Highlights
► Most viral fitness work examines replicative fitness within hosts or in cultured cells. ► There is increasing study of transmission fitness and epidemiologic fitness. ► A wide diversity of viruses are studied for fitness in vitro, in vivo, and ex vivo. ► A recent advance for vertebrate viruses is more assessment of fitness in vivo. ► Major topics include fitness in drug resistance, immune escape, and viral emergence.
Abstract
Viral fitness is an active area of research, with recent work involving an expanded number of human, non-human vertebrate, invertebrate, plant, and bacterial viruses. Many publications deal with RNA viruses associated with major disease emergence events, such as HIV-1, influenza virus, and Dengue virus. Study topics include drug resistance, immune escape, viral emergence, host jumps, mutation effects, quasispecies diversity, and mathematical models of viral fitness. Important recent trends include increasing use of in vivo systems to assess vertebrate virus fitness, and a broadening of research beyond replicative fitness to also investigate transmission fitness and epidemiologic fitness. This is essential for a more integrated understanding of overall viral fitness, with implications for disease management in the future.
Current Opinion in Virology 2012, 2:538–545
This review comes from a themed issue on Virus evolution
Edited by Raul Andino and Marco Vignuzzi
For a complete overview see the Issue and the Editorial
Available online 15th September 2012
1879-6257/$ – see front matter, Published by Elsevier Ltd.
Introduction
Fitness is a complex concept at the foundation of all ecology and evolution. For viruses fitness was originally defined as “the capacity of a virus to produce infectious progeny in a given environment” [16]. This definition is still in wide use today, referred to more specifically as replicative fitness [15••], measured in cultured cells, tissue explants, or within individual hosts. This standard definition is founded in, but does not exactly match, the more general Darwinian definition of overall fitness, which is the amount of genetic material passed on to the next generation. Due to host immune clearance of viruses and finite host lifespan, viruses must be transmitted to new hosts to survive. As such, transmission fitness is an important component of overall fitness. Ultimately, replication and transmission contribute to the prevalence of viral genetic material at a population level in the field over time. The capacity of a virus (a serotype, clade, or variant) to become dominant in the field, relative to other serotypes, clades, or variants of the same virus has been defined as epidemiologic fitness [15••].
In this review we focus on viral fitness work published from 2009 through early 2012. Literature searches found nearly 750 publications involving viral fitness during this period, and we have focused on a subset of 98 papers selected to represent the current breadth of work in the field. In the interest of brevity these papers are not all cited here, but they were used to discern the general trends described below.
Viral fitness study systems
The field of viral fitness was originally developed through studies of a relatively small number of bacteriophage, animal, and plant viruses. With increasing recognition of the importance of viral fitness, there is now a wide array of study systems as detailed in Table 1 . The majority of viral fitness study systems are based on RNA viruses, and the highest numbers of publications in recent years involves human pathogens associated with major disease emergence events, such as human immunodeficiency virus-1 (HIV-1), influenza virus, and dengue virus (DENV).
Table 1.
Overview of viral fitness study systems reported in the literature from January 2009 to April 2012
| Host | Virusesa (No. refs/98)b | Host systems | Topics addressed |
|---|---|---|---|
| Human | HIV-1 (27), influenza (10), HCV (6), HBV (2), SARS (1) |
cultured cell lines, ex vivo PBMC, other ex vivo tissue explants, in vivo nonhuman primates, mice, waterfowl, guinea pig, ferrets, field sample surveys | drug resistance, immune escape, vaccine escape, epistasis, role of quasispecies, virulence evolution |
| Animal and insect (Arboviruses) | DENV (5), VSV (5), WNV (4), CHIKV (2) |
cultured cell lines, mosquitoes, chickens, mice, nonhuman primates, field sample surveys | host specificity, alternating host cycles, generalist vs. specialist, drug resistance, virulence determinants, virulence evolution, replication fidelity, role of quasispecies, single cell infection, environmental change, field displacements |
| Vertebrate animals (nonhuman) | SHIV (1), SIV (2), FMDV (1), CPV (1), EIAV (1), IHNV (3), Friend virus complex (1) |
cultured cell lines, ex vivo PBMC, other ex vivo tissue explants, mice, fish, nonhuman primates | immune escape, superinfection, virulence evolution, host specificity, host switching, virus infection cycle, viral load variation, field displacements, MHC evolution |
| Invertebrate animals | baculovirus (1), entomopoxvirus (1), WSSV (1) | insects, shrimp, crab | genetic modifications, genetic diversity, insect biocontrol, virulence |
| Plants | PVY (4), TEV (2), tobamovirus (1), MSV (1), CMVsat (1) |
pepper, tobacco, potato, maize, melon, aphids, leafhoppers | virulence evolution, host specificity, host adaptation, host resistance, mutation effects, mutation interactions, recombination, transmission fitness, transmission tradeoffs |
| Bacteriophage | Qβ (2), ΦX174 (2), F1 (2), Φ6 (2), MS2 (1), SP (1), SP6 (1), G4 (1), Φ2 (1), cyanophage (1) | E. coli, Pseudomonas, Salmonella, cyanobacteria databases (in silico) | mutation effects, host shifts, host range expansion, abiotic environment, antiviral resistance, in silico viral fitness model |
Virus abbreviations: HIV-1, human immunodeficiency virus-1; HCV, hepatitis C virus; HBV, hepatitis B virus; SARS, severe acute respiratory syndrome virus; DENV, dengue virus; VSV, vesicular stomatitis virus; WNV, West Nile virus; CHIKV, chikungunya virus; SHIV, simian/human immunodeficiency virus; SIV, simian immunodeficiency virus; FMDV, foot and mouth disease virus; CPV, canine parvovirus; EIAV, equine infectious anemia virus; IHNV, infectious hematopoietic necrosis virus; Friend complex virus, mixed infection with Friend murine leukemia virus and spleen focus forming virus (defective); WSSV, white spot shrimp virus; PYV, potato Y virus; TEV, tobacco etch virus; MSV, maize streak virus; CMVsat, cucumber mosaic virus satellite RNA.
Number of fitness-related papers published in scientific journals during 2009–2012 using each viral study system, from a total set of 98 viral fitness papers selected to represent breadth of activity in the field. Due to space limitations several papers counted for this Table are not cited in the text, but the full list is available from the authors upon request.
Replicative fitness
During development of the viral fitness field most research has assessed replicative fitness of viral variants within individual hosts (in vivo) or in cultured cells (in vitro). This trend has continued during recent years, likely due to the fact that replicative fitness is most readily measured in the laboratory. Typically replicative fitness studies compare the replication of two or more virus isolates that are variants of the same viral species. Under the general umbrella of replicative fitness there are many variations that will be described here and in later sections.
Replicative fitness is sometimes assessed by comparing viral replication in parallel hosts or cell cultures infected with single viral variants. However, it has long been recognized that assessment of fitness in mixed infections of two viral variants is a more sensitive and valid measure of viral fitness differences [16]. Therefore competitive fitness is often examined in growth competition assays, in which cells or hosts are co-infected with mixtures of two viral variants and the replication of each variant is determined in a competitive environment. Variables in these assays include the use of different input ratios, use of a standard reference strain or head-to-head competitions between test variants, and varied timing for analysis of progeny populations. For example, replicative fitness is sometimes examined at one time point post-infection, but it is often assessed at multiple time points [17, 18, 33, 42, 45, 46, 60] or in serial passage experiments [25, 26, 63].
There are a variety of methods used to measure replicative fitness. The traditional method quantifies individual viral variants as plaque forming units (PFU). Recent advances in molecular techniques have led to the use of methodologies that measure viral DNA, RNA, or protein levels, such as quantitative PCR, ELISA, fluorescent probe labeling, and next generation sequencing. These molecular methods estimate viral load, and have the advantage that they typically allow for higher throughput as well as sensitivity, and can be used to distinguish viral genotypes in mixed infections. However, they do not quantify infectious virus as PFU assays do, and the relationships between PFU and molecular quantifications of viral load remain to be defined in many systems.
Viral fitness is expressed in various ways, such as comparing viral loads statistically [58•], using a fitness parameter equation [1, 48, 53], or determining the slope of a fitness vector in comparison with a constant reference strain after multiple sampling times or serial passage [37]. In the past fitness data were generally presented as relative fitness values, reported as a fitness ratio between two viral variants. The use of appropriate standards with qRT-PCR quantification has recently made it possible to determine absolute fitness values in terms of average RNA copy numbers per μg of host tissue, for each viral variant [58•].
A major variable in replicative fitness work is the in vitro, ex vivo, or in vivo nature of the environments used for viral replication (Figure 1 ). For viruses of bacteria, insects, and plants there is a long record of sophisticated in vivo fitness studies using controlled laboratory populations of living hosts, and in vivo work in these systems has remained prolific to date [3•, 6, 10, 11•, 14, 19, 25, 27, 35, 36••, 39, 41, 54]. Viruses of vertebrates have traditionally been studied in cultured cell lines, and this work also continues [1, 7, 11•, 12, 23, 24, 31, 34, 37, 38, 47, 49••, 51, 61, 62]. However, over the last three years there has been an expansion of vertebrate virus fitness studies in vivo using systems such as influenza, DENV, West Nile virus (WNV), and infectious hematopoietic necrosis virus (IHNV) [13••, 17, 18, 20, 21, 28, 32•, 33, 46, 56, 58•]. For human viruses such as HIV-1, where in host experimental studies are difficult or impossible, ex vivo systems in primary tissue explants have been developed using peripheral blood mononuclear cells (PBMC) [53] and other tissues [1].
Figure 1.
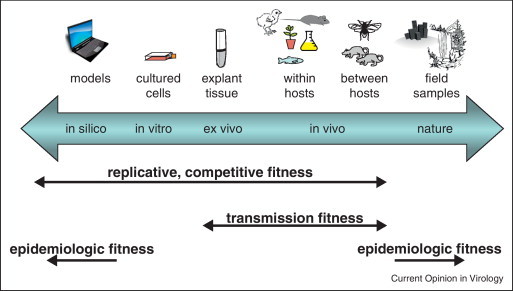
Diversity of research systems used to investigate different aspects of overall viral fitness.
Replicative fitness studies typically measure the average fitness of populations of multiple virus particles in populations of in vitro cultured cells or in vivo host tissues. An exciting recent advance has been analysis of the replicative fitness of individual virus particles, defined as the total number of virus progeny produced when one virus infects an individual susceptible cell [62]. Work with vesicular stomatitis virus (VSV) has revealed dramatic variation ranging from 50 to 8000 progeny virus particles per host cell, with additional experiments indicating the host cell cycle stage as a major influence on this variability.
Another component of competitive co-infection fitness is superinfection fitness, in which infection with one viral variant is established before exposure to the second variant. Despite the clear relevance of superinfection to natural viral infections in the field, there are few studies of controlled superinfections [60]. In general, viruses isolated from natural superinfections have been analyzed in simultaneous co-infection assays, often indicating that the superinfecting strains have higher fitness [55].
Transmission fitness
Due to the major role of transmission in overall viral fitness, transmission fitness is a research area that is currently expanding. The majority of work on transmission fitness has been conducted in plant viruses [3•, 35] and arboviruses such as WNV, DENV, and chikungunya virus (CHIKV) [18, 28] that are transmitted via arthropod vectors. These studies often quantify transmission and replicative fitness through the vector as well as in the plant or vertebrate host [3•, 8••, 28, 35, 56], and differences in transmission fitness have been observed. For viruses that are transmitted directly, between-host transmission fitness is investigated by exposing naive hosts to infected hosts and examining the absolute or relative quantities of viral variants in source and recipient hosts. This has been described in several studies of H1N1 influenza virus using in vivo guinea pig and ferret models [17, 32•, 46]. Alternative methods for generating inferences about transmission fitness include quantification of shed virus as an indication of transmission potential [58•], and ex vivo fitness assays assessing replication or transmission efficiency in tissues specifically associated with transmission [1].
Transmission fitness studies in plant systems often have additional levels of complexity such as examining multiple rounds of transmission [25], with accurate quantification of the timing of transmission and exposure dosage. Vertebrate virus studies of transmission are beginning to approach the sophistication of plant virus studies, but this remains a major research area for future development. One of the intriguing findings from many systems is that replicative fitness does not always match transmission fitness in that viral genotypes with the greatest replication success are not always those that have the greatest transmission to new hosts [17, 46].
Epidemiologic fitness
In many cases the ultimate goal of replicative and transmission fitness studies is to understand the epidemiology and population level processes governing viral evolution, emergence, and displacement in the field. As such there has been an increased effort in recent years to quantify epidemiologic fitness. Quantification of epidemiologic fitness is based largely on observational data and examines changes in distribution, prevalence, and composition of viral genotypes over time to infer their relative fitness. This is particularly valuable for systems where a wealth of epidemiological data is available, and the ability to conduct in vivo experimental studies is limited. It is of no surprise therefore that most recent work in this area was conducted in human virus systems such as HIV, and DENV [1, 21, 30, 51].
Topics addressed
-
(I)
Drug resistance
Of all topics addressed in recent viral fitness literature the resistance fitness of drug resistant variants is by far the most common, defined as the ability of a virus to replicate in the presence of antiviral drugs or compounds. The majority of this work was conducted in influenza and HIV, but other systems include hepatitis B (HBV), hepatitis C (HCV), and WNV [2, 23, 48, 51, 52, 57]. Largely this work characterized the replicative and competitive fitness of drug resistant mutants compared to wild type drug sensitive virus types. In the majority of cases drug resistant mutants were found to have fitness costs in that they showed reduced replication ability compared to wild-type strains in the absence of the selective drug. Interestingly, many systems also revealed the occurrence of compensatory mutations, which caused drug resistant mutants to partially or even completely regain their fitness [20, 23, 61]. A study on bacteriophage even found that the virus became metabolically dependent on the drug and experienced enhanced growth when the drug was present [6]. Numerous laboratory studies of H1N1 influenza (only a few cited here) demonstrated that a drug resistant strain that originally emerged in the field had a fitness cost in laboratory in vivo studies, whereas a later resistant mutant that became more widespread did not [17, 33, 38, 42, 46].
-
(II)
Escape from host defenses
Other examples of resistance fitness involve viral variants that escape surveillance and destruction by host immune responses, notably T cells or neutralizing antibodies, or host genetic resistance mechanisms. Largely this work was conducted in human or primate viruses such as HIV, HCV, severe acute respiratory syndrome virus (SARS) and simian immunodeficiency virus (SIV) [12, 26, 44, 53, 60, 61], but there were also some studies of plant virus variants that overcome host genetic resistance [19, 25]. As with drug resistance, immune escape mutations varied in their fitness costs, and the occurrence of compensatory mutations was common [12, 44, 53, 60]. Implication for vaccine development was a heavy focus, demonstrating that immune escape presents a major challenge to vaccine efficacy for some viruses.
-
(III)
Field emergence and displacement
There has been increasing interest in the role of viral fitness in viral emergence and displacement events in the field [15••]. DENV is the most active system for this inquiry and there is now substantial evidence that viral replication and transmission fitness as measured in the laboratory correlates with the prevalence and displacement of dominant dengue virus genotypes in the field [28]. Similar approaches are being used for HIV as well as foot-and-mouth disease virus (FMDV), yielding similar results where dominant genotypes in the field seem to have greater fitness in laboratory assays [1, 34, 45]. This may be partially driven by superinfection fitness, in that a study of HIV-1 variants from two natural superinfection cases showed that the superinfecting strains had higher fitness than the primary infecting strains [55]. Viral emergence has also been explored in a bacteriophage system, showing that genetic diversity from source populations drives host range expansion or host shifts [14].
-
(IV)
Viral fitness in different hosts
Several studies addressed questions related to the influence of different hosts on viral fitness. For arboviruses there is a well developed body of literature investigating the fitness consequences of the natural cycle between vertebrate and insect vector hosts. This has continued recently in experimental evolution studies in which viruses are serially passaged, in vitro or in vivo, in single or alternating host types, and then assayed for fitness gains or losses in each host. Studies with WNV and CHIKV both found that fitness increases were greater upon passage in vertebrate hosts than insect hosts [9, 13••]. Such studies are often designed to test theoretical predictions associated with host tradeoff or generalist-specialist hypotheses. Results vary with different viral systems, and it is clear that viral fitness assayed in one host type has low predictive power as to fitness assayed in a different host type [9, 13••]. An experimental evolution study with VSV attributed much of this difference to incongruent fitness landscapes in different hosts rather than tradeoffs.
Beyond arboviruses, fitness aspects of host jumps into new hosts have been studied in different systems. A bacteriophage system was used to demonstrate that that multiple mutational pathways led to very similar increased fitness levels during adaptation to a novel host, suggesting a maximum fitness limit [36••]. In the fish virus IHNV, in vivo competitive fitness assays demonstrated a host tradeoff in which virus adapted to rainbow trout as a new host had lost fitness in the previous host, sockeye salmon [40]. In the canine parvovirus (CPV) system an investigation of the mechanism of a well documented host jump found reduced fitness for all possible transitional mutants, and used fitness levels to suggest the most likely path for evolutionary adaptation [49••].
-
(V)
Mutational diversity and quasispecies
Interest in how mutations impact viral fitness has continued in recent years with analyses of Mutational Fitness Effects (MFE) in various bacteriophage species. As expected, the majority of mutations have negative impacts on fitness, often resulting in nonviable viruses. However, mutations with positive fitness effects were also noted [10, 41]. Interestingly, work on epistasis, that is, the combined effects of multiple mutations on fitness, revealed that mutational effects are not always additive [11•, 31] and may come with fitness tradeoffs [35, 59].
The quasispecies nature of populations of RNA viruses, retroviruses, and hepadnaviruses means they exist as dynamic distributions of mutant viruses related to a consensus sequence [16]. A positive relationship between viral quasispecies diversity and fitness has been observed in three very different studies. For HIV-1, increases in quasispecies heterogeneity were associated with fitness recovery in vitro, even in the absence of changes in the consensus sequence [4]. For WNV, artificial quasispecies of high complexity had higher fitness than less complex populations when tested in mosquitoes, but not in chicks, so the fitness advantage was host-specific [18]. Finally, a high fidelity polymerase mutant of CHIKV has been described that generates low diversity quasispecies, and has reduced fitness in both mosquitos and mice [8••].
Mathematical modeling of fitness
In recent years there has been an increased effort to mathematically quantify viral fitness. This includes standard statistical package approaches as well as system-specific mathematical model development. Various formulas and methods are being explored to put fitness into mathematical terms. Most of these approaches are based on a comparison of absolute or relative viral loads between viral variants to generate fitness parameters that can be compared analytically. The basic formula typically takes the form: w = f o/i o, where f o equals the final ratio of virus genotypes, and i o equals the initial ratio of genotypes [45]. For influenza [32•] and tobacco etch virus [27], more detailed population level transmission models have recently been developed incorporating a variety of parameters such as relative fitness, transmission rates, host recovery rates, viral virulence, and virus stability. The aim of these models is to track the spread and infection dynamics of the virus within the host or at the level of host populations. The models are then used to explore fitness factors that influence genotype prevalence, or make predictive inferences about evolutionary trajectories. Recently mathematical approaches have been used to make inferences about influenza virus inoculum sizes [32•], drug resistance evolution in HCV [2], DENV field displacement events [30], and the evolutionary impact of incorporating new traits into the viral genome such as photosynthesis genes in a cyanophage [22]. Parameters in mathematical models are typically defined with in vivo laboratory and field data, and recent growth in these areas has made modeling approaches more tractable.
New technologies
Exciting advances in the field of viral fitness have largely been driven by the rapid expansion in molecular technologies and computing power. For example molecular barcoding microarray technologies have been employed to create quasispecies swarms in the laboratory and simultaneously characterize the fitness of numerous mutants of poliovirus [29]. Next generation sequencing is also being used to rapidly sequence entire viral genomes and determine how genome wide mutation accumulation impacts fitness [37]. Likewise, large scale site directed mutagenesis has recently been used as a tool to determine the molecular interactions that regulate fitness [31]. New bioinformatics tools are being employed to explore large HIV field sequence databases and determine the fitness landscapes [52], and we have entered the age of purely in silico studies that use advanced agent based mathematical models parameterized from published literature to make inferences about viral evolution [22]. Ultimately these advances have made understanding the genetic regulators of fitness within arms grasp, and not surprisingly, revealed that the story is likely to be more complex than previously assumed [11•, 31].
Current challenges: merging in vitro and in vivo work, and addressing virulence
For vertebrate viruses the recent increase of in vivo virus fitness research is encouraging, but the majority of studies still remain in vitro. This is likely due to the ethical and practical constraints of conducting in vivo research in many vertebrate systems, particularly human viruses. Furthermore in vitro work has numerous advantages, most notably it allows for a higher level of control of variables than in vivo work and is useful for understanding phenomena at a molecular or cellular level [62]. The drawback is that it is unclear how well in vitro results reflect natural phenomena in vivo. Few systems have addressed this question and those that have in recent years show varying levels of consistency between in vitro and in vivo findings [21, 44, 56, 59]. More controlled studies comparing in vitro and in vivo fitness assays with the same viruses are needed to define the relevance and limitations of each approach.
The relationship between viral fitness and virulence has been of interest for decades, but surprisingly little has been published recently. Many recent publications assume that viral replicative fitness and virulence are positively correlated and thus use the terms interchangeably. For example, the term ‘attenuation’ is often used to describe a virus with reduced replicative fitness [46]. In actuality, the relationship between viral fitness and virulence remains poorly characterized in many systems, and both agreement and exception to this assumption have been reported [3•, 5, 24, 27, 40, 44, 58•]. Some of the discrepancies in this area may come from the multitude of ways virulence is defined or measured, often beyond the standard definition of morbidity and mortality caused to the host due to infection [3•, 19, 25, 26, 27, 43, 50, 58•]. More focus on virulence, and definition of system-specific correlations of virulence with fitness would be a benefit to future work.
Conclusions
Viral fitness continues to be an active area of research [7]. Given the increase in issues such as drug resistance evolution, vaccine escape, virulence evolution, viral emergence, and host jumps, the understanding of viral fitness has become essential. For decades viral fitness has been primarily defined by replication capacity in the host. This definition is now broadening as researchers attempt to understand the population level evolutionary implications of overall viral fitness in natural infections. Making inferences about population level processes requires integration of replicative, competitive, transmission, and epidemiologic fitness measures (Figure 1). Perhaps the most critical need for the field of viral fitness is the further development of integrative approaches, with the ultimate goals of making accurate predictive inferences and informing long-term management or control of disease. Many the tools to achieve this goal are now available, and we are collectively faced with the task of putting them together.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was funded by the US Geological Survey, Western Fisheries Research Center and by National Science Foundation Ecology of Infectious Diseases grant 0812603. Mention of trade names does not imply endorsement by the US Government.
References
- 1.Abraha A., Nankya I.L., Gibson R., Demers K., Tebit D.M., Johnston E., Katzenstein D., Siddiqui A., Herrera C., Fischetti L. CCR5-and CXCR4-tropic subtype C human immunodeficiency virus type 1 isolates have a lower level of pathogenic fitness than other dominant group M subtypes: implications for the epidemic. Journal of Virology. 2009;83:5592–5605. doi: 10.1128/JVI.02051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adiwijaya B.S., Kieffer T.L., Henshaw J., Eisenhauer K., Kimko H., Alam J.J., Kauffman R.S., Garg V. A viral dynamic model for treatment regimens with direct-acting antivirals for chronic hepatitis C infection. PLoS Computational Biology. 2012;8:11. doi: 10.1371/journal.pcbi.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Betancourt M., Fraile A., Garcia-Arenal F. Cucumber mosaic virus satellite RNAs that induce similar symptoms in melon plants show large differences in fitness. Journal of General Virology. 2011;92:1930–1938. doi: 10.1099/vir.0.032359-0. [DOI] [PubMed] [Google Scholar]; This paper provides an example of the sophistication of plant virus fitness work, using two groups of ten plant viral satellite RNAs that differ in fitness and virulence on a tomato host. On a melon host these satellite RNAs differ in multiple measures of viral fitness including replication in single or mixed infections and aphid transmissibility, but they do not differ in virulence, demonstrating both host-specific fitness traits and a lack of correlation between replicative fitness and virulence.
- 4.Borderia A.V., Lorenzo-Redondo R., Pernas M., Casado C., Alvaro T., Domingo E., Lopez-Galindez C. Initial fitness recovery of HIV-1 is associated with quasispecies heterogeneity and can occur without modifications in the consensus sequence. PLoS ONE. 2010;5:8. doi: 10.1371/journal.pone.0010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh J., Meuleman P., Tellier R., Engle R.E., Feinstone S.M., Eder G., Satterfield W.C., Govindarajan S., Krawczynski K., Miller R.H. Challenge pools of hepatitis C virus genotypes 1-6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. Journal of Infectious Diseases. 2010;201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherwa J.E., Sanchez-Soria P., Wichman H.A., Fane B.A. Viral adaptation to an antiviral protein enhances the fitness level to above that of the uninhibited wild type. Journal of Virology. 2009;83:11746–11750. doi: 10.1128/JVI.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementi M., Lazzarin A. Human immunodeficiency virus type 1 fitness and tropism: concept, quantification, and clinical relevance. Clinical Microbiology and Infection. 2010;16:1532–1538. doi: 10.1111/j.1469-0691.2010.03335.x. [DOI] [PubMed] [Google Scholar]
- 8••.Coffey L.L., Beeharry Y., Borderia A.V., Blanc H., Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper presents a new high fidelity variant of the chikungunya arbovirus (CHIKV) that generates virus populations with reduced genetic diversity. The variant shows reduced fitness in both mosquitos and mice, providing a new system for studies of the impact of high fidelity viral replication in vivo.
- 9.Coffey L.L., Vignuzzi M. Host alternation of chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. Journal of Virology. 2011;85:1025–1035. doi: 10.1128/JVI.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuevas J.M., Domingo-Calap P., Sanjuan R. The fitness effects of synonymous mutations in DNA and RNA viruses. Molecular Biology and Evolution. 2012;29:17–20. doi: 10.1093/molbev/msr179. [DOI] [PubMed] [Google Scholar]
- 11•.da Silva J., Coetzer M., Nedellec R., Pastore C., Mosier D.E. Fitness epistasis and constraints on adaptation in a human immunodeficiency virus type 1 protein region. Genetics. 2010;185 doi: 10.1534/genetics.109.112458. 293-U430. [DOI] [PMC free article] [PubMed] [Google Scholar]; Fitness epistasis was investigated here by creating seven mutations, singly and in combination, in HIV-1 glycoprotein and testing effects on viral infectivity. Epistatic effects were found to be common, complex, and often very strong, providing insights into the barriers and probable pathways of evolution of co-receptor usage.
- 12.Dazert E., Neumann-Haefelin C., Bressanelli S., Fitzmaurice K., Kort J., Timm J., McKiernan S., Kelleher D., Gruener N., Tavis J.E. Loss of viral fitness and cross-recognition by CD8(+) T cells limit HCV escape from a protective HLA-B27-restricted human immune response. Journal of Clinical Investigation. 2009;119:376–386. doi: 10.1172/JCI36587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Deardorff E.R., Fitzpatrick K.A., Jerzak G.V.S., Shi P.Y., Kramer L.D., Ebel G.D. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathogens. 2011;7:8. doi: 10.1371/journal.ppat.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work explores host tradeoff hypothesis associated with the alternate cycling of the arbovirus West Nile Virus between insect vector and bird hosts. WNV adapted to serial passage only in chicks gained fitness in chicks and in one of two insect vector species, while virus serially passaged in insects lost fitness in chicks without any gain in either insect host. Thus fitness measured in birds supported the tradeoff hypothesis, but measurements in insects did not.
- 14.Dennehy J.J., Friedenberg N.A., McBride R.C., Holt R.D., Turner P.E. Experimental evidence that source genetic variation drives pathogen emergence. Proceedings of the Royal Society B-Biological Sciences. 2010;277:3113–3121. doi: 10.1098/rspb.2010.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Domingo E. Mechanisms of viral emergence. Veterinary Research. 2010;41:14. doi: 10.1051/vetres/2010010. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an excellent review article presenting an updated synthesis of numerous fitness-related publications, as well as clear definitions of the difference between replicative fitness and epidemiologic fitness. Relevance of RNA viral quasispecies to viral fitness and viral emergence is also nicely summarized.
- 16.Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annual Review of Microbiology. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 17.Duan S.S., Boltz D.A., Seiler P., Li J.A., Bragstad K., Nielsen L.P., Webby R.J., Webster R.G., Govorkova E.A. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathogens. 2010;6:10. doi: 10.1371/journal.ppat.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzpatrick K.A., Deardorff E.R., Pesko K., Brackney D.E., Zhang B., Bedrick E., Shi P.Y., Ebel G.D. Population variation of West Nile virus confers a host-specific fitness benefit in mosquitoes. Virology. 2010;404:89–95. doi: 10.1016/j.virol.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraile A., Pagan I., Anastasio G., Saez E., Garcia-Arenal F. Rapid genetic diversification and high fitness penalties associated with pathogenicity evolution in a plant virus. Molecular Biology and Evolution. 2011;28:1425–1437. doi: 10.1093/molbev/msq327. [DOI] [PubMed] [Google Scholar]
- 20.Govorkova E.A., Ilyushina N.A., Marathe B.M., McClaren J.L., Webster R.G. Competitive fitness of oseltamivir-sensitive and -resistant highly pathogenic H5N1 influenza viruses in a ferret model. Journal of Virology. 2010;84:8042–8050. doi: 10.1128/JVI.00689-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hang V.T.T., Holmes E.C., Veasna D., Nguyen T.Q., Tran T.H., Quail M., Churcher C., Parkhill J., Cardosa J., Farrar J. Emergence of the Asian 1 genotype of dengue virus serotype 2 in Viet Nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Neglected Tropical Diseases. 2010;4:11. doi: 10.1371/journal.pntd.0000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellweger F.L. Carrying photosynthesis genes increases ecological fitness of cyanophage in silico. Environmental Microbiology. 2009;11:1386–1394. doi: 10.1111/j.1462-2920.2009.01866.x. [DOI] [PubMed] [Google Scholar]
- 23.Hu Z.X., Kuritzkes D.R. Effect of raltegravir resistance mutations in HIV-1 integrase on viral fitness. JAIDS-Journal of Acquired Immune Deficiency Syndromes. 2010;55:148–155. doi: 10.1097/QAI.0b013e3181e9a87a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang K.H.G., Goedhals D., Carlson J.M., Brockman M.A., Mishra S., Brumme Z.L., Hickling S., Tang C.S.W., Miura T., Seebregts C. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS ONE. 2011;6:20. doi: 10.1371/journal.pone.0019018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janzac B., Montarry J., Palloix A., Navaud O., Moury B. A point mutation in the polymerase of potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Molecular Plant-Microbe Interactions. 2010;23:823–830. doi: 10.1094/MPMI-23-6-0823. [DOI] [PubMed] [Google Scholar]
- 26.Kubinak J.L., Ruff J.S., Hyzer C.W., Slev P.R., Potts W.K. Experimental viral evolution to specific host MHC genotypes reveals fitness and virulence trade-offs in alternative MHC types. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3422–3427. doi: 10.1073/pnas.1112633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafforgue G., Sardanyes J., Elena S.F. Differences in accumulation and virulence determine the outcome of competition during tobacco etch virus coinfection. PLoS ONE. 2011;6:8. doi: 10.1371/journal.pone.0017917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambrechts L., Fansiri T., Pongsiri A., Thaisomboonsuk B., Klungthong C., Richardson J.H., Ponlawat A., Jarman R.G., Scott T.W. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. Journal of Virology. 2012;86:1853–1861. doi: 10.1128/JVI.06458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauring A.S., Andino R. Exploring the fitness landscape of an RNA virus by using a universal barcode microarray. Journal of Virology. 2011;85:3780–3791. doi: 10.1128/JVI.02217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lourenco J., Recker M. Viral and epidemiological determinants of the invasion dynamics of novel dengue genotypes. PLoS Neglected Tropical Diseases. 2010;4:12. doi: 10.1371/journal.pntd.0000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez J.P., Bocharov G., Ignatovich A., Reiter J., Dittmar M.T., Wain-Hobson S., Meyerhans A. Fitness ranking of individual mutants drives patterns of epistatic interactions in HIV-1. PLoS ONE. 2011;6:9. doi: 10.1371/journal.pone.0018375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.McCaw J.M., Arinaminpathy N., Hurt A.C., McVernon J., McLean A.R. A mathematical framework for estimating pathogen transmission fitness and inoculum size using data from a competitive mixtures animal model. PLoS Computational Biology. 2011;7:11. doi: 10.1371/journal.pcbi.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors introduce a mathematical framework for quantitative estimation of the relative transmissibility of two viral types in co-infection in vivo, and the inoculum size associated with transmission events. The model is tested using data from in vivo influenza virus co-infection studies in ferrets, providing one of the first rigorous investigations of transmission fitness for a virus that is directly transmitted between vertebrate hosts (i.e. non-arbovirus).
- 33.Memoli M.J., Davis A.S., Proudfoot K., Chertow D.S., Hrabal R.J., Bristol T., Taubenberger J.K. MultiDrug-resistant 2009 pandemic influenza A(H1N1) viruses maintain fitness and transmissibility in ferrets. Journal of Infectious Diseases. 2011;203:348–357. doi: 10.1093/infdis/jiq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohapatra J.K., Subramaniam S., Singh N.K., Sanyal A., Pattnaik B. Experimental evidence for competitive growth advantage of genotype VII over VI: implications for foot-and-mouth disease virus serotype A genotype turnover in nature. Research in Veterinary Science. 2012;92:317–319. doi: 10.1016/j.rvsc.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Moury B., Simon V. dN/dS-based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of potato virus Y. Molecular Biology and Evolution. 2011;28:2707–2717. doi: 10.1093/molbev/msr105. [DOI] [PubMed] [Google Scholar]
- 36••.Nguyen A.H., Molineux I.J., Springman R., Bull J.J. Multiple genetic pathways to similar fitness limits during viral adaptation to a new host. Evolution. 2012;66:363–374. doi: 10.1111/j.1558-5646.2011.01433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study investigates the existence of fitness limits using a bacteriophage of Salmonella being adapted to an E. coli host under varying conditions. Although the nucleotide changes associated with adaptation differed dramatically, four independent lines achieved similar absolute fitness increases, demonstrating a fitness limit that could be attained by multiple genetic pathways.
- 37.Novella I.S., Presloid J.B., Zhou T., Smith-Tsurkan S.D., Ebendick-Corpus B.E., Dutta R.N., Lust K.L., Wilke C.O. Genomic evolution of vesicular stomatitis virus strains with differences in adaptability. Journal of Virology. 2010;84:4960–4968. doi: 10.1128/JVI.00710-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parry C.M., Kohli A., Boinett C.J., Towers G.J., McCormick A.L., Pillay D. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. Journal of Virology. 2009;83:9094–9101. doi: 10.1128/JVI.02356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascua L.L., Gandon S., Buckling A. Abiotic heterogeneity drives parasite local adaptation in coevolving bacteria and phages. Journal of Evolutionary Biology. 2012;25:187–195. doi: 10.1111/j.1420-9101.2011.02416.x. [DOI] [PubMed] [Google Scholar]
- 40.Penaranda M.M.D., Wargo A.R., Kurath G. In vivo fitness correlates with host-specific virulence of Infectious hematopoietic necrosis virus (IHNV) in sockeye salmon and rainbow trout. Virology. 2011;417:312–319. doi: 10.1016/j.virol.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 41.Peris J.B., Davis P., Cuevas J.M., Nebot M.R., Sanjuan R. Distribution of fitness effects caused by single-nucleotide substitutions in bacteriophage f1. Genetics. 2010;185 doi: 10.1534/genetics.110.115162. 603-U308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizzorno A., Abed Y., Bouhy X., Beaulieu E., Mallett C., Russell R., Boivin G. Impact of mutations at residue I223 of the neuraminidase protein on the resistance profile, replication level, and virulence of the 2009 pandemic influenza virus. Antimicrobial Agents and Chemotherapy. 2012;56:1208–1214. doi: 10.1128/AAC.05994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pradeep B., Karunasagar I. Fitness and virulence of different strains of white spot syndrome virus. Journal of Fish Diseases. 2009;32:801–805. doi: 10.1111/j.1365-2761.2009.01053.x. [DOI] [PubMed] [Google Scholar]
- 44.Rockx B., Donaldson E., Frieman M., Sheahan T., Corti D., Lanzavecchia A., Baric R.S. Escape from human monoclonal antibody neutralization affects in vitro and in vivo fitness of severe acute respiratory syndrome coronavirus. Journal of Infectious Diseases. 2010;201:946–955. doi: 10.1086/651022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez M.A., Ding M., Ratner D., Chen Y., Tripathy S.P., Kulkarni S.S., Chatterjee R., Tarwater P.M., Gupta P. High replication fitness and transmission efficiency of HIV-1 subtype C from India: implications for subtype C predominance. Virology. 2009;385:416–424. doi: 10.1016/j.virol.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Seibert C.W., Kaminski M., Philipp J., Rubbenstroth D., Albrecht R.A., Schwalm F., Stertz S., Medina R.A., Kochs G., Garcia-Sastre A. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza a virus are not attenuated in the guinea pig and ferret transmission models. Journal of Virology. 2010;84:11219–11226. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith-Tsurkan S.D., Wilke C.O., Novella I.S. Incongruent fitness landscapes, not tradeoffs, dominate the adaptation of vesicular stomatitis virus to novel host types. Journal of General Virology. 2010;91:1484–1493. doi: 10.1099/vir.0.017855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soares E.A., Santos A., Gonzalez L.M., Lalonde M.S., Tebit D.M., Tanuri A., Arts E.J., Soares M.A. Mutation T74S in HIV-1 subtype B and C proteases resensitizes them to ritonavir and indinavir and confers fitness advantage. Journal of Antimicrobial Chemotherapy. 2009;64:938–944. doi: 10.1093/jac/dkp315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Stucker K.M., Pagan I., Cifuente J.O., Kaelber J.T., Lillie T.D., Hafenstein S., Holmes E.C., Parrish C.R. The role of evolutionary intermediates in the host adaptation of canine parvovirus. Journal of Virology. 2012;86:1514–1521. doi: 10.1128/JVI.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors explored the mechanism of a documented host switch in canine parvovirus (CPV) by constructing viruses with all possible intermediate genomes and assessing their fitness in feline cells. They show that host adaptation involves complex interactions between mutations and most transition intermediates have lower fitness, allowing them to infer the most probable pathway of the viral host switch.
- 50.Takatsuka J., Okuno S., Ishii T., Nakai M., Kunimi Y. Fitness-related traits of entomopoxviruses isolated from Adoxophyes honmai (Lepidoptera: Tortricidae) at three localities in Japan. Journal of Invertebrate Pathology. 2010;105:121–131. doi: 10.1016/j.jip.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Tebit D.M., Lobritz M., Lalonde M., Immonen T., Singh K., Sarafianos S., Herchenroder O., Krausslich H.G., Arts E.J. Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. Journal of Virology. 2010;84:9817–9830. doi: 10.1128/JVI.00991-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theys K., Deforche K., Beheydt G., Moreau Y., van Laethem K., Lemey P., Camacho R.J., Rhee S.Y., Shafer R.W., Van Wijngaerden E., Vandamme A.M. Estimating the individualized HIV-1 genetic barrier to resistance using a nelfinavir fitness landscape. BMC Bioinformatics. 2010;11:9. doi: 10.1186/1471-2105-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Troyer R.M., McNevin J., Liu Y., Zhang S.C., Krizan R.W., Abraha A., Tebit D.M., Zhao H., Avila S., Lobritz M.A. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathogens. 2009;5:13. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner P.E., Draghi J.A., Wilpiszeski R. High-throughput analysis of growth differences among phage strains. Journal of Microbiological Methods. 2012;88:117–121. doi: 10.1016/j.mimet.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 55.van der Kuyl A.C., Kozaczynska K., Arien K.K., Gali Y., Balazs V.R., Dekker S.J., Zorgdrager F., Vanham G., Berkhout B., Cornelissen M. Analysis of infectious virus clones from two HIV-1 superinfection cases suggests that the primary strains have lower fitness. Retrovirology. 2010;7:15. doi: 10.1186/1742-4690-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Slyke G.A., Ciota A.T., Willsey G.G., Jaeger J., Shi P.Y., Kramer L.D. Point mutations in the West Nile virus (Flaviviridae; Flavivirus) RNA-dependent RNA polymerase alter viral fitness in a host-dependent manner in vitro and in vivo. Virology. 2012;427:18–24. doi: 10.1016/j.virol.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Villet S., Billioud G., Pichoud C., Lucifora J., Hantz O., Sureau C., Deny P., Zoulim F. In vitro characterization of viral fitness of therapy-resistant hepatitis B variants. Gastroenterology. 2009;136:168–176. doi: 10.1053/j.gastro.2008.09.068. [DOI] [PubMed] [Google Scholar]
- 58•.Wargo A.R., Kurath G. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. Journal of Virology. 2011;85:3959–3967. doi: 10.1128/JVI.01891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first study to quantify how viral infection cycle traits correlate with viral fitness and virulence, using a fish rhabdovirus in rainbow trout as an in vivo system. Although within-host replication had the largest impact on fitness, host entry, competitive fitness, and shedding also contributed, indicating fitness as a complex trait
- 59.Warter L., Cohen L., Benureau Y., Chavez D., Yang Y., Bodola F., Lemon S.M., Traboni C., Lanford R.E., Martin A. A cooperative interaction between nontranslated RNA sequences and NS5A protein promotes in vivo fitness of a chimeric hepatitis C/GB virus B. PLoS ONE. 2009;4:14. doi: 10.1371/journal.pone.0004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinfurter J.T., May G.E., Soma T., Hessell A.J., Leon E.J., MacNair C.E., Piaskowski S.M., Weisgrau K., Furlott J., Maness N.J. Macaque long-term nonprogressors resist superinfection with multiple CD8(+) T cell escape variants of simian immunodeficiency virus. Journal of Virology. 2011;85:530–541. doi: 10.1128/JVI.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu W.W., Blythe D.C., Loyd H., Mealey R.H., Tallmadge R.L., Dorman K.S., Carpenter S. Decreased infectivity of a neutralization-resistant equine infectious anemia virus variant can be overcome by efficient cell-to-cell spread. Journal of Virology. 2011;85:10421–10424. doi: 10.1128/JVI.05349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y., Yongky A., Yin J. Growth of an RNA virus in single cells reveals a broad fitness distribution. Virology. 2009;385:39–46. doi: 10.1016/j.virol.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zwart M.P., van der Werf W., van Oers M.M., Hemerik L., van Lent J.M.V., de Visser J., Vlak J.M., Cory J.S. Mixed infections and the competitive fitness of faster-acting genetically modified viruses. Evolutionary Applications. 2009;2:209–221. doi: 10.1111/j.1752-4571.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


