Highlights
-
•
Host genes play an important role in the pathogenesis of influenza virus infection.
-
•
High-throughput technologies accelerate the discovery of susceptibility genes.
-
•
Host susceptibility studies facilitate the development of host-targeted therapy.
-
•
Host-targeted therapy may allow individualized treatment for influenza.
Abstract
The emergence of the pandemic influenza virus A(H1N1)pdm09 in 2009 and avian influenza virus A(H7N9) in 2013 provided unique opportunities for assessing genetic predispositions to severe disease because many patients did not have any underlying risk factor or neutralizing antibody against these agents, in contrast to seasonal influenza viruses. High-throughput screening platforms and large human or animal databases from international collaborations allow rapid selection of potential candidate genes for confirmatory functional studies. In the last 2 years, at least seven new human susceptibility genes have been identified in genetic association studies. Integration of knowledge from genetic and phenotypic studies is essential to identify important gene targets for treatment and prevention of influenza virus infection.
Current Opinion in Virology 2015, 14:7–15
This review comes from a themed issue on Engineering for viral resistance
Edited by Albrecht von Brunn
For a complete overview see the Issue and the Editorial
Available online 14th June 2015
http://dx.doi.org/10.1016/j.coviro.2015.04.010
1879-6257/© 2015 Elsevier B.V. All rights reserved.
Introduction
Influenza virus is one of the most common seasonal respiratory viruses affecting humans, leading to 250 000–500 000 deaths every year [1]. The 2009 pandemic A(H1N1) virus (A[H1N1]pdm09 virus) was estimated to cause 201 000 respiratory deaths in the first 12 months, with most deaths occurring in patients aged <65 years [2, 3•]. The avian influenza viruses A(H5N1), A(H7N9), A(H10N8), A(H5N6), together with the severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses, are the most virulent respiratory viruses affecting humans [4•, 5, 6, 7]. Most patients with influenza virus infection develop mild upper respiratory tract infection. Life-threatening complications include severe viral pneumonia or secondary bacterial pneumonia, acute respiratory distress syndrome, pulmonary embolism, myocarditis, encephalopathy, Reye's syndrome, hemophagocytic syndrome, multiorgan dysfunction and exacerbation of underlying chronic cardiovascular and respiratory diseases [3•, 4•]. Currently available antivirals and vaccines have limited efficacy in the treatment and prevention of influenza virus infection [8]. Understanding the pathogenesis of influenza virus infection in human is important in designing novel strategies to improve the management of influenza.
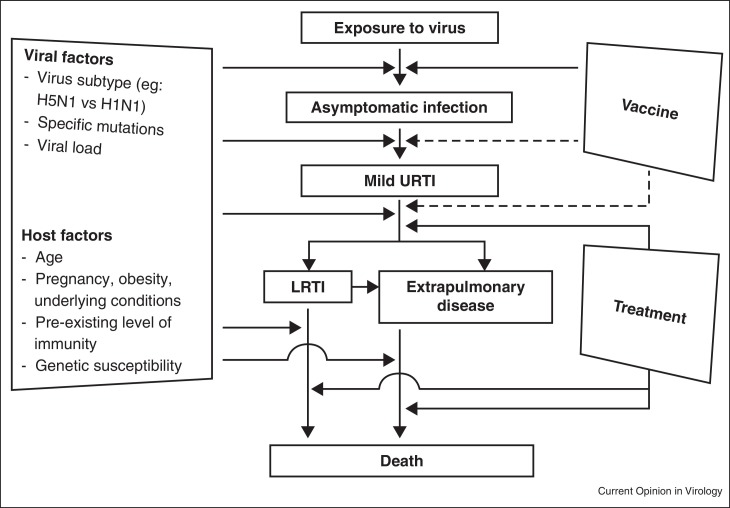
To cause infection, influenza virus needs to evade the host immunity, enter and replicate inside host cells, and disseminate to other cells or organs. Host damage can be a result of direct virus-induced damage, immune-mediated damage and/or secondary bacterial infection [9, 10, 11]. Virologists have extensively studied the role of viral components in the viral life cycle and in the pathogenesis of influenza virus infection in humans [12]. Specific amino acid changes in the viral proteins have been associated with increased disease severity in humans or adaptation of avian influenza viruses in humans [13]. Clinical risk factors for severe influenza have been well described (Figure 1 ) [3•]. However, specific human genes regulating the influenza virus life cycle or virus-induced inflammatory responses are less well described.
Figure 1.
Natural course of influenza virus infection. LRTI, lower respiratory tract infection; URTI, upper respiratory tract infection.
Traditionally, the study of host genes depends on prior knowledge of a candidate gene. The importance of the candidate gene is verified using single gene knockout/knockdown or gain-of-function studies with reverse genetics in vitro or in vivo. For example, Chinese and Japanese patients with influenza-associated encephalopathy (IAE) were found to have elevated (C16:0 + C18:1)/C2 acylcarnitines ratios. Carnitine palmitoyltransferase 2 (CPT2) variants F352C and V368I were found to be over-represented in these IAE patients when compared with the general population. These mutations render this enzyme susceptible to inactivation at high temperature occurring in febrile patients [14, 15].
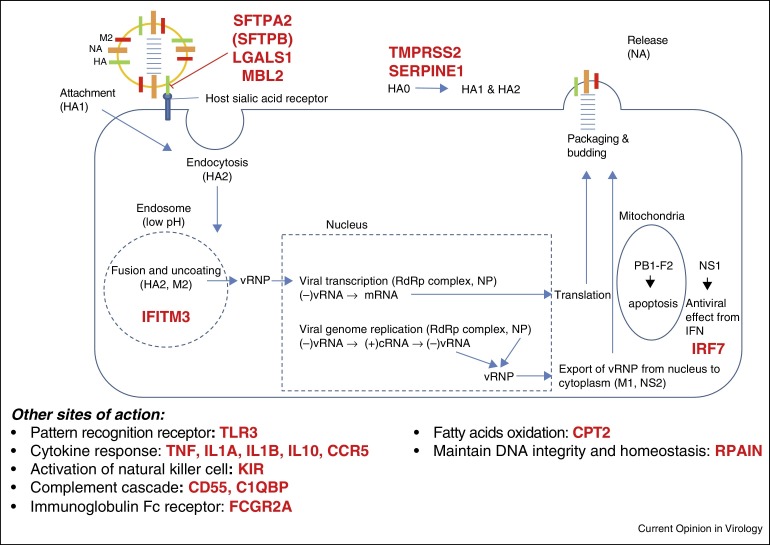
High-throughput screening platforms have allowed researchers to systematically screen a large number of genes associated with influenza virus infection in vitro, in animals or in humans. The availability of deep sequencing data of the human genome allows researchers to compare genetic variations between influenza patients and the general population [16•]. These technological advances, combined with in vitro or bioinformatics analysis, have revealed several genes associated with severe influenza (Figure 2 and Table 1 ). In this review, we focus on specific host genes that have been shown to be directly related to the pathogenesis of human influenza identified or with updated knowledge in the past 2 years.
Figure 2.
Host genetic determinants of influenza virus disease severity identified in humans. Host genes that have been associated with severe influenza are highlighted in red.
Table 1.
Host genes associated with severe influenza virus in humans
| Gene | Human studies |
Animal studies | Phenotypic studies in cell lines in vitro |
References | ||
|---|---|---|---|---|---|---|
| Genetic association study from large patient cohorts | Genetic variants identified from small patient cohorts with severe disease | Comparison of different genetic variants from human cells | Confirmation of functional significance of identified genetic variant | |||
| IFITM3 | +a,b | + | − | + | + | [25, 26•, 27, 28, 29, 30, 32, 33, 35, 79] |
| IRF7 | − | + | − | + | + | [38, 39, 40••] |
| SERPINE1 | − | − | + | + | + | [47••] |
| TMRPSS2 | + | − | − | + | + | [42, 43, 44, 45, 46, 80] |
| LGALS1 | + | − | − | + | + | [23, 24•] |
| MBL2 | +b | − | − | − | +c | [18, 54, 81] |
| SFTPA2 | + | − | − | − | + | [18] |
| SFTPB | +a | − | − | − | − | [20••] |
| CD55 | +a | − | − | − | + | [82] |
| C1QBP | + | − | − | − | − | [58] |
| FCGR2A | +b | − | − | − | − | [58, 59] |
| CPT2 | + | + | − | − | + | [14, 15, 83] |
| TNF | +b | − | − | + | + | [48, 49, 50, 52, 53, 54, 84] |
| IL-1A, IL-1B | + | − | − | − | + | [56, 57] |
| TLR3 | + | + | − | + | + | [63, 64, 65] |
| KIR | +a | − | − | − | − | [85, 86] |
| CCR5 | +b | + | − | − | − | [87, 88, 89, 90] |
| RPAIN | + | − | − | − | − | [58] |
Abbreviations: C1QBP, complement component 1 Q subcomponent-binding protein; CCR5, chemokine (C-C motif) receptor 5; CD55, CD55 molecule, decay accelerating factor for complement; CPT2, carnitine palmitoyltransferase 2; FCGR2A, Fc fragment of IgG, low affinity IIa, receptor; IFITM3, interferon-induced transmembrane protein 3; IL, interleukin; IRF7, interferon regulatory factor 7; ISG, interferon stimulated genes; KIR, killer-cell immunoglobulin-like receptors; LGALS1, lectin, galactoside-binding, soluble, 1; MBL2, mannose binding lectin 2; RPAIN, RPA interacting protein; SFTPA2, surfactant protein A2; SFTPB, surfactant protein B; TLR3, toll like receptor 3; TMPRSS2, transmembrane protease, serine 2; TNF, tumor necrosis factor.
Significant association demonstrated in more than one genetic association study or in more than one cohort of patients.
At least one study showing no statistical significance between severe cases and controls.
Does not inhibit A(H1N1)pdm09 because this virus lacks a paucity of glycan.
Antimicrobial molecules in the airway
Four types of surfactant proteins are found in human pulmonary surfactant. Both surfactant protein A2 (SFTPA2) and surfactant protein D (SFTPD) are collectins and exhibit antiviral activity against influenza virus [17]. SFTPA2 variants (rs1965708-C, rs1059046-A, and haplotype 1A[0]) have been associated with more severe respiratory deterioration [18]. Surfactant protein B (SFTPB) is a hydrophobic protein responsible for the structural integrity of the pulmonary alveoli [19]. In a cohort of 84 Chinese patients with severe and mild A(H1N1)pdm09 infection who were matched for age, sex and underlying conditions, the SNP rs1130866-C was significantly associated with severe disease. The association between rs1130866-C and severe disease was verified in a second cohort patients with A(H1N1)pdm09 infection, in which multivariate analysis was performed to adjust for potential confounding factors [20••]. The SNP rs1130866-C has been associated with glycosylation at the amino acid residue 129 of SFTPB, which reduces secretion of SFTPB [21]. It remains to be determined whether SFTPB has direct antiviral activity against influenza virus. Surfactant protein C (SFTPC) is a lipoprotein with antiviral activity against respiratory syncytial virus [22], but its antiviral activity against influenza virus or its genetic variants associated with severe influenza has not yet been reported.
Lectin, galactoside-binding, soluble, 1 (LGALS1), also known as galectin 1, can bind to influenza viruses and inhibit viral replication [23]. Carriage of LGALS1 rs4820294/rs2899292 haplotype GG was associated with protection from A(H7N9) virus infection in humans. Furthermore, rs4820294/rs2899292 haplotype GG was correlated with higher levels of LGALS1 mRNA and protein expression in lymphoblast cell lines [24•]. Therefore, the differential LGALS1 expression may contribute to the distinct susceptibility of some individuals to human A(H7N9) influenza.
The interferon pathway
Upon influenza virus infection, type I interferons are produced by host cells to limit viral replication. The function of type I interferons is mediated via the expression of many interferon stimulated genes (ISGs). Several ISGs are especially important in the pathogenesis of influenza virus infection. Interferon-induced transmembrane proteins (IFITM), including IFITM1, IFITM2 and IFITM3, were identified to have direct antiviral activity against influenza virus in a genomewide siRNA screen [25]. A human long noncoding RNA, which reduces IFITM3 gene expression, enhanced influenza virus replication in a transgenic mouse model [26•]. IFITM3, which is located in endosome, inhibits viral replication by blocking the fusion of the viral and the host membrane [27, 28]. Interestingly, amphotericin B, an antifungal, was found to increase viral replication by interfering with the blockage of membrane fusion by IFITM3 [29]. The C/C genotype of rs12252 is associated with a 21-amino-acid truncation at the N-terminal of the IFITM3 protein, which alters the localization of the IFITM3 from endosomal compartment to the cell periphery [30]. Several studies have looked into IFITM3 SNP rs12252 in the susceptibility to severe influenza in humans. The genotype rs12252-C was over-represented in Caucasian and Chinese patients with severe A(H1N1)pdm09 virus infection [31••, 32]. In patients with A(H7N9) infection, those carrying rs12252-C/C genotype had higher mortality [33]. However, the importance of rs12252-C allele in severe influenza has been challenged. Firstly, in a study using pseudotyped influenza A viruses, transfection of A549 cells with plasmids expressing the truncated form of IFITM3 could reduce viral replication similarly to those with plasmids carrying the full length IFITM3 [34]. Secondly, in a study comparing 34 patients with severe A(H1N1)pdm09 virus infection and >5000 controls, no significant difference in the rs12252 genotype frequency was found [35].
Interferon regulatory factor 7 (IRF7) is a transcription factor regulating the expression of type I interferons [36]. Influenza A virus replicates to high titers in Madin-Darby canine kidney cells with knockdown of the IRF7 gene [37]. Murine tracheal epithelial cells deficient in IRF7 have impaired expression of interferons after influenza virus infection [38]. In a study comparing the genetic variants in 534 healthy individuals, IRF7 SNP rs12805435 was found to be associated with the induction of antiviral genes in dendritic cells in response to influenza virus infection [39]. In a 7-year-old girl without known immunodeficiency who suffered from severe influenza virus infection, IRF7 mutation was identified using whole exome sequencing [40••]. The induction of type I and type III interferon genes in dendritic cells and pulmonary epithelial cells were found to be impaired in this girl.
Proteolytic activation of viral hemagglutinin
Post-translational cleavage of influenza virus hemagglutinin by host protease is a prerequisite for the virus-host membrane fusion and thereby, for virus infectivity, tissue tropism and virus pathogenicity [41]. Transmembrane protease, serine 2 (TMPRSS2), a type II transmembrane serine protease, cleaves and activates the viral hemagglutinin during influenza virus infection [42]. Three independent studies assessed the role of TMPRSS2 in influenza-infected mice. TMPRSS2 knockout mice were resistant to influenza A(H7N9) and A(H1N1) virus infection, but there were discrepant results regarding susceptibility to A(H3N2) virus infection [43, 44, 45]. By integration of a pilot genomewide association study and the lung expression quantitative trait loci (eQTL) dataset, a TMPRSS2 intronic SNP rs2070788 was prioritized for further studies [46]. The genetic predisposition of rs2070788 to severe A(H1N1)pdm09 was validated in 409 A(H1N1)pdm09 patients including 162 severe cases and 247 mild controls. In the functional study, a regulatory SNP rs383510 in high linkage disequilibrium with rs2070788 was uncovered as the causal variant underlying the genetic association. Genetic predispositions of rs2070788 and rs383510 to severe influenza were also validated in an A(H7N9) patient cohort.
SERPINE1, an ISG, encodes plasminogen activator inhibitor 1 (PAI-1). A549 cells over-expressing SERPINE1 inhibited the spread of influenza A virus when compared to cells without SERPINE1 overexpression [47••]. PAI-1 inhibits the extracellular cleavage of hemagglutinin from HA0 into HA1 and HA2. Serpine1 knockout mice had higher viral titers and more severe disease than wild type mice. In human fibroblast cell lines derived from patients carrying rs6092-A allele which results in intracellular retention of PAI-1, influenza virus replication was enhanced when compared with infection in cell lines deriving from patients carrying rs6092-T/T genotype.
Pro-inflammatory and anti-inflammatory cytokines
Cytokines are important in the defense against influenza virus infection. However, excessive cytokine response is associated with severe influenza [9, 32]. Tumor necrosis factor-α (TNF-α) has been considered a proinflammatory cytokine, and treatment with anti-TNF-α improved the survival of A(H1N1)-infected mice [48]. However, TNF-knockout mice had more severe disease [49]. Furthermore, the soluble form of TNF-α has been shown to be required for the control of CD8+ T cell response [50], which suggests that TNF-α is required for the control of infection. A network based approach combining mouse, human and in vitro data showed that the TNF pathway is important in influenza virus infection [51•]. TNF-238A and TNF-308G alleles have been associated with severe influenza in studies comparing A(H1N1)pdm09 patients and healthy controls [52, 53]. However, these associations were not found in another study comparing fatal cases and the general population [54].
Interleukin-10 (IL-10) is persistently elevated in patients with severe influenza virus infection [9]. IL-10 knockout mice were shown to have more rapid viral clearance and improved survival [55]. Blockage of IL-1β could ameliorate inflammation of influenza-virus infected human pulmonary endothelial cells and lung fibroblasts [56]. IL-10-592C, IL-10-1082A allele and IL-10-1082 A/A genotype have been associated with severe disease [53]. In another study involving 167 patients with A(H1N1)pdm09 virus infection and 192 healthy controls, rs17561 of IL1A and rs1143627 of IL1B gene were associated with susceptibility to infection [57]. However, the associations of SNPs in IL-10, IL-1A and IL-1B genes with influenza virus disease severity were based on single cohorts, and further studies are necessary to confirm the significance of these polymorphisms in influenza.
Immunoglobulin Fc receptor
The interaction between the host antibody Fc region and Fc receptors is important in the immune defense against influenza virus infection. Fc fragment of IgG, low affinity IIa, receptor (FCGR2A) gene encodes Fcγ receptor IIA (FcγRIIA), which binds immune complexes. The association between FCGR2A polymorphism and severe A(H1N1)pdm09 virus infection was first identified in a European cohort [58], although such association was not found in a Chinese cohort [59]. FcγRIIA signaling was found to be important in immune complex-mediated platelet activation during influenza virus infection [60]. Interestingly, in recent studies of broadly neutralizing antibodies against influenza viruses, the ability to form immune complex by the interaction between Fc region of the antibody and Fcγ receptors may mediate antibody-dependent cellular cytotoxicity, which is important for in vivo efficacy of the antibody [61, 62].
Pattern recognition receptor
TLR3 is a major sensor of viral double stranded RNA. F303S mutation of TLR3 was found to be associated with influenza-associated encephalopathy [63], and SNP rs5743313-C/T genotype was associated with A(H1N1)pdm09 patients with pneumonia [64]. The importance of TLR3 has been subsequently confirmed in a knockout mice study [65].
The future on the study of host genes in humans
This review has summarized host genes that have been shown to be important for the pathogenesis of influenza in humans. With high-throughput assays, a large number of genes or genetic variants were found to be associated with viral replication or disease severity in animals or in humans. The current challenge is how we can select specific host genes that are important for the treatment or prevention of influenza virus infection. Discrepancies in the results from similar studies also raise the suspicion whether certain genes are indeed important. For example, out of 925 host factors which affect influenza virus replication that were identified during in vitro genomewide siRNA knockdown screening, only 69 genes were present in at least two of these screens [66]. In human genetic association studies, many genetic variants identified to be associated with disease severity in one study could not be reproduced in other studies (Table 1).
Several strategies can be used to improve our ability to find genes that are relevant to influenza pathogenesis in humans. In human genetic association studies, it is important to limit confounding factors as much as possible. For example, the cases and controls should be matched for age, sex, ethnic group and comorbidities, and multivariate analysis can be utilized to assess whether the genetic variant is an independent risk factor. The identified genetic variants should be validated in other independent cohorts. The functional significance of these genetic variants can be refined by eQTL analysis [67]. Pathway analysis helps to identify pathways that may not be apparent when analyzing individual genes. Interactome screens, which identify host genes that interact with viral proteins, may increase the likelihood of identifying genes that are potential host targets for treatment [68]. The establishment of genetically diverse mice population has allowed the identification of susceptibility genes by correlating genetic variations to disease phenotypes [69•]. In addition to genetic data, host response to influenza has also been assessed using transcriptomic, proteomic, metabolomic and lipidomic analysis of cell cultures, animals or humans [70•, 71]. These data reflect biological regulation of host factors at different levels. An integration of these ‘omics’ data will improve the accuracy of identifying the susceptibility genes [72••].
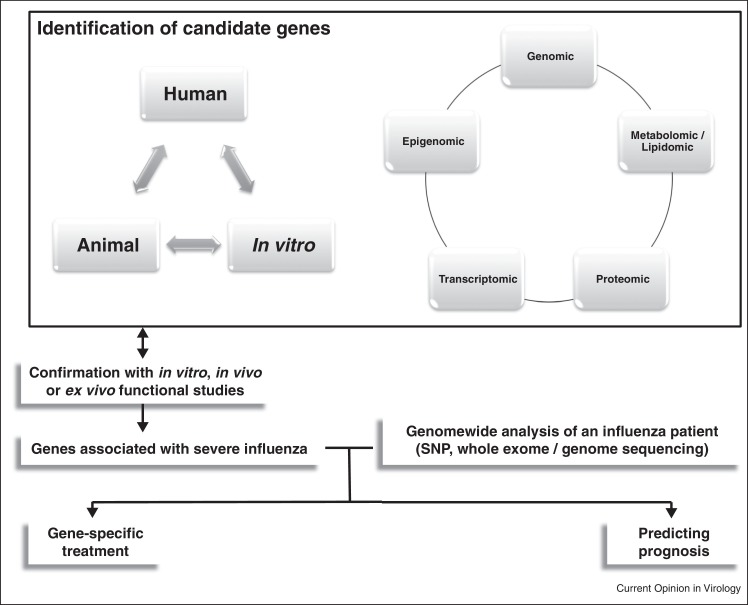
Although studies have identified several genetic polymorphic genes that predispose to severe influenza, it is possible that these susceptible individuals may have very different susceptibility gene combinations. In the future, it is possible that whole genome sequencing will allow more accurate identification of susceptibility gene combinations in any particular individual (Figure 3 ).
Figure 3.
Pathways of identifying and characterizing susceptibility genes in human and potential clinical application.
In addition to comparing severe and mild influenza cases, another approach to identify host susceptibility genes is to examine patients who are resistant to influenza virus infection. In a study of a patient with a congenital disorder of glycosylation but without serological evidence of influenza virus infection, virus production was only found in 1 of 3 macrophage cultures derived from this patient [73]. The authors of this study postulated that the lack of glycosylation of viral surface proteins may affect virus production.
Conclusion
Understanding the role of host factors in virus infections have led to host-targeted antivirals. DAS181 removes sialic-acid containing receptors from respiratory epithelial cells and prevents virus attachment to host cells. In a phase II clinical trial, DAS181 has been shown to reduce nasal or pharyngeal viral load in influenza patients [74]. Antivirals and anti-inflammatory agents targeting host factors and vaccine strategies improving host response have shown promise in animal models. For example, sphingosine-1-phosphate agonist, protectin D1 and anti-leptin antibody have been shown to protect mice against lethal influenza virus infection [70•, 75, 76]. Imiquimod, a TLR7 agonist, has been shown to expedite and augment antibody response after influenza virus vaccine in both animal and human studies [77, 78]. An integration of in vitro, animal and clinical data will allow researchers to search for other host targets for treatment and prevention of influenza virus infection.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Our studies presented in this review are supported by the Health and Medical Research Fund of the Food and Health Bureau of the Hong Kong SAR Government [Ref. No. 13120842, 12111412, RRG-05 and HKM-15-M03], National Key Program for Infectious Diseases of China (Ref. No. 2012ZX10004210), the Providence Foundation Limited in memory of the late Dr Lui Hac Minh, and a donation from Larry Chi-Kin Yung.
References
- 1.2014. World Health Organization: Influenza (seasonal) (available at http://www.who.int/mediacentre/factsheets/fs211/en/; accessed on 29 September 2014) [Google Scholar]
- 2.Dawood F.S., Iuliano A.D., Reed C., Meltzer M.I., Shay D.K., Cheng P.Y., Bandaranayake D., Breiman R.F., Brooks W.A., Buchy P. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 3•.Cheng V.C., To K.K., Tse H., Hung I.F., Yuen K.Y. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev. 2012;25:223–263. doi: 10.1128/CMR.05012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review of the 2009 A(H1N1) pandemic.
- 4•.To K.K., Chan J.F., Yuen K.Y. Viral lung infections: epidemiology, virology, clinical features, and management of avian influenza A(H7N9) Curr Opin Pulm Med. 2014;20:225–232. doi: 10.1097/MCP.0000000000000047. [DOI] [PubMed] [Google Scholar]; A review of A(H7N9) infection in humans.
- 5.To K.K., Tsang A.K., Chan J.F., Cheng V.C., Chen H., Yuen K.Y. Emergence in China of human disease due to avian influenza A(H10N8) — cause for concern? J Infect. 2014;68:205–215. doi: 10.1016/j.jinf.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To K.K., Chan J.F., Chen H., Li L., Yuen K.Y. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect Dis. 2013;13:809–821. doi: 10.1016/S1473-3099(13)70167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muthuri S.G., Venkatesan S., Myles P.R., Leonardi-Bee J., Al Khuwaitir T.S., Al Mamun A., Anovadiya A.P., Azziz-Baumgartner E., Baez C., Bassetti M. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med. 2014;2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To K.K., Hung I.F., Li I.W., Lee K.L., Koo C.K., Yan W.W., Liu R., Ho K.Y., Chu K.H., Watt C.L. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis. 2010;50:850–859. doi: 10.1086/650581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J., Yao H., Wo J., Fang Q., Cui D. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L., Wang Z., Chen Y., Ding W., Jia H., Chan J.F., To K.K., Chen H., Yang Y., Liang W. Clinical, virological, and histopathological manifestations of fatal human infections by avian influenza A(H7N9) virus. Clin Infect Dis. 2013;57:1449–1457. doi: 10.1093/cid/cit541. [DOI] [PubMed] [Google Scholar]
- 12.Medina R.A., Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J.F., To K.K., Chen H., Yuen K.Y. Cross-species transmission and emergence of novel viruses from birds. Curr Opin Virol. 2015;10:63–69. doi: 10.1016/j.coviro.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Mizuguchi H., Yao D., Ide M., Kuroda Y., Shigematsu Y., Yamaguchi S., Yamaguchi M., Kinoshita M., Kido H. Thermolabile phenotype of carnitine palmitoyltransferase II variations as a predisposing factor for influenza-associated encephalopathy. FEBS Lett. 2005;579:2040–2044. doi: 10.1016/j.febslet.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 15.Mak C.M., Lam C.W., Fong N.C., Siu W.K., Lee H.C., Siu T.S., Lai C.K., Law C.Y., Tong S.F., Poon W.T. Fatal viral infection-associated encephalopathy in two Chinese boys: a genetically determined risk factor of thermolabile carnitine palmitoyltransferase II variants. J Hum Genet. 2011;56:617–621. doi: 10.1038/jhg.2011.63. [DOI] [PubMed] [Google Scholar]
- 16•.Galperin M.Y., Rigden D.J., Fernandez-Suarez X.M. The 2015 Nucleic Acids Research Database Issue and molecular biology database collection. Nucleic Acids Res. 2015;43:D1–D5. doi: 10.1093/nar/gku1241. [DOI] [PMC free article] [PubMed] [Google Scholar]; This annual report summarizes important databases relevant to human genes.
- 17.Han S., Mallampalli R.K. The role of surfactant in lung disease and host defense against pulmonary infections. Ann Am Thorac Soc. 2015 doi: 10.1513/AnnalsATS.201411-507FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera-Ramos E., Lopez-Rodriguez M., Ruiz-Hernandez J.J., Horcajada J.P., Borderias L., Lerma E., Blanquer J., Perez-Gonzalez M.C., Garcia-Laorden M.I., Florido Y. Surfactant protein A genetic variants associate with severe respiratory insufficiency in pandemic influenza A virus infection. Crit Care. 2014;18:R127. doi: 10.1186/cc13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitsett J.A., Wert S.E., Weaver T.E. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.To K.K., Zhou J., Song Y.Q., Hung I.F., Ip W.C., Cheng Z.S., Chan A.S., Kao R.Y., Wu A.K., Chau S. Surfactant protein B gene polymorphism is associated with severe influenza. Chest. 2014;145:1237–1243. doi: 10.1378/chest.13-1651. [DOI] [PubMed] [Google Scholar]; In this hypothesis-drive genetic association study comparing severe and mild influenza cases in 2 independent cohorts, several features were incorporated into the study design to minimize the interference by confounding factors.
- 21.Taponen S., Huusko J.M., Petaja-Repo U.E., Paananen R., Guttentag S.H., Hallman M., Haataja R. Allele-specific N-glycosylation delays human surfactant protein B secretion in vitro and associates with decreased protein levels in vivo. Pediatr Res. 2013;74:646–651. doi: 10.1038/pr.2013.151. [DOI] [PubMed] [Google Scholar]
- 22.Glasser S.W., Senft A.P., Maxfield M.D., Ruetschilling T.L., Baatz J.E., Page K., Korfhagen T.R. Genetic replacement of surfactant protein-C reduces respiratory syncytial virus induced lung injury. Respir Res. 2013;14:19. doi: 10.1186/1465-9921-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M.L., Chen Y.H., Wang S.W., Huang Y.J., Leu C.H., Yeh N.C., Chu C.Y., Lin C.C., Shieh G.S., Chen Y.L. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J Virol. 2011;85:10010–10020. doi: 10.1128/JVI.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Chen Y., Zhou J., Cheng Z., Yang S., Chu H., Fan Y., Li C., Wong B.H., Zheng S., Zhu Y. Functional variants regulating LGALS1 (Galectin 1) expression affect human susceptibility to influenza A(H7N9) Sci Rep. 2015;5:8517. doi: 10.1038/srep08517. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified a novel susceptibility gene for A(H7N9) infection.
- 25.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Ouyang J., Zhu X., Chen Y., Wei H., Chen Q., Chi X., Qi B., Zhang L., Zhao Y., Gao G.F. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated the importance of epigenetics in antiviral response in humans.
- 27.Desai T.M., Marin M., Chin C.R., Savidis G., Brass A.L., Melikyan G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin T.Y., Chin C.R., Everitt A.R., Clare S., Perreira J.M., Savidis G., Aker A.M., John S.P., Sarlah D., Carreira E.M. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 2013;5:895–908. doi: 10.1016/j.celrep.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia R., Pan Q., Ding S., Rong L., Liu S.L., Geng Y., Qiao W., Liang C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study has demonstrated the importance of IFITM3 using in vitro and animal experiments, and identified a IFITM3 genetic variant to be over-represented in hospitalized influenza patients when compared to the general population.
- 32.Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H., Yan H.P., Wu H., Liu J.H., Liu N. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams D.E., Wu W.L., Grotefend C.R., Radic V., Chung C., Chung Y.H., Farzan M., Huang I.C. IFITM3 polymorphism rs12252-C restricts influenza A viruses. PLoS ONE. 2014;9:e110096. doi: 10.1371/journal.pone.0110096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills T.C., Rautanen A., Elliott K.S., Parks T., Naranbhai V., Ieven M.M., Butler C.C., Little P., Verheij T., Garrard C.S. IFITM3 and susceptibility to respiratory viral infections in the community. J Infect Dis. 2014;209:1028–1031. doi: 10.1093/infdis/jit468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikushima H., Negishi H., Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 37.Hamamoto I., Takaku H., Tashiro M., Yamamoto N. High yield production of influenza virus in Madin Darby canine kidney (MDCK) cells with stable knockdown of IRF7. PLoS ONE. 2013;8:e59892. doi: 10.1371/journal.pone.0059892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crotta S., Davidson S., Mahlakoiv T., Desmet C.J., Buckwalter M.R., Albert M.L., Staeheli P., Wack A. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M., Imboywa S.H., Chipendo P.I., Ran F.A., Slowikowski K. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Ciancanelli M.J., Huang S.X., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015 doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of the use of whole exome sequencing in identifying genes associated with susceptibility to severe influenza.
- 41.Bottcher-Friebertshauser E., Garten W., Matrosovich M., Klenk H.D. The hemagglutinin: a determinant of pathogenicity. Curr Top Microbiol Immunol. 2014;385:3–34. doi: 10.1007/82_2014_384. [DOI] [PubMed] [Google Scholar]
- 42.Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pohlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakai K., Ami Y., Tahara M., Kubota T., Anraku M., Abe M., Nakajima N., Sekizuka T., Shirato K., Suzaki Y. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol. 2014;88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarnow C., Engels G., Arendt A., Schwalm F., Sediri H., Preuss A., Nelson P.S., Garten W., Klenk H.D., Gabriel G. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol. 2014;88:4744–4751. doi: 10.1128/JVI.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatesuer B., Bertram S., Mehnert N., Bahgat M.M., Nelson P.S., Pohlman S., Schughart K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9:e1003774. doi: 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Z., Zhou J., To K.K., Chu H., Li C., Wang D., Yang D., Zheng S., Hao K., Bosse Y. The identification of TMPRSS2 as the susceptible gene for severe illness of 2009 pandemic A(H1N1) influenza and infection of A(H7N9) influenza. J Infect Dis. 2015 doi: 10.1093/infdis/jiv246. (Apr 22; pii: jiv246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Dittmann M., Hoffmann H.H., Scull M.A., Gilmore R.H., Bell K.L., Ciancanelli M., Wilson S.J., Crotta S., Yu Y., Flatley B. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell. 2015;160:631–643. doi: 10.1016/j.cell.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated the importance of SERPINE1 gene in influenza virus pathogenesis by showing the differences in the phenotype of cells isolated from patients with different genetic variant in SERPINE1 gene.
- 48.Shi X., Zhou W., Huang H., Zhu H., Zhou P., Zhu H., Ju D. Inhibition of the inflammatory cytokine tumor necrosis factor-alpha with etanercept provides protection against lethal H1N1 influenza infection in mice. Crit Care. 2013;17:R301. doi: 10.1186/cc13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damjanovic D., Divangahi M., Kugathasan K., Small C.L., Zganiacz A., Brown E.G., Hogaboam C.M., Gauldie J., Xing Z. Negative regulation of lung inflammation and immunopathology by TNF-alpha during acute influenza infection. Am J Pathol. 2011;179:2963–2976. doi: 10.1016/j.ajpath.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeBerge M.P., Ely K.H., Enelow R.I. Soluble, but not transmembrane, TNF-alpha is required during influenza infection to limit the magnitude of immune responses and the extent of immunopathology. J Immunol. 2014;192:5839–5851. doi: 10.4049/jimmunol.1302729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Bao S., Zhou X., Zhang L., Zhou J., To K.K., Wang B., Wang L., Zhang X., Song Y.Q. Prioritizing genes responsible for host resistance to influenza using network approaches. BMC Genomics. 2013;14:816. doi: 10.1186/1471-2164-14-816. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study incorporated in vitro, animal and human data to prioritize genes for future research on genetic susceptibility to severe influenza.
- 52.Antonopoulou A., Baziaka F., Tsaganos T., Raftogiannis M., Koutoukas P., Spyridaki A., Mouktaroudi M., Kotsaki A., Savva A., Georgitsi M. Role of tumor necrosis factor gene single nucleotide polymorphisms in the natural course of 2009 influenza A H1N1 virus infection. Int J Infect Dis. 2012;16:e204–e208. doi: 10.1016/j.ijid.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Ocana J., Olivo-Diaz A., Salazar-Dominguez T., Reyes-Gordillo J., Tapia-Aquino C., Martinez-Hernandez F., Manjarrez M.E., Antonio-Martinez M., Contreras-Molina A., Figueroa-Moreno R. Plasma cytokine levels and cytokine gene polymorphisms in Mexican patients during the influenza pandemic A(H1N1)pdm09. J Clin Virol. 2013;58:108–113. doi: 10.1016/j.jcv.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 54.Ferdinands J.M., Denison A.M., Dowling N.F., Jost H.A., Gwinn M.L., Liu L., Zaki S.R., Shay D.K. A pilot study of host genetic variants associated with influenza-associated deaths among children and young adults. Emerg Infect Dis. 2011;17:2294–2302. doi: 10.3201/eid1712.111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun K., Torres L., Metzger D.W. A detrimental effect of interleukin-10 on protective pulmonary humoral immunity during primary influenza A virus infection. J Virol. 2010;84:5007–5014. doi: 10.1128/JVI.02408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim K.S., Jung H., Shin I.K., Choi B.R., Kim D.H. Induction of interleukin-1 beta (IL-1beta) is a critical component of lung inflammation during influenza A (H1N1) virus infection. J Med Virol. 2015 doi: 10.1002/jmv.24138. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y., Li S., Zhang G., Nie G., Meng Z., Mao D., Chen C., Chen X., Zhou B., Zeng G. Genetic variants in IL1A and IL1B contribute to the susceptibility to 2009 pandemic H1N1 influenza A virus. BMC Immunol. 2013;14:37. doi: 10.1186/1471-2172-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuniga J., Buendia-Roldan I., Zhao Y., Jimenez L., Torres D., Romo J., Ramirez G., Cruz A., Vargas-Alarcon G., Sheu C.C. Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur Respir J. 2012;39:604–610. doi: 10.1183/09031936.00020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan J.F., To K.K., Tse H., Lau C.C., Li I.W., Hung I.F., Chan K.H., Cheng V.C., Lai T.S., Woo P.C. The lower serum immunoglobulin G2 level in severe cases than in mild cases of pandemic H1N1 2009 influenza is associated with cytokine dysregulation. Clin Vaccine Immunol. 2011;18:305–310. doi: 10.1128/CVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boilard E., Pare G., Rousseau M., Cloutier N., Dubuc I., Levesque T., Borgeat P., Flamand L. Influenza virus H1N1 activates platelets through FcgammaRIIA signaling and thrombin generation. Blood. 2014;123:2854–2863. doi: 10.1182/blood-2013-07-515536. [DOI] [PubMed] [Google Scholar]
- 61.Tharakaraman K., Subramanian V., Cain D., Sasisekharan V., Sasisekharan R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe. 2014;15:644–651. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hidaka F., Matsuo S., Muta T., Takeshige K., Mizukami T., Nunoi H. A missense mutation of the Toll-like receptor 3 gene in a patient with influenza-associated encephalopathy. Clin Immunol. 2006;119:188–194. doi: 10.1016/j.clim.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Esposito S., Molteni C.G., Giliani S., Mazza C., Scala A., Tagliaferri L., Pelucchi C., Fossali E., Plebani A., Principi N. Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol J. 2012;9:270. doi: 10.1186/1743-422X-9-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leung Y.H., Nicholls J.M., Ho C.K., Sia S.F., Mok C.K., Valkenburg S.A., Cheung P., Hui K.P., Chan R.W., Guan Y. Highly pathogenic avian influenza A H5N1 and pandemic H1N1 virus infections have different phenotypes in Toll-like receptor 3 knockout mice. J Gen Virol. 2014;95:1870–1879. doi: 10.1099/vir.0.066258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Chassey B., Meyniel-Schicklin L., Aublin-Gex A., Andre P., Lotteau V. Genetic screens for the control of influenza virus replication: from meta-analysis to drug discovery. Mol Biosyst. 2012;8:1297–1303. doi: 10.1039/c2mb05416g. [DOI] [PubMed] [Google Scholar]
- 67.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe T., Kawakami E., Shoemaker J.E., Lopes T.J., Matsuoka Y., Tomita Y., Kozuka-Hata H., Gorai T., Kuwahara T., Takeda E. Influenza virus–host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014;16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Ferris M.T., Aylor D.L., Bottomly D., Whitmore A.C., Aicher L.D., Bell T.A., Bradel-Tretheway B., Bryan J.T., Buus R.J., Gralinski L.E. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used genetically diverse inbred mice to identify genetic susceptibility to severe influenza.
- 70•.Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153:112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]; This study showed that treatment targeting lipid mediators can improve the survival of mice with severe influenza.
- 71.Hoang L.T., Tolfvenstam T., Ooi E.E., Khor C.C., Naim A.N., Ho E.X., Ong S.H., Wertheim H.F., Fox A., Van Vinh Nguyen C. Patient-based transcriptome-wide analysis identify interferon and ubiquination pathways as potential predictors of influenza A disease severity. PLoS ONE. 2014;9:e111640. doi: 10.1371/journal.pone.0111640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Ritchie M.D., Holzinger E.R., Li R., Pendergrass S.A., Kim D. Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]; This review summarizes the methods in integrating data obtained from different experimental systems.
- 73.Sadat M.A., Moir S., Chun T.W., Lusso P., Kaplan G., Wolfe L., Memoli M.J., He M., Vega H., Kim L.J. Glycosylation, hypogammaglobulinemia, and resistance to viral infections. N Engl J Med. 2014;370:1615–1625. doi: 10.1056/NEJMoa1302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moss R.B., Hansen C., Sanders R.L., Hawley S., Li T., Steigbigel R.T. A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J Infect Dis. 2012;206:1844–1851. doi: 10.1093/infdis/jis622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teijaro J.R., Walsh K.B., Cahalan S., Fremgen D.M., Roberts E., Scott F., Martinborough E., Peach R., Oldstone M.B., Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146:980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang A.J., To K.K., Li C., Lau C.C., Poon V.K., Chan C.C., Zheng B.J., Hung I.F., Lam K.S., Xu A. Leptin mediates the pathogenesis of severe 2009 pandemic influenza A(H1N1) infection associated with cytokine dysregulation in mice with diet-induced obesity. J Infect Dis. 2013;207:1270–1280. doi: 10.1093/infdis/jit031. [DOI] [PubMed] [Google Scholar]
- 77.To K.K., Zhang A.J., Chan A.S., Li C., Cai J.P., Lau C.C., Li C.G., Jahan A.S., Wu W.L., Li L. Recombinant influenza A virus hemagglutinin HA2 subunit protects mice against influenza A(H7N9) virus infection. Arch Virol. 2015;160:777–786. doi: 10.1007/s00705-014-2314-x. [DOI] [PubMed] [Google Scholar]
- 78.Hung I.F., Zhang A.J., To K.K., Chan J.F., Li C., Zhu H.S., Li P., Li C., Chan T.C., Cheng V.C. Immunogenicity of intradermal trivalent influenza vaccine with topical imiquimod: a double blind randomized controlled trial. Clin Infect Dis. 2014;59:1246–1255. doi: 10.1093/cid/ciu582. [DOI] [PubMed] [Google Scholar]
- 79.Bailey C.C., Huang I.C., Kam C., Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bottcher E., Matrosovich T., Beyerle M., Klenk H.D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verma A., White M., Vathipadiekal V., Tripathi S., Mbianda J., Ieong M., Qi L., Taubenberger J.K., Takahashi K., Jensenius J.C. Human H-ficolin inhibits replication of seasonal and pandemic influenza A viruses. J Immunol. 2012;189:2478–2487. doi: 10.4049/jimmunol.1103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou J., To K.K., Dong H., Cheng Z.S., Lau C.C., Poon V.K., Fan Y.H., Song Y.Q., Tse H., Chan K.H. A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J Infect Dis. 2012;206:495–503. doi: 10.1093/infdis/jis378. [DOI] [PubMed] [Google Scholar]
- 83.Yao D., Mizuguchi H., Yamaguchi M., Yamada H., Chida J., Shikata K., Kido H. Thermal instability of compound variants of carnitine palmitoyltransferase II and impaired mitochondrial fuel utilization in influenza-associated encephalopathy. Hum Mutat. 2008;29:718–727. doi: 10.1002/humu.20717. [DOI] [PubMed] [Google Scholar]
- 84.Seo S.H., Webster R.G. Tumor necrosis factor alpha exerts powerful anti-influenza virus effects in lung epithelial cells. J Virol. 2002;76:1071–1076. doi: 10.1128/JVI.76.3.1071-1076.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.La D., Czarnecki C., El-Gabalawy H., Kumar A., Meyers A.F., Bastien N., Simonsen J.N., Plummer F.A., Luo M. Enrichment of variations in KIR3DL1/S1 and KIR2DL2/L3 among H1N1/09 ICU patients: an exploratory study. PLoS ONE. 2011;6:e29200. doi: 10.1371/journal.pone.0029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aranda-Romo S., Garcia-Sepulveda C.A., Comas-Garcia A., Lovato-Salas F., Salgado-Bustamante M., Gomez-Gomez A., Noyola D.E. Killer-cell immunoglobulin-like receptors (KIR) in severe A (H1N1) 2009 influenza infections. Immunogenetics. 2012;64:653–662. doi: 10.1007/s00251-012-0623-3. [DOI] [PubMed] [Google Scholar]
- 87.Keynan Y., Juno J., Meyers A., Ball T.B., Kumar A., Rubinstein E., Fowke K.R. Chemokine receptor 5 big up tri, open32 allele in patients with severe pandemic (H1N1) 2009. Emerg Infect Dis. 2010;16:1621–1622. doi: 10.3201/eid1610.100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sironi M., Cagliani R., Pontremoli C., Rossi M., Migliorino G., Clerici M., Gori A. The CCR5Delta32 allele is not a major predisposing factor for severe H1N1pdm09 infection. BMC Res Notes. 2014;7:504. doi: 10.1186/1756-0500-7-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodriguez A., Falcon A., Cuevas M.T., Pozo F., Guerra S., Garcia-Barreno B., Martinez-Orellana P., Perez-Brena P., Montoya M., Melero J.A. Characterization in vitro and in vivo of a pandemic H1N1 influenza virus from a fatal case. PLoS ONE. 2013;8:e53515. doi: 10.1371/journal.pone.0053515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falcon A., Cuevas M.T., Rodriguez-Frandsen A., Reyes N., Pozo F., Moreno S., Ledesma J., Martinez-Alarcon J., Nieto A., Casas I. CCR5 deficiency predisposes to fatal outcome in influenza virus infection. J Gen Virol. 2015 doi: 10.1099/vir.0.000165. [DOI] [PubMed] [Google Scholar]