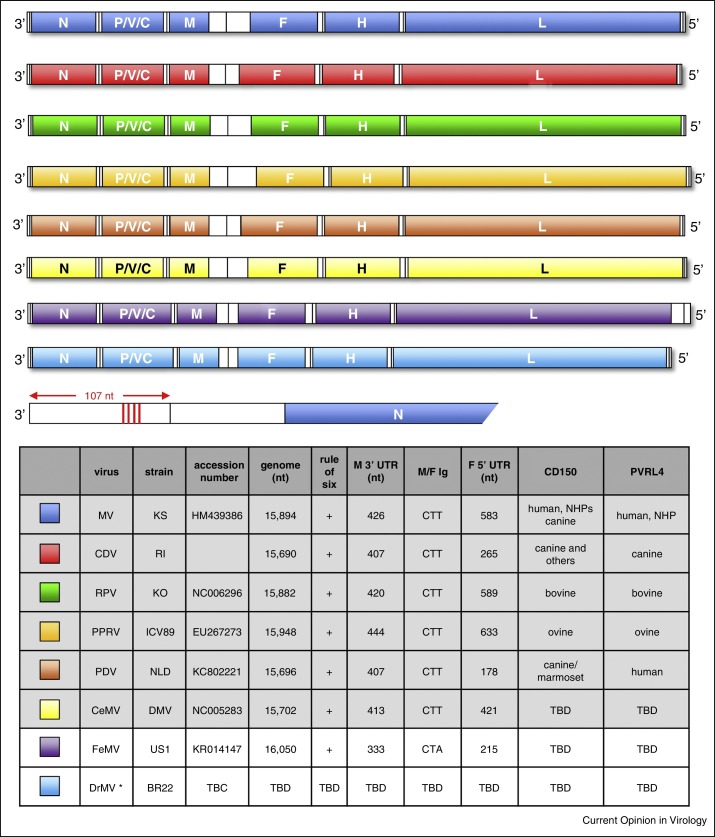
At present six members of the Morbillivirus genus are recognized by ICTV (Figure 1 ). Measles virus (MV) is the prototype species and the others are Canine distemper virus (CDV), Rinderpest virus (RPV), Peste des petits ruminants virus (PPRV), Phocine distemper virus (PDV) and Cetacean morbillivirus (CeMV). When categorizing morbillivirus attributes it is essential to focus exclusively on wild-type morbillivirus strains as overarching generalizations cannot be made if vaccine viruses and laboratory-adapted strains are included since adaptation during in vitro passage in inappropriate cell lines is common. This is best illustrated by the first, and least clinically-relevant receptor identified for MV, CD46 [1, 2] a cell surface molecule used in vitro by laboratory-adapted and vaccine strains but not by wild-type viruses [3]. Immunization of macaques with a recombinant (r) MV derived from the Edmonston-Zagreb (EZ) vaccine strain grown in CD46 expressing MRC-5 cells demonstrated that even though the vaccine virus uses CD46 in vitro this is not the case in vivo [4]. Following intramuscular injection only cells of the immune system were infected and the virus was not detected in neighboring muscle cells which express CD46. Therefore, for the purposes of generalization in this review we have selected six representative strains of known provenance, with confirmed virulence in natural hosts for which reliable full-length genomic sequences are available. These are the Khartoum Sudan (MVKS), Rhode Island US/2012 (CDVRI), Kabete ‘O’ Kenya/1910 (RPVKO), Wadden Sea NLD/1988 (PDV), Mediterranean Sea ESP/1990 (CeMV) and Côte d’Ivoire/1989 (PPRV) strains [5, 6•, 7, 8, 9]. These are well characterized viruses used in many in vitro and in vivo studies, which are representative of other wild-type strains of known provenance and we simply use them to provide specific examples of key morbillivirus molecular signatures.
Figure 1.
Schematic representation of the genomic organization of known and proposed morbilliviruses. MV = measles virus (blue), CDV = canine distemper virus (red), RPV = rinderpest virus (green), PPRV = peste des petits ruminants virus (light orange), PDV = phocine distemper (dark orange), CeMV = cetacean morbilliviruses (yellow), FeMV = feline morbillivirus (purple) and DrMV = vampire bat morbillivirus (light blue). Genomes are drawn to relative scale for comparison of the 3′ and 5′ termini, open reading frames and intergenic sequence length. Open reading frames represent the nucleocapsid (N), phospho- (P), matrix (M), fusion (F), hemagglutinin (H) and large (L) proteins which are present in the virions. The P/V/C gene encodes the non-structural V and C proteins. Four conserved hexamers in the 107 nucleotide (nt) 3′ leader sequence of MV are indicated (vertical red lines). The table lists known (gray) and proposed (white) morbilliviruses. Standard genome lengths, representative strains, proven CD150 and PVRL4 receptor use, 3′ M gene untranslated region (UTR) lengths, the sequence of the non-transcribed intergenic (Ig) trinucleotide spacer between the M and F genes, the length of the 5′ UTR of the F gene and available accession numbers on which the schematic diagrams, genome lengths etc. are based. Colors from this plate are used equivalently in figure 2 for clarity and comparability. * DrMV has not been isolated as a replicating virus.
Genotypically, morbilliviruses are negative sense, non-segmented, single-stranded, RNA viruses, with genomes ranging from 15 690 to 16 050 nucleotides containing six transcription units (Figure 1). Full-length sequences have been obtained and although they share many common features seen in viruses from other Paramyxoviridae, Filoviridae, Bornaviridae and Rhabdoviridae families, they have several defining features. Morbillivirus genomes all conform to the ‘rule of six’ [10] meaning that the total genome length is divisible by 6 and have a large untranslated region located between the open reading frames encoding the matrix (M) protein and fusion (F) glycoprotein. The 3′ non-coding termini are 107 nucleotides in length and these contain the genome sense (CN5)3 motif at nucleotide positions 79, 85 and 91 [11], the corresponding position in the 17th hexamer (nucleotide 97) is also a conserved C nucleotide (Figure 1, red lines). These key molecular signatures, along with the presence of six transcription units, and a phospho- (P) protein gene containing an overlapping open reading frame encoding the non-structural C protein and an RNA editing site which leads to the incorporation of non-templated guanosine nucleotides into mRNAs which encode the non-structural V protein [12], are sufficient to recognize a genomic sequence as that of a morbillivirus. Interestingly these criteria were not applicable to ‘equine morbillivirus’ which emerged in Australia in 1994 [13]. Even though the genome organization is similar and the rule of six is obeyed the genome is considerably longer, the M/F untranslated region is much shorter, the 3′ non-coding terminus is 112 nucleotides long and there is an A nucleotide at position 97. Shortly after its discovery this previously unidentified zoonotic pathogen, which caused a fatal disease in people and horses, was renamed Hendra virus [14] and it is now a biosafety level (BSL)-4 agent grouped in the Henipavirus genus. This episode highlights the need to pay close attention to the genomic organization of emerging pathogens and judiciously select a name which ensures clarity. It also emphasizes the weaknesses of relying on small sequences from individual genes with a high level of conservation for taxonomic purposes.
Phenotypically, morbilliviruses share a number of biological features which also should be considered when novel viruses are discovered in clinical samples during ecological surveys. First and foremost, morbilliviruses are highly lymphotropic and CD150 (also known as signaling lymphocyte activation molecule/F1) is the universal receptor which is essential to initiate an infection [15••, 16, 17, 18, 19]. The molecule is present on the cell surface of subsets of T- and B-lymphocytes, dendritic cells and macrophages. Dendritic cells and macrophages in the respiratory tract have been identified as early target cells when MV is transmitted to a new host [6•, 20]. In keeping with receptor mediated, host tropism human (h) MV uses hCD150 and bovine (b) RPV uses bCD150. This receptor is used by the vaccine virus in vivo even though CD46 can be used in vitro (see above). Although MV is first and foremost a human virus, species specificity is not exclusive, as illustrated by the fact that the wild-type readily infects marmoset (mar) B95a cells which express high levels of marCD150 [21]. Wild-type CDV uses canine (c) CD150 in vitro in Vero cells overexpressing the receptor although a change in the H glycoprotein is required for the virus to use marCD150 [22]. It could be argued that experimental confirmation of CD150-usage is non-negotiable prior to designating any closely related, novel virus as a morbillivirus. Poliovirus receptor-like 4 (PVRL4) is the second clinically-relevant morbillivirus receptor [23••, 24, 25••, 26]. The HUGO Gene Nomenclature Committee is proposing to change the gene name to NECTIN4 (nectin cell adhesion molecule 4) since the current name does describe the function of adequately. Nectin-4/PVRL4 is present at the basolateral side of epithelial cells in the adherens junction and is critical in the later stages of the morbillivirus infections, being vital for transmission to susceptible hosts [27, 28]. Significant levels of virus are present in the upper respiratory tract during the peak of infection and this contributes to the release of transmissible MV into the air [29]. Whether PVRL4/nectin-4 use should be considered as a defining characteristic of a morbillivirus in addition to CD150 use is open to question.
Where did morbilliviruses come from?
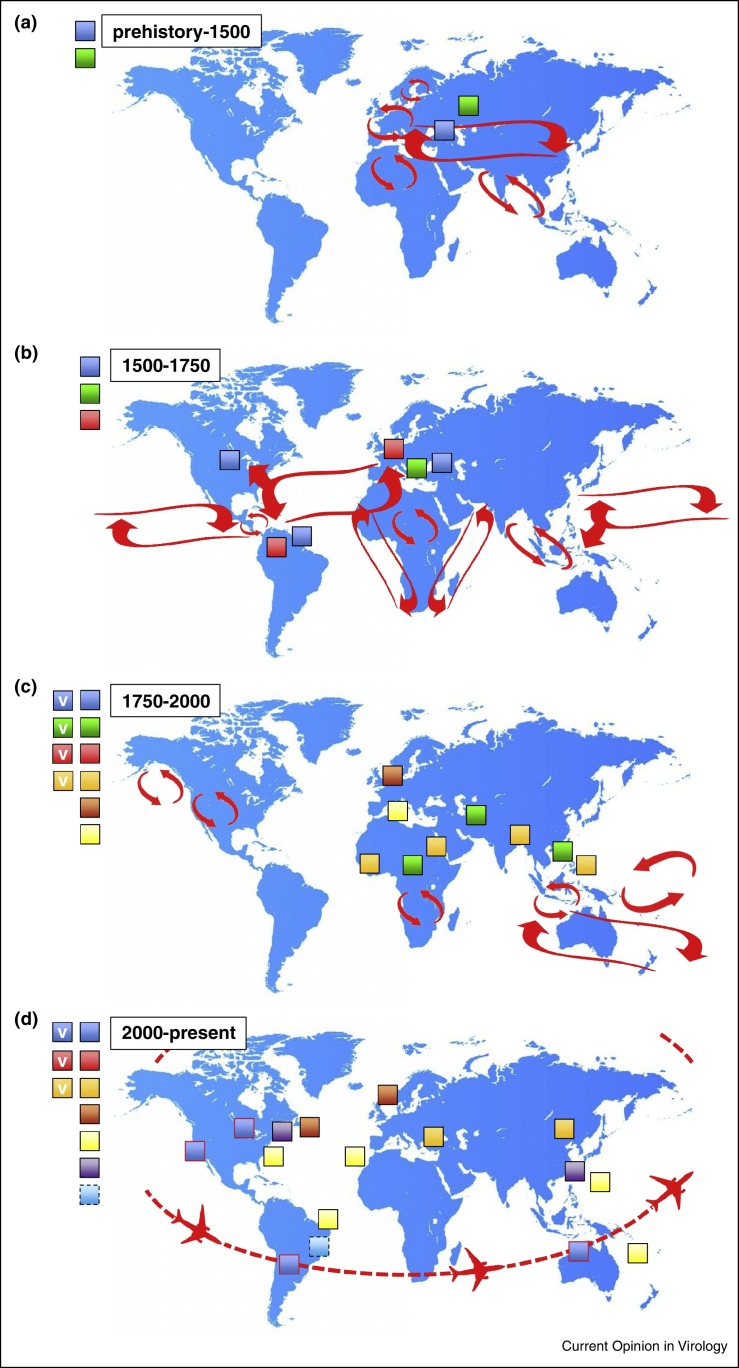
Due to the extreme transmissibility of morbilliviruses and the fact that infection leads to lifelong immunity, critical community sizes of 250 000–400 000 individuals are required to maintain endemic transmission [30]. It is likely that morbillivirus-infectable ungulates were present in larger population densities earlier in history than humans leading to the assumption that RPV, or more probably an ancient precursor, predates MV [31]. What is certain is that in terms of the written historical record these are the two ‘oldest’ morbilliviruses. Rinderpest is thought to have originated in Asia (Figure 2a) long before cattle plagues were described in the Middle East, Africa and Europe in the first millennium BC. As such rinderpest can be considered as a disease of domestication. Although cities such as Rome and Alexandria had populations of around one million at the end of the 1st Century BC Hippocrates (ca. 460–370 BC) failed to list measles in his otherwise comprehensive categorization of childhood diseases including, for example, poliomyelitis and shigellosis [32]. The first written account of the disease is in a book by Muhammad ibn Zakariyā Rāzī, also known as Rhazes of Baghdad (ca. 850–932 AD). Based on thorough clinical descriptions Rāzī discriminated measles from smallpox as the ‘smaller disease’ hence ‘morbilli’, the diminutive of morbus [33]. Therefore even though measles is often referred to as a disease of civilization there is a disconnection between the development and growth of complex human societies from around 3000 BC to the first definitive description of measles close to the end of the first millennium. These observations and genome sequence similarities have led to the suggestion that MV originated from an ancestral cattle virus which jumped species and RPV is the modern descendant of this ancient morbillivirus of ungulates (Figure 2a). Rinderpest was a devastating disease and mortality rates approach 100% in immunologically naïve herds [31, 34]. It is reasonable to assume that some degree of adaptation to humans ensued after the first putative zoonotic transmission events and it is possible that initial mortality rates were higher in immunologically naïve populations. However, evolution could have increased person-to-person transmissibility and endemicity in the Old World would have driven the development of resistance in humans which in turn should decrease morbidity and mortality rates. Such post-zoonotic viral adaptation likely facilitates endemic diseases in established populations where person-to-person transmission is mediated by direct contact. Long established trade routes between the Mediterranean, the Indian subcontinent and the rest of Asia such as the Silk Road enabled both the transportation of cargo, and disease (Figure 2a). As new westerly trade routes opened to the New World in the 16th Century large, morbillivirus-naïve populations likely encountered well-adapted, highly transmissible viruses (Figure 2b). Much has been written about the origins and devastating effect of smallpox in the New World during the Age of Exploration [35]. Likewise the arrival of MV, an even more transmissible pathogen was catastrophic and measles decimated thriving pre-Columbian populations in North, Central and South America and many Caribbean Islands, possibly by up to 95%. Mortality rates of 65% were reported as the disease spread rapidly and by the end of the 17th Century the virus was endemic in the New World. In 1657 Boston, Massachusetts experienced the first recorded outbreak in the 13 colonies and the disease seems to have been regularly reimported from Europe [36]. The city was the epicenter for frequent outbreaks and officials developed extensive contingency plans. For example, a ship arriving from Ireland in 1724 with measles patients was quarantined in Massachusetts Bay and an epidemic was avoided [36].
Figure 2.
Representation of the approximate global distribution of morbilliviruses throughout history. (a) MV (blue) and RPV (green) are the oldest morbilliviruses which were spread along ancient trade routes (red arrows). (b) Importation of MV to the New World and CDV (red) to the Old World during the Age of Exploration. (c) Spread of RPV to Africa and Asia due to the trans-border movement of cattle and establishment of MV as the first globally distributed morbillivirus (MV and CDV are not indicated for the purposes of clarity). Discovery of PPRV (light orange), PDV (dark orange) and CeMVs (yellow) in small ruminants, seals and cetaceans respectively. Development of MV, RPV, CDV and PPRV live attenuated vaccines (v). (d) Discovery FeMV (purple) a proposed novel member of the genus in Asia and the United States. Determination of the sequence of DrMV (light blue with dashed line) from clinical material obtained in Brazil. PPRV expands geographical range in Asia and Africa and is commonly isolated in Turkey and China. Resurgence of MV (blue with red line) in regions of the world where endemic transmission had ceased via air travel, areas where MV is endemic are omitted for clarity. Detection of CeMV (yellow) in a broader range of more widely distributed marine mammals. Eradication of RPV and discontinuation of vaccine use in cattle. CDV remains globally distributed and is not indicated for the purposes of clarity.
Dog distemper was first described in 1746 by Antonio de Ulloa y de la Torre-Giral (1716–1795 AD) following his travels in South America. Interestingly this is around the time a reciprocal, retrograde morbillivirus importation from the New to the Old World occurred as the ships which carried MV to the Americas brought CDV to Europe (Figure 2b). Mirroring the spread of measles in people in the Americas, CDV was reported first in Spain in the 1760s, described in England and Italy in 1764 and was present in Russia by 1770 [37•]. In 1809, Edward Jenner wrote ‘That disease among dogs which has familiarly been called ‘the distemper,’ has not hitherto, I believe, been much noticed by medical men’ and therefore he made a comprehensive description of the clinical signs and symptoms [38]. Jenner compared its transmissibility to measles, realized that puppies where particularly susceptible and recognized that survivors are protected from subsequent infection. Although the virus was recognized in dogs it infects many more species and is the most promiscuous of all known morbilliviruses, infecting ferrets, raccoons, tigers, lions, pandas and even primates [39, 40, 41, 42]. This significant zoonotic potential makes CDV an ideal model to explore barriers which restrict cross species infections, for example dissecting the role of host-specific entry receptors and assessing the impact virus assembly in and virus egress from infected cells has on intra-host spread and inter-host transmission [22, 43, 44, 45, 46, 47].
Over the following 250 years RPV, MV and CDV continued to expand their geographical ranges as new trade routes emerged and large scale transportation of livestock became possible due to the development of railroad networks. Rinderpest laid waste to massive numbers of cattle in Africa, India and Asia (Figure 2c) but the virus never made it to the New World. Measles killed significant proportions of Pacific Island dwellers and became the first verifiable global morbillivirus. However, even at the start of the 20th Century large numbers of adolescents had not been exposed to MV, demonstrating the importance of population movements and transportation in the global spread [48]. Distemper was identified as being caused by a filterable agent in 1905 by Henri Carré [49] and the English love of the domestic dog, particularly the foxhound, spurred the development of the first morbillivirus vaccine and introduced the ferret as a tractable small animal model for respiratory viruses [50••]. This in turn spawned the development of other morbillivirus vaccines in the middle of the 20th Century [51] (Figure 2c). Three new morbilliviruses were encountered, PPRV in 1942 [52], PDV in 1988 [53] and CeMV in 1991 [54] doubling the size of the genus (Figure 2c). Given the widespread distribution, multi-host infections and availability of many CDV isolates comprehensive phylogenetic relationships and nine genetic lineages have been described based on the sequence of the H gene [55]. Unsurprisingly these groups cluster geographically although their precise origins remain uncertain. Molecular phylogenetics has been used to examine how these historical clinical descriptions link to evolutionary predictions and the time to the most recent common ancestor (TMRCA) has been calculated as 1880 [56]. However, these calculations also suggest current CDV strains emerged from the United States, which is at variance with the description of the virus in Europe in the 18th Century. This highlights the challenges of modeling the evolutionary history of a virus with a highly labile RNA genome since many sequences are lost from the paleovirological record and questions have been raised about the utility of current TMRCA calculations [57]. A similar approach using whole genome sequences has been used to examine the molecular evolution of MV, RPV and PPRV [58]. This analysis suggests a TMRCA of 1904 (highest posterior density range 1730–1966) for PPRV strains and a TRMCA of 1666 (highest posterior density range 1072–1859) for MV/RPV/PPRV. An alternative molecular clock approach suggests 1074 (highest posterior density range 437–1576) as the date of divergence of RPV and MV [59]. No such analysis exists for PDV or the CeMVs although the course and outcome of the 1988 epizootic in seals suggests that this was a ‘virgin soil’ event [53]. Where the virus resides between outbreaks remains a mystery and sequence analysis suggests a reintroduction event into seals in the North Sea [60, 61]. It is likely these were spillover events and it is thus reasonable to propose the virus is maintained in other marine mammal species during the intervening period. For example interspecies transmission of CeMV from a dolphin to a captive seal has been demonstrated [62]. Atlantic harp seal populations exceed 7 million which is well in excess of the numbers to maintain endemic transmission of a morbillivirus.
In the last 15 years there have been historic changes in morbillivirus global epidemiological patterns (Figure 2d). First, the use of a highly efficacious live attenuated vaccine has led to the eradication of rinderpest, with the last case diagnosed in Kenya in 2001 [63], and formal certification in 2011 by the Food and Agriculture Agency [64]. Second, vaccination led to the elimination of measles from the Americas in 2002, defined as the absence of endemic transmission in a specific geographic area for ≥12 months in the presence of a well-performing surveillance system [65]. There has been a consistent reduction in global mortality due to measles from 562 400 in 2001 to 122 000 in 2012 and it is estimated that vaccination prevented 13.8 million deaths. The impact of the Measles and Rubella Initiative has been significant, 215 million children were vaccinated in 2014 and ambitious goals have been set for the elimination of the virus in all other WHO regions (Measles and Rubella Initiative Annual Report). Third, PPRV has expanded both its geographical range and the hosts it infects. It has spread from goats to cattle [66], has been detected in camels [67] and the Asian lineage has spread to Africa [68]. It has been speculated that RPV eradication may be driving these changes as PPRV spreads eastward to China, northward to Turkey and southward throughout Africa [58]. However, increased surveillance cannot be excluded as a potential bias of the apparently higher PPRV incidence. Fourth, CeMVs are now known to be globally distributed with viruses being isolated from dolphins in Australia and Brazil, and whales in Hawaii and the Canary Islands [53]. Fifth, CDV still circulates globally and has the potential to wreak havoc in endangered species, for example lethal infections have recently been diagnosed in Amur tigers making it a serious and emerging threat [69]. Outbreaks of CDV in China from 2006 [41] and Japan in 2008 [42] are particularly worrisome as this illustrates the potential for cross species jumps to primates. The implications of such a zoonotic infection occurring in a post-eradication MV-free world where vaccination during childhood has been discontinued could be catastrophic.
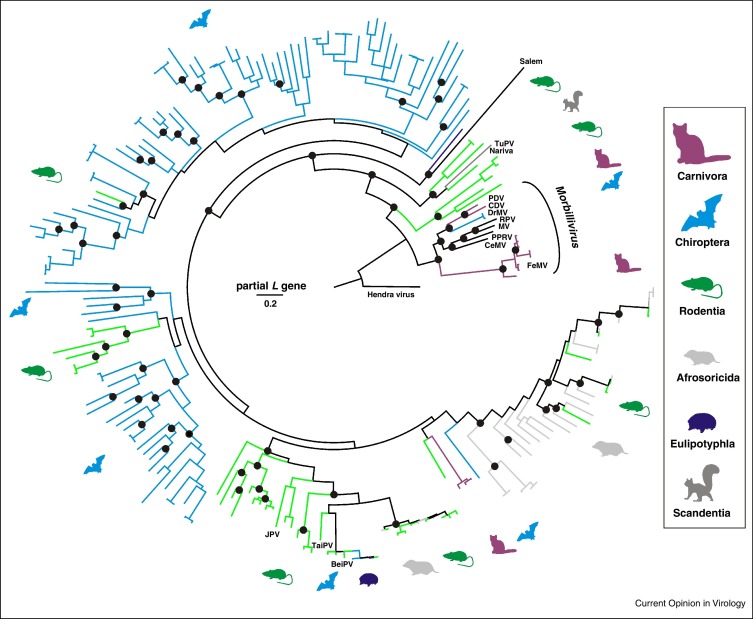
The recent description of several hundred viruses from small mammals which are phylogenetically related to the morbilliviruses, tentatively termed unclassified morbilli-related viruses (UMRV) has enabled new perspective on the origins of the genus. Most UMRV originate from bats (Figure 3 ), which is consistent with the prominent role of chiropteran hosts in paramyxovirus evolution and spread [70•]. Detection of genetically closely related viruses in members of different mammalian orders is in keeping with a low degree of species specificity of at least some of these viruses [71]. The recent descriptions of feline paramyxoviruses clustering phylogenetically within the UMRV in an intermediate position between the genus Morbillivirus sensu strictu may hint at the acquisition of UMRVs by carnivore predators from small mammals. The relevance of small mammals and their carnivorous predators is consistent with their representation as morbillivirus hosts in the form of CDV, PDV and a partially characterized Brazilian vampire bat (Desmodus rotundus) morbillivirus (DrMV) [70•]. The genetic diversity of UMRV outnumbers that of morbilliviruses many fold and it is tempting to speculate that all morbilliviruses have ancestral origins in small mammals. The ecological scenario of carnivores acquiring viruses from their prey is in line with the evolutionary origins of rabies virus in chiropteran hosts followed by an introduction into canids [72] and the acquisition of SARS-coronavirus by civets from bats [73•]. Whether carnivores function as an entry point for viruses previously restricted to small mammals to infect a wider range of mammalian hosts is unclear. However, this is not without precedent as alteration of the SARS-CoV glycoprotein during passage in civets has been hypothesized [74] and rabies viruses acquired from insectivorous bats have established endemic circulation in several canids after the initial host switch [75]. Within the order Mononegavirales, the relevance of small mammal hosts in general and chiropteran hosts in particular, is demonstrated by the evolutionary origins of the closely related family Filoviridae in bats [76, 77].
Figure 3.
Phylogenetic relationships of morbilliviruses and related small mammal viruses. Bayesian phylogenetic reconstruction done using MrBayes V3.1 with 2 000 000 tree iterations sampled every 100 steps, corresponding to 20 000 trees of which 25% were discarded as burn-in. The final tree was annotated using TreeAnnotator and visualized using FigTree from the BEAST package. Hendra virus was used as an outgroup. Host orders are indicated by color and pictograms to the right. Prototype morbilliviruses and related viruses are indicated next to branches (JPV, J virus; BeiPV, Beilong virus; TuPV, Tupaia paramyxovirus; see text for other abbreviations). All available unclassified morbilli-related viruses (UMRV) that differed by more than 5% in their partial L gene sequences from any other virus, were from different hosts or different sampling countries were included into the analysis. The final dataset comprised 205 partial L gene sequences of 438 nucleotides generated by the RT-PCR assay described by [93] commonly used in paramyxovirus field studies.
Whither morbilliviruses might go?
Knowing what morbilliviruses are, where they originated, how they have spread globally and that eradication is possible drives us to consider future evolutionary trajectories as new members of the genus are discovered and old members are eliminated. Successful eradication of RPV provides an impetus to eliminate MV and PPRV [64, 78•]. However, eradication of pathogens which have coexisted with humans and animals for thousands of years could also lead to unforeseen risks. Morbilliviruses are antigenically similar enough to permit dogs vaccinated with attenuated MV, to be protected from clinical signs of CDV following challenge [79, 80]. Concerns have been raised that circulating morbilliviruses could spill over into naïve populations, for example by broadening their CD150 usage [39, 78•]. Adaptation studies to understand the likelihood of switches in viral tropism under accelerated evolutionary pressures would help address this issue. Even though RPV, PPRV and CDV infect multiple species there seems to be a significant barrier for these modern morbilliviruses to infect people and cause clinical disease. Clearly, CDV was present in South America before MV was imported from Europe and for whatever reason the virus failed to jump species to morbillivirus-naïve humans. Likewise PPRV causes subclinical infections of large ruminants in regions where RPV was eliminated and the virus failed to increase transmission or virulence. This indicates that CD150 use is not the only hurdle and intracellular factors are likely to play a significant role stopping cross species transmission events. This has been demonstrated experimentally by naturally infecting macaques with virulent CDV [45]. Although the virus infected many immune cells of the monkeys it was cleared rapidly and neurological signs were not observed. Why thousands of non-human primates succumbed to CDV infection in China [41] remains an enigma which needs to be addressed. A major challenge for MV eradication stems from resurgence of the virus in regions of the world where endemic transmission has been stopped (Figure 2d) due to the failure to use what is a safe and highly efficacious vaccine in some communities. Unsubstantiated links between the vaccine and a host of medical conditions alongside an underestimation of the severity of the disease has led to large outbreaks in the developed world [81] with the inevitable concomitant measles-related deaths.
At the same time RPV was certified as eradicated another candidate morbillivirus was discovered [82•]. Feline morbillivirus (FeMV) is present in urine and blood samples from domestic cats in Hong Kong (Figure 2d), full-length genome sequences have been reported in Japan [83, 84] and we have shown it is present in the United States and causes a chronic infection [85•]. However, if the criteria for ‘what makes a morbillivirus?’ are applied to FeMV only some are met, for example although the virus obeys the rule of six the M/F non-coding sequence is much smaller, there is an unusually long 3′ untranslated region in the L gene and importantly usage of feline CD150 has not been demonstrated. As noted above, FeMV clusters phylogenetically in basal sister relationship to classical morbilliviruses, exceeding the phylogenetic diversity within the genus. Lack of diversification within FeMV strains may suggest a limited evolutionary history within feline hosts after a putative host switch from small mammals.
A correlation with tubulointerstitial nephritis (TIN) has been suggested although Koch's postulates have not been tested for FeMV in morbillivirus-naïve cats. CDV infects the bladder [86] and although throat swabs are the optimal specimen for detecting MV in clinical samples, the virus is also present in urine [87]. Acute renal failure concomitant with MV isolation in three patients with neurological complications has been reported although correlation does not equate to causation [88]. To date TIN has not been associated with any morbillivirus infection making the purported FeMV pathogenesis very different from the other viruses in the genus. Efforts should be made to address these important questions given TIN has a major impact on the quality of life of the 74–96 million domestic cats owned in the United States. This population size is more than sufficient to maintain endemic transmission of a morbillivirus even if lifelong immunity follows the acute infection. Whether this proposed morbillivirus makes it into the genus remains to be seen.
Ecological studies, degenerate reverse transcription PCR amplification and unbiased next generation sequencing approaches are being used to identify evolutionary ancestral viruses in a diversity of wildlife species from across the globe [70•]. These approaches have radically altered our view of the number of potential viruses in the global ecosystem. However, given the challenges obtaining and storing clinical material, phylogenetic analyses often rely on short, highly conserved sequences which although useful for assessing the breadth and diversity tend not to be accompanied by biological isolates. Sampling tends to be focused on rodents and bats which could skew the phylogenetic analysis considerably and ideally the types of species examined should be expanded. Vampire bats sampled in Brazil [70•] contained viruses which were very closely related to CDV and PDV (Figure 2, Figure 3). It is attractive to speculate that CDV and DrMV might share a common South American ancestor. However, in the absence of a DrMV vampire bat isolate and a complete genome sequence this hypothesis is challenging to test. Likewise, applying the ‘what makes a morbillivirus?’ test in terms of genome organization and receptor usage is not yet possible for DrMV. This highlights a major challenge in virus discovery, the gap between the identification of genomic fragments in clinical samples and understanding the biological properties of pathogens which could be impossible to isolate [89]. Synthetic biology offers an opportunity to bridge this gap and given the success with assembling reverse genetics systems for paramyxoviruses directly from unpassaged clinical material [90] it should be possible to develop completely synthetic reverse genetics systems as have been generated for non-segmented positive strand RNA viruses [91••] and segmented negative strand RNA viruses [92•]. For putative morbilliviruses such as DrMV this might be the only tractable option to obtain a cultivatable isolate. Piloting these technologies with a view to developing a pipeline to connect virus discovery and biological analyses will have a significant impact on understanding virus evolution and the propensity for cross-species jumps in the wild.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We would like to thank Bert Rima, Linda Rennick and Rik de Swart for helpful suggestions during the preparation of the manuscript, David Wilkinson for advice on UMRV diversity and Ashley Banyard and Dalan Bailey for their insights into the dynamic interactions between PPRV and RPV in the developing world. This work was financially supported by a Defense Advanced Research Projects Agency (DARPA) Prophecy Program award to WPD and JFD (HR0011-13-2-0020).
References
- 1.Dörig R.E., Marcil A., Chopra A., Richardson C.D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 2.Naniche D., Varior-Krishnan G., Cervoni F., Wild T.F., Rossi B., Rabourdin-Combe C., Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi K., Nagata N., Kato S.I., Ami Y., Suzaki Y., Suzuki T., Sato Y., Tsunetsugu-Yokota Y., Mori K., Van Nguyen N. Wild-type measles virus with the hemagglutinin protein of the edmonston vaccine strain retains wild-type tropism in macaques. J Virol. 2012;86:3027–3037. doi: 10.1128/JVI.06517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rennick L.J., de Vries R.D., Carsillo T.J., Lemon K., van Amerongen G., Ludlow M., Nguyen D.T., Yuksel S., Verburgh R.J., Haddock P. Live-attenuated measles virus vaccine targets dendritic cells and macrophages in muscle of nonhuman primates. J Virol. 2015;89:2192–2200. doi: 10.1128/JVI.02924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rima B.K., Collin A.M., Earle J.A. Completion of the sequence of a cetacean morbillivirus and comparative analysis of the complete genome sequences of four morbilliviruses. Virus Genes. 2005;30:113–119. doi: 10.1007/s11262-004-4588-7. [DOI] [PubMed] [Google Scholar]
- 6•.Lemon K., de Vries R.D., Mesman A.W., McQuaid S., van Amerongen G., Yuksel S., Ludlow M., Rennick L.J., Kuiken T., Rima B.K. Early target cells of measles virus after aerosol infection of non-human primates. PLoS Pathogens. 2011;7:e1001263. doi: 10.1371/journal.ppat.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identifies the primary target cells for wild-type measles virus in vivo.
- 7.Chard L.S., Bailey D.S., Dash P., Banyard A.C., Barrett T. Full genome sequences of two virulent strains of peste-des-petits ruminants virus, the Cote d’Ivoire 1989 and Nigeria 1976 strains. Virus Res. 2008;136:192–197. doi: 10.1016/j.virusres.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Plowright W., Ferris R.D. Studies with rinderpest virus in tissue culture. III. The stability of cultured virus and its use in virus neutralization tests. Arch Gesamte Virusforsch. 1962;11:516–533. doi: 10.1007/BF01241304. [DOI] [PubMed] [Google Scholar]
- 9.de Vries R.D., Verburgh R.J., van de Bildt M.W., Osterhaus A.D., de Swart R.L. Complete genome sequence of phocine distemper virus isolated from a Harbor Seal (Phoca vitulina) during the 1988 North Sea Epidemic. Genome Announc. 2013:1. doi: 10.1128/genomeA.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calain P., Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tapparel C., Maurice D., Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rima B.K., Duprex W.P. The measles virus replication cycle. Curr Top Microbiol Immunol. 2009;329:77–102. doi: 10.1007/978-3-540-70523-9_5. [DOI] [PubMed] [Google Scholar]
- 13.Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 14.Murray K.A., Eaton B.T., Hooper P., Wang L., Williamson M., Young P. In: Flying foxes, horses, and humans: a zoonosis caused by a new member of the Paramyxoviridae. Scheld W.M., Armstrong D., Hughes J.M., editors. ASM Press; Washington, D.C.: 1998. [Google Scholar]
- 15••.Tatsuo H., Ono N., Tanaka K., Yanagi Y. SLAM (CDw150) is a cellular receptor for measles virus. Nature. 2000;406:893–897. doi: 10.1038/35022579. [DOI] [PubMed] [Google Scholar]; Identification of CD150 as the major cellular receptor for morbilliviruses.
- 16.Erlenhoefer C., Wurzer W.J., Loffler S., Schneider-Schaulies S., Ter M.V., Schneider-Schaulies J. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J Virol. 2001;75:4499–4505. doi: 10.1128/JVI.75.10.4499-4505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu E.C., Iorio C., Sarangi F., Khine A.A., Richardson C.D. CDw150(SLAM) is a receptor for a lymphotropic strain of measles virus and may account for the immunosuppressive properties of this virus. Virology. 2001;279:9–21. doi: 10.1006/viro.2000.0711. [DOI] [PubMed] [Google Scholar]
- 18.Tatsuo H., Ono N., Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J Virol. 2001;75:5842–5850. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baron M.D. Wild-type Rinderpest virus uses SLAM (CD150) as its receptor. J Gen Virol. 2005;86:1753–1757. doi: 10.1099/vir.0.80836-0. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira C.S., Frenzke M., Leonard V.H., Welstead G.G., Richardson C.D., Cattaneo R. Measles virus infection of alveolar macrophages and dendritic cells precedes spread to lymphatic organs in transgenic mice expressing human signaling lymphocytic activation molecule (SLAM, CD150) J Virol. 2010;84:3033–3042. doi: 10.1128/JVI.01559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobune F., Sakata H., Sugiura A. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J Virol. 1990;64:700–705. doi: 10.1128/jvi.64.2.700-705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki F., Ono N., Yamaguchi R., Yanagi Y. Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine SLAM (CD150) and their adaptability to marmoset B95a cells. J Virol. 2003;77:9943–9950. doi: 10.1128/JVI.77.18.9943-9950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Muhlebach M.D., Mateo M., Sinn P.L., Prufer S., Uhlig K.M., Leonard V.H., Navaratnarajah C.K., Frenzke M., Wong X.X., Sawatsky B. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes the identification of PVRL4 (Nectin 4) as a cellular receptor for MV on epithelial cells.
- 24.Mateo M., Navaratnarajah C.K., Syed S., Cattaneo R. The measles virus hemagglutinin beta-propeller head beta4-beta5 hydrophobic groove governs functional interactions with nectin-4 and CD46 but not those with the signaling lymphocytic activation molecule. J Virol. 2013;87:9208–9216. doi: 10.1128/JVI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Noyce R.S., Bondre D.G., Ha M.N., Lin L.T., Sisson G., Tsao M.S., Richardson C.D. Tumor cell marker PVRL4 (Nectin 4) is an epithelial cell receptor for measles virus. PLoS pathogens. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes the identification of PVRL4 (nectin 4) as a cellular receptor for MV on epithelial cells.
- 26.Birch J., Juleff N., Heaton M.P., Kalbfleisch T., Kijas J., Bailey D. Characterization of ovine Nectin-4, a novel peste des petits ruminants virus receptor. J Virol. 2013;87:4756–4761. doi: 10.1128/JVI.02792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludlow M., Lemon K., de Vries R.D., McQuaid S., Millar E.L., van Amerongen G., Yuksel S., Verburgh R.J., Osterhaus A.D., de Swart R.L. Measles virus infection of epithelial cells in the macaque upper respiratory tract is mediated by sub-epithelial immune cells. J Virol. 2013 doi: 10.1128/JVI.03258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frenzke M., Sawatsky B., Wong X.X., Delpeut S., Mateo M., Cattaneo R., von Messling V. Nectin-4-dependent measles virus spread to the cynomolgus monkey tracheal epithelium: role of infected immune cells infiltrating the lamina propria. J Virol. 2013;87:2526–2534. doi: 10.1128/JVI.03037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludlow M., de Vries R.D., Lemon K., McQuaid S., Millar E., van Amerongen G., Yuksel S., Verburgh R.J., Osterhaus A.D., de Swart R.L. Infection of lymphoid tissues in the macaque upper respiratory tract contributes to the emergence of transmissible measles virus. J Gen Virol. 2013;94:1933–1944. doi: 10.1099/vir.0.054650-0. [DOI] [PubMed] [Google Scholar]
- 30.Keeling M.J., Grenfell B.T. Disease extinction and community size: modeling the persistence of measles. Science. 1997;275:65–67. doi: 10.1126/science.275.5296.65. [DOI] [PubMed] [Google Scholar]
- 31.Barrett T., Rossiter P.B. Rinderpest: the disease and its impact on humans and animals. Adv Virus Res. 1999;53:89–110. doi: 10.1016/s0065-3527(08)60344-9. [DOI] [PubMed] [Google Scholar]
- 32.Pappas G., Kiriaze I.J., Falagas M.E. Insights into infectious disease in the era of Hippocrates. Int J Infect Dis. 2008;12:347–350. doi: 10.1016/j.ijid.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch A. New Syndenham Society; London: 1883. Handbook of Geographical and Historical Pathology: Acute Infective Diseases. [Google Scholar]
- 34.Barrett T. Morbillivirus infections, with special emphasis on morbilliviruses of carnivores. Vet Microbiol. 1999;69:3–13. doi: 10.1016/s0378-1135(99)00080-2. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Carroll D.S., Gardner S.N., Walsh M.C., Vitalis E.A., Damon I.K. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caulfield E. Early measles epidemics in America. Yale J Biol Med. 1943;15:531–556. [PMC free article] [PubMed] [Google Scholar]
- 37•.Blancou J. Dog distemper: imported into Europe from South America? Historia medicinae veterinariae. 2004;29:35–41. [PubMed] [Google Scholar]; A comprehensive overview of the history and possible origins of CDV.
- 38.Jenner E. Observations on the Distemper in Dogs. Med Chir Trans. 1809;1:265–270. [PMC free article] [PubMed] [Google Scholar]
- 39.Ludlow M., Rennick L.J., Nambulli S., de Swart R.L., Duprex W.P. Using the ferret model to study morbillivirus entry, spread, transmission and cross-species infection. Curr Opin Virol. 2014;4:15–23. doi: 10.1016/j.coviro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z., Li A., Ye H., Shi Y., Hu Z., Zeng L. Natural infection with canine distemper virus in hand-feeding Rhesus monkeys in China. Vet Microbiol. 2010;141:374–378. doi: 10.1016/j.vetmic.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 41.Qiu W., Zheng Y., Zhang S., Fan Q., Liu H., Zhang F., Wang W., Liao G., Hu R. Canine distemper outbreak in rhesus monkeys, China. Emerging Infectious Diseases. 2011;17:1541–1543. doi: 10.3201/eid1708.101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai K., Nagata N., Ami Y., Seki F., Suzaki Y., Iwata-Yoshikawa N., Suzuki T., Fukushi S., Mizutani T., Yoshikawa T. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol. 2013;87:1105–1114. doi: 10.1128/JVI.02419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pratakpiriya W., Seki F., Otsuki N., Sakai K., Fukuhara H., Katamoto H., Hirai T., Maenaka K., Techangamsuwan S., Lan N.T. Nectin4 is an epithelial cell receptor for canine distemper virus and involved in neurovirulence. J Virol. 2012;86:10207–10210. doi: 10.1128/JVI.00824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawatsky B., Wong X.X., Hinkelmann S., Cattaneo R., von Messling V. Canine distemper virus epithelial cell infection is required for clinical disease but not for immunosuppression. J Virol. 2012;86:3658–3666. doi: 10.1128/JVI.06414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Vries R.D., Ludlow M., Verburgh R.J., van Amerongen G., Yuksel S., Nguyen D.T., McQuaid S., Osterhaus A.D., Duprex W.P., de Swart R.L. Measles vaccination of nonhuman primates provides partial protection against infection with canine distemper virus. J Virol. 2014;88:4423–4433. doi: 10.1128/JVI.03676-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohishi K., Suzuki R., Maeda T., Tsuda M., Abe E., Yoshida T., Endo Y., Okamura M., Nagamine T., Yamamoto H. Recent host range expansion of canine distemper virus and variation in its receptor, the signaling lymphocyte activation molecule, in carnivores. J Wildl Dis. 2014;50:596–606. doi: 10.7589/2013-09-228. [DOI] [PubMed] [Google Scholar]
- 47.Bieringer M., Han J.W., Kendl S., Khosravi M., Plattet P., Schneider-Schaulies J. Experimental adaptation of wild-type Canine Distemper Virus (CDV) to the human entry receptor CD150. PLoS One. 2013;8:e57488. doi: 10.1371/journal.pone.0057488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morens D.M., Taubenberger J.K. A forgotten epidemic that changed medicine: measles in the US Army, 1917-18. Lancet Infect Dis. 2015;15:852–861. doi: 10.1016/S1473-3099(15)00109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffin D.E. Measles Virus. In: Knipe D.M., Howley P.M., editors. 6th edition. vol 1. Williams & Wilkins; Lippincott: 2013. pp. 1042–1069. (Fields Virology). [Google Scholar]
- 50••.Bresalier M., Worboys M. ‘Saving the lives of our dogs’: the development of canine distemper vaccine in interwar Britain. Br J Hist Sci. 2014;47:305–334. doi: 10.1017/S0007087413000344. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive account of the development of the first morbillivirus vaccine in the first part of the 20th Century demonstrating the utilty of public private partnerships.
- 51.Buczkowski H., Muniraju M., Parida S., Banyard A.C. Morbillivirus vaccines: recent successes and future hopes. Vaccine. 2014;32:3155–3161. doi: 10.1016/j.vaccine.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banyard A.C., Parida S., Batten C., Oura C., Kwiatek O., Libeau G. Global distribution of peste des petits ruminants virus and prospects for improved diagnosis and control. J Gen Virol. 2010;91:2885–2897. doi: 10.1099/vir.0.025841-0. [DOI] [PubMed] [Google Scholar]
- 53.Duignan P.J., Van Bressem M.F., Baker J.D., Barbieri M., Colegrove K.M., De Guise S., de Swart R.L., Di Guardo G., Dobson A., Duprex W.P. Phocine distemper virus: current knowledge and future directions. Viruses. 2014;6:5093–5134. doi: 10.3390/v6125093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Bressem M.F., Duignan P.J., Banyard A., Barbieri M., Colegrove K.M., De Guise S., Di Guardo G., Dobson A., Domingo M., Fauquier D. Cetacean morbillivirus: current knowledge and future directions. Viruses. 2014;6:5145–5181. doi: 10.3390/v6125145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolt G., Jensen T.D., Gottschalck E., Arctander P., Appel M.J., Buckland R., Blixenkrone-Moller M. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol. 1997;78:367–372. doi: 10.1099/0022-1317-78-2-367. [DOI] [PubMed] [Google Scholar]
- 56.Panzera Y., Sarute N., Iraola G., Hernandez M., Perez R. Molecular phylogeography of canine distemper virus: Geographic origin and global spreading. Mol Phylogenet Evol. 2015;92:147–154. doi: 10.1016/j.ympev.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 57.Sharp P.M., Simmonds P. Evaluating the evidence for virus/host co-evolution. Curr Opin Virol. 2011;1:436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Muniraju M., Munir M., Parthiban A.R., Banyard A.C., Bao J., Wang Z., Ayebazibwe C., Ayelet G., El Harrak M., Mahapatra M. Molecular evolution of peste des petits ruminants virus. Emerging Infect Dis. 2014;20:2023–2033. doi: 10.3201/eid2012.140684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furuse Y., Suzuki A., Oshitani H. Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virol J. 2010;7:52. doi: 10.1186/1743-422X-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nielsen L., Arctander P., Jensen T.H., Dietz H.H., Hammer A.S., Banyard A.C., Barrett T., Blixenkrone-Moller M. Genetic diversity and phylogenetic analysis of the attachment glycoprotein of phocine distemper viruses of the 2002 and 1988 epizootics. Virus Res. 2009;144:323–328. doi: 10.1016/j.virusres.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 61.Bodewes R., Morick D., van de Bildt M.W., Osinga N., Rubio Garcia A., Sanchez Contreras G.J., Smits S.L., Reperant L.A., Kuiken T., Osterhaus A.D. Prevalence of phocine distemper virus specific antibodies: bracing for the next seal epizootic in north-western Europe. Emerg Microbes Infect. 2013;2:e3. doi: 10.1038/emi.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazzariol S., Peletto S., Mondin A., Centelleghe C., Di Guardo G., Di Francesco C.E., Casalone C., Acutis P.L. Dolphin morbillivirus infection in a captive harbor seal (Phoca vitulina) J Clin Microbiol. 2013;51:708–711. doi: 10.1128/JCM.02710-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roeder P.L. Rinderpest: the end of cattle plague. Prev Vet Med. 2011;102:98–106. doi: 10.1016/j.prevetmed.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 64.de Swart R.L., Duprex W.P., Osterhaus A.D. Rinderpest eradication: lessons for measles eradication? Curr Opin Virol. 2012;2:330–334. doi: 10.1016/j.coviro.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 65.Perry R.T., Gacic-Dobo M., Dabbagh A., Mulders M.N., Strebel P.M., Okwo-Bele J.M., Rota P.A., Goodson J.L. Global control and regional elimination of measles, 2000-2012. MMWR. Morbidity and mortality weekly report. 2014;63:103–107. [PMC free article] [PubMed] [Google Scholar]
- 66.Lembo T., Oura C., Parida S., Hoare R., Frost L., Fyumagwa R., Kivaria F., Chubwa C., Kock R., Cleaveland S. Peste des petits ruminants infection among cattle and wildlife in northern Tanzania. Emerging Infect Diseases. 2013;19:2037–2040. doi: 10.3201/eid1912.130973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saeed I.K., Ali Y.H., AbdulRahman M.B., Mohammed Z.A., Osman H.M., Taha K.M., Musa M.Z., Khalafalla A.I. Mixed infection of peste des petits ruminants virus (PPRV) and other respiratory viruses in dromedary camels in Sudan, an abattoir study. Trop Anim Health Prod. 2015;47:995–998. doi: 10.1007/s11250-015-0798-3. [DOI] [PubMed] [Google Scholar]
- 68.Kwiatek O., Ali Y.H., Saeed I.K., Khalafalla A.I., Mohamed O.I., Obeida A.A., Abdelrahman M.B., Osman H.M., Taha K.M., Abbas Z. Asian lineage of peste des petits ruminants virus, Africa. Emerging Infect Diseases. 2011;17:1223–1231. doi: 10.3201/eid1707.101216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilbert M., Miquelle D.G., Goodrich J.M., Reeve R., Cleaveland S., Matthews L., Joly D.O. Estimating the potential impact of canine distemper virus on the Amur tiger population (Panthera tigris altaica) in Russia. PLoS One. 2014;9:e110811. doi: 10.1371/journal.pone.0110811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Drexler J.F., Corman V.M., Muller M.A., Maganga G.D., Vallo P., Binger T., Gloza-Rausch F., Rasche A., Yordanov S., Seebens A. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes the identification a large number of negative strand RNA viruses in bats and rodents and provides evidence for a vampire bat morbillivirus.
- 71.Wilkinson D.A., Melade J., Dietrich M., Ramasindrazana B., Soarimalala V., Lagadec E., le Minter G., Tortosa P., Heraud J.M., de Lamballerie X. Highly diverse morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean Islands: evidence of exchange between introduced and endemic small mammals. J Virol. 2014;88:8268–8277. doi: 10.1128/JVI.01211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Badrane H., Tordo N. Host switching in Lyssavirus history from the Chiroptera to the Carnivora orders. J Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]; This work describes the identification the animal resevior for SARS.
- 74.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuzmin I.V., Shi M., Orciari L.A., Yager P.A., Velasco-Villa A., Kuzmina N.A., Streicker D.G., Bergman D.L., Rupprecht C.E. Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001-2009. PLoS Pathogens. 2012;8:e1002786. doi: 10.1371/journal.ppat.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 77.Swanepoel R., Smit S.B., Rollin P.E., Formenty P., Leman P.A., Kemp A., Burt F.J., Grobbelaar A.A., Croft J., Bausch D.G. Studies of reservoir hosts for Marburg virus. Emerging Infect Diseases. 2007;13:1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78•.Morens D.M., Holmes E.C., Davis A.S., Taubenberger J.K. Global rinderpest eradication: lessons learned and why humans should celebrate too. J Infect Dis. 2011;204:502–505. doi: 10.1093/infdis/jir327. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review on rinderpest erradication.
- 79.Strating A. Measles vaccine in dogs: efficacy against aerosol challenge with virulent canine distemper virus. J Am Vet Med Assoc. 1975;167:59–62. [PubMed] [Google Scholar]
- 80.Appel M.J., Shek W.R., Shesberadaran H., Norrby E. Measles virus and inactivated canine distemper virus induce incomplete immunity to canine distemper. Arch Virol. 1984;82:73–82. doi: 10.1007/BF01309369. [DOI] [PubMed] [Google Scholar]
- 81.Ludlow M., McQuaid S., Milner D., de Swart R.L., Duprex W.P. Pathological consequences of systemic measles virus infection. J Pathol. 2015;235:253–265. doi: 10.1002/path.4457. [DOI] [PubMed] [Google Scholar]
- 82•.Woo P.C., Lau S.K., Wong B.H., Fan R.Y., Wong A.Y., Zhang A.J., Wu Y., Choi G.K., Li K.S., Hui J. Feline morbillivirus, a previously undescribed paramyxovirus associated with tubulointerstitial nephritis in domestic cats. Proc Natl Acad Sci U S A. 2012;109:5435–5440. doi: 10.1073/pnas.1119972109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of a virus which is very similar to known morbilliviruses in domestic cats.
- 83.Furuya T., Sassa Y., Omatsu T., Nagai M., Fukushima R., Shibutani M., Yamaguchi T., Uematsu Y., Shirota K., Mizutani T. Existence of feline morbillivirus infection in Japanese cat populations. Arch Virol. 2014;159:371–373. doi: 10.1007/s00705-013-1813-5. [DOI] [PubMed] [Google Scholar]
- 84.Sakaguchi S., Nakagawa S., Yoshikawa R., Kuwahara C., Hagiwara H., Asai K., Kawakami K., Yamamoto Y., Ogawa M., Miyazawa T. Genetic diversity of feline morbilliviruses isolated in Japan. J Gen Virol. 2014;95:1464–1468. doi: 10.1099/vir.0.065029-0. [DOI] [PubMed] [Google Scholar]
- 85•.Sharp C.R., Nambulli S., Acciardo A.S., Rennick L.J., Drexler J.F., Rima B.K., Williams T., Duprex W.P. Chronic infection of domestic cats with feline morbillivirus, United States. Emerging Infect Diseases. 2016;22 doi: 10.3201/eid2204.151921. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that cats can be chronically infected with feline morbillivirus.
- 86.von Messling V., Milosevic D., Cattaneo R. Tropism illuminated: Lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc Natl Acad Sci U S A. 2004;101:14216–14421. doi: 10.1073/pnas.0403597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Riddell M.A., Chibo D., Kelly H.A., Catton M.G., Birch C.J. Investigation of optimal specimen type and sampling time for detection of measles virus RNA during a measles epidemic. J Clin Microbiol. 2001;39:375–376. doi: 10.1128/JCM.39.1.375-376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wairagkar N.S., Gandhi B.V., Katrak S.M., Shaikh N.J., Parikh P.R., Wadia N.H., Gadkari D.A. Acute renal failure with neurological involvement in adults associated with measles virus isolation. Lancet. 1999;354:992–995. doi: 10.1016/s0140-6736(98)10101-0. [DOI] [PubMed] [Google Scholar]
- 89.Drosten C. Virus ecology: a gap between detection and prediction. Emerg Microbes Infect. 2013;2:e31. doi: 10.1038/emi.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lemon K., Nguyen D.T., Ludlow M., Rennick L.J., Yuksel S., van Amerongen G., McQuaid S., Rima B.K., de Swart R.L., Duprex W.P. Recombinant subgroup B human respiratory syncytial virus expressing enhanced green fluorescent protein efficiently replicates in primary human cells and is virulent in cotton rats. J Virol. 2015;89:2849–2850. doi: 10.1128/JVI.03587-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91••.Cello J., Paul A.V., Wimmer E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]; Generation of first recombinant positive strand virus by synthetic biology.
- 92•.Dormitzer P.R., Suphaphiphat P., Gibson D.G., Wentworth D.E., Stockwell T.B., Algire M.A., Alperovich N., Barro M., Brown D.M., Craig S. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Science Translational Med. 2013;5 doi: 10.1126/scitranslmed.3006368. 185ra168. [DOI] [PubMed] [Google Scholar]; Rapid generation of recombinant negative strand virus by synthetic biology based on database sequences.
- 93.Tong S., Chern S.W., Li Y., Pallansch M.A., Anderson L.J. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–2658. doi: 10.1128/JCM.00192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]