Highlights
-
•
Pathogens that transmit via the fecal-oral route are generally very stable.
-
•
Different pathogens have similar requirements to become human-to-human transmissible.
-
•
Research strategies for zoonotic pathogens should include mechanisms of transmission.
Abstract
Bacterial, viral and parasitic zoonotic pathogens that transmit via the fecal-oral route have a major impact on global health. However, the mechanisms underlying the emergence of such pathogens from the animal reservoir and their persistence in the human population are poorly understood. Here, we present a framework of human-to-human transmission of zoonotic pathogens that considers the factors relevant for fecal-oral human-to-human transmission route at the levels of host, pathogen, and environment. We discuss current data gaps and propose future research directions.
Current Opinion in Virology 2017, 22:1–6
This review comes from a themed issue on Emerging viruses: intraspecies transmission
Edited by Ron A.M Fouchier and Lin-Fa Wang
For a complete overview see the Issue and the Editorial
Available online 23rd November 2016
http://dx.doi.org/10.1016/j.coviro.2016.11.001
1879-6257/© 2016 The Authors. Published by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Problem setting
In recent years there have been many examples of pathogens crossing the species barrier and infecting humans, although the vast majority of these zoonotic events did not result in sustained human-to-human transmission [1, 2, 3]. Nevertheless, the continuing emergence of zoonotic pathogens is a cause of concern globally, especially due to the high morbidity and mortality of pathogens like MERS-CoV and A/H5N1 influenza virus [4, 5]. Human-to-human transmission of microorganisms generally occurs via one or multiple transmission routes, including the fecal-oral, airborne, direct contact, or vector-borne route. Whilst pathogens including bacteria, parasites and viruses have very different biological properties, they can employ similar routes of transmission and emergence. Identification of the mechanisms underlying the effective human-to-human transmission of emerging zoonotic pathogens and their commonalities across different pathogens, may help design of interventions aimed at reducing the risk of sustained human-to-human transmission after a zoonotic event.
Expert opinion meeting
As part of the activities of the ANTIGONE consortium on the emergence of zoonotic pathogens, an expert opinion meeting was organized. Using a comparative approach including parasites, bacteria and viruses that transmit via the fecal-oral route, the meeting aimed at identifying the key drivers of sustained human-to-human transmission after a zoonotic event, taking into account the host, the pathogen and the interface (transmission amplifiers). In addition, major knowledge gaps were identified that require future research in order to better control emerging zoonotic pathogens that potentially are transmitted through a fecal-oral route. The main conclusions of this meeting are presented in this perspective.
A framework of fecal-oral transmission
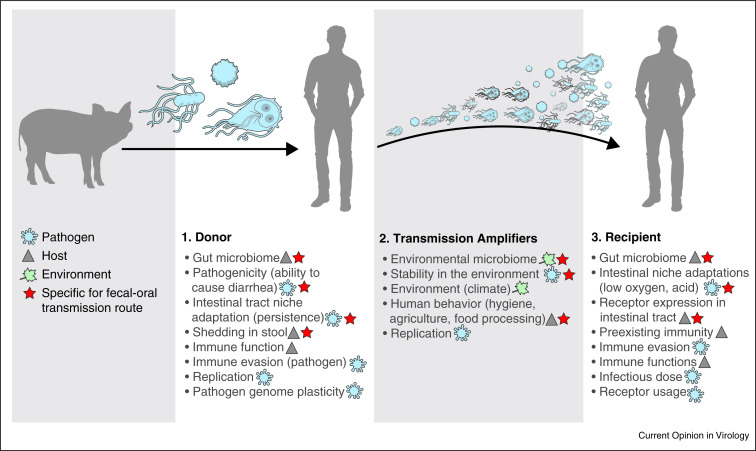
Enteric pathogens can be transmitted between humans by the fecal-oral route via direct contact or indirect contact via contaminated fluids, including surface water, food, and carriers such as fomites (Figure 1 ). The risk of a zoonotic pathogen becoming human-to-human transmissible depends on its adaptation to the human host and the environment. To analyze this process, we considered fecal-oral transmission of a zoonotic pathogen between two human hosts as follows; the human host that is infected with a zoonotic pathogen after a zoonotic event is defined as the donor while the susceptible human host that is subsequently infected by the first human host is considered the recipient. The transmission interface is the environment that the pathogen encounters after release from the donor and before infecting the recipient. For sustained fecal-oral human-to-human transmission certain elements in this transmission cycle, which we will refer to as transmission amplifiers, appear essential whereas other elements are not an absolute requirement, but increase the likelihood of transmission. Transmission amplifiers may interact and their presence may or may not depend on conditions under which transmission occurs, including for example socio-economic conditions and cultural and behavioral variation. We designed a framework of human-to-human transmission that includes the transmission amplifiers relevant for the fecal-oral transmission route at the levels of host, pathogen, and environment (Figure 2 ).
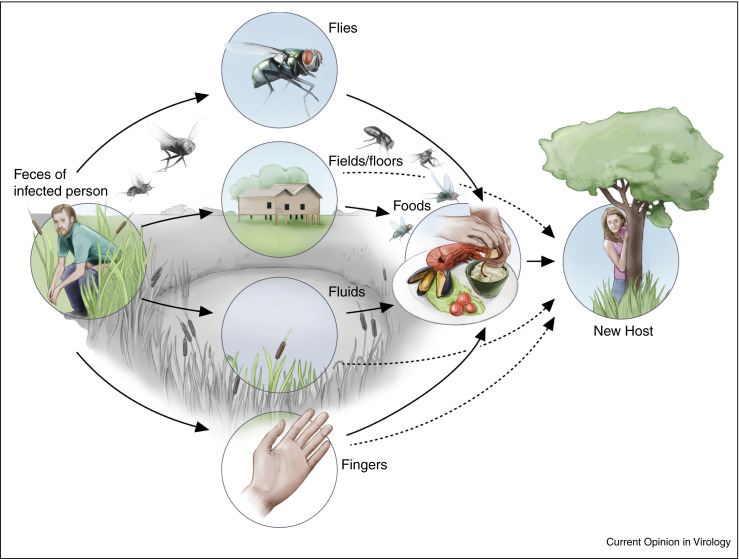
Figure 1.
Fecal-oral transmission between humans. After shedding from the host enteric pathogens can be transmitted between humans by the fecal-oral route via direct contact between humans, or via indirect contact via contaminated fluids, including surface water, food, and carriers such as fomites.
Figure 2.
Framework for human-to-human transmission after a zoonotic event showing the key transmission amplifiers from the host (triangle), pathogen (blue) and environmental transmission amplifiers (green), respectively. The transmission amplifiers that are specific to the fecal-oral route are indicated with a red star.
Transmission amplifiers specific for fecal-oral transmission
Several key transmission amplifiers are specific for fecal-oral transmission (Figure 2), such as the intestinal microbiomes of the donor and recipient hosts. Individuals with a healthy intestine are less likely to become infected or colonized by opportunistic pathogens, although the resistance provided by a healthy colonization (microbiome) can, in principle, be disrupted by a pathogenic species depending on its pathogenic potential (virulence) [6, 7]. The composition of the human intestinal microbiome is, amongst others dependent on the presence of a functional immune system [8, 9, 10]. Changes in microbiome composition, in addition to the impaired immunity itself, may impact the outcome of infection and subsequent transmission.
Clinical symptoms such as diarrhea and vomiting can increase the likelihood of fecal-oral transmission as they can facilitate the spread of a pathogen into the environment and onto fomites [11]. Remarkably, most pathogens that transmit via the fecal-oral route are very stable and can survive under various conditions, which may be related to the fact that these pathogens have to pass the hostile conditions of the gastrointestinal tract. Zoonotic pathogens need to adapt to factors specific to this niche, such as the acidic conditions in the stomach and low oxygen in the large intestine, the temperature and the availability of specific sugars and nutrients. For example, comparative genomics of Cryptosporidium parvum genotype IIc suggests that the ability to establish an infection in a particular host species may depend in part on the presence of transporters controlling the exchange of metabolites between the host cell and the pathogen [12].
Fecal shedding of a pathogen does not necessarily require replication in the intestine. For example, the hepatitis E virus (HEV) is shed via the feces despite its liver tropism [13]. However, the presence of receptors and the tissue distribution of these receptors is a crucial element for tropism of infection, the shedding of microorganisms in stool and subsequent human-to-human transmission. In addition, it should be noted that not all pathogens that are shed via the feces transmit via the fecal-oral route. Several respiratory viruses of zoonotic origin, that are capable of human-to-human transmission, are shed in feces. During SARS-CoV, MERS-CoV and influenza A infection, viral RNA can be detected in stool. However, for these pathogens there is currently no evidence of fecal-oral transmission resulting in disease [14, 15, 16, 17, 18, 19]. Similarly, the enteric pathogen Campylobacter subspecies jejuni was transmitted from human-to-human via sexual contact following a zoonotic event [20].
Once a donor is shedding the pathogen, environmental factors at the transmission interface can have a large impact on transmission efficiency. Contamination of the surface water after flooding can magnify the size of an outbreak via waterborne and foodborne routes [21]. Food sources can be contaminated by irrigation with sewage-contaminated water or the use of manure that contains traces of human feces, or on site by food-handlers. Anecdotally, even food preservation measures can impact transmission as some additives to preserve lettuce were shown to also increase the stability of hepatitis A virus [22]. Transmission via food can have a major impact on the global spread of pathogens. In 2011 nearly 4000 people were infected during an Escherichia coli O104:H4 outbreak in Europe, resulting in 54 deaths. Epidemiological and trace-back investigations pointed to salad sprouts as the possible contaminated food source [23].
Transmission amplifiers not specific for fecal-oral transmission route
Several factors that are important transmission amplifiers of the likelihood of fecal-oral transmission are generic to most human-to-human transmission routes.
The immune system of the human host is an important factor that has to be confronted for sustained human-to-human transmission. Most pathogens that successfully transmit, have acquired genes that can counteract or evade the adaptive and or innate immune responses. Pathogens may have adapted through altered domains that are recognized by the cellular or humoral immune system, to evade pre-existing immunity based on previously infecting pathogens. Loss of genes or gene function may also be associated with adaptation to the human host. A Salmonella enterica serotype Typhimurium clone which causes bloodstream infection amongst children and HIV-infected adults in Sub-Saharan Africa, has adapted to these immuno-compromised hosts through loss of gene functions enabling bacterial survival outside the host, whilst retaining the ability to cause enteritis in multiple host species [24••].
With the general population aging and technologies becoming more invasive, medical interventions can become an amplifier of human-to-human transmission. Although medical interventions do not necessarily select for human-to-human transmissible pathogens, they can increase the duration of infection, and thereby the likelihood of evolution and adaptation [25]. For instance, the application of extracorporeal membrane oxygenation prolongs and increases survival in patients with otherwise acute fatal infectious disease [26]. The use of immune modulatory and suppressive drugs has created a human population that is more susceptible to prolonged pathogen proliferation and shedding [27, 28, 29, 30, 31]. An immunodeficient individual was found to excrete a vaccine-derived poliovirus for twenty years. During this time the virus became virulent and changed antigenically [32]. The unrestrained use of antimicrobial drugs in medical and veterinary care and in agriculture in low-income, middle-income, and high-income countries creates an unprecedented selective pressure that may select for pathogens that are more transmissible. Salmonella enterica serotype Typhimurium has the ability to develop a super shedder phenotype, that can be induced by antibiotic treatment in mice [33]. A human reservoir for non-typhoid Salmonella (NTS) transmission of multiple serotypes was demonstrated in a study of NTS-infected patients who continued to shed NTS for months up to years, and strains of these patients acquired antimicrobial resistance genes and virulence genes that possibly affected host–pathogen interactions [34••].
The infectious dose, replication kinetics and the number of pathogens being shed can have a major impact on the efficiency of fecal-oral transmission. For example, norovirus and Shigella spp. require a low infectious dose and can be transmitted via hands and fomites [35, 36], whereas Listeria monocytogenes infections require a high infectious dose [37], making these transmission routes less likely. However, surprisingly little is known about the infectious dose for many fecal-orally transmitted human pathogens. As for shedding in the environment, several strategies can be successful, such as shedding of lower amounts of microorganisms over a long period (chronic/persistent infection) and thus a long period of transmission associated with mild clinical symptoms , or shedding of high loads of microorganisms for a relatively short period with significant clinical symptoms [36].
Receptor usage is a key element for successful human-to-human transmission. Not only are the site of the expression of these receptors in the host and the receptor specificity of the pathogen of importance, but also the prevalence of these receptors in the human population can potentially contribute to the likelihood of efficient transmission. For group A rotaviruses attachment to histobloodgroup antigens is an essential step for infection. Interestingly, strains of the P[9], P[14] and P[25] subtypes that are generally found in cattle but can also infect humans, attach to the blood group A epitope [38]. However, since the frequency of blood group A in human populations ranges from 20% to 40%, the majority of individuals is expected to be resistant to infection by these strains, which would constitute a barrier to transmission. Sustained human-to-human transmission for a virus with a relatively low R 0 would thus require an adaptation of the glycan binding capacity to the human HBGA genetic polymorphism [39]. HEV genotype 1 and 2 exclusively infect humans while genotypes 3 and 4 infect pigs but occasionally infect and transmit between humans [40]. The high conservation of HEV attachment and entry factors may explain the observed cross-species transmission while factors limiting efficient human-to-human transmission are thought to include regulation of subgenomic translation and specific virus–host receptor interactions [41, 42].
Inherent differences between viruses, bacteria, and parasites
Surprisingly few differences exist between the basic requirements of the different types of pathogens to become human-to-human transmissible. However, bacteria can replicate in the environment whereas viruses and parasites cannot. As the transmittable stages of parasites are environmentally very resistant, and can withstand water treatment processes, parasites are probably more likely to be transmitted via food and water compared to direct fecal-oral-transmission [43, 44••]. Genome plasticity is an important factor for all pathogens but while parasites, viruses and bacteria can all adapt by mutations, recombination and lateral gene transfer, viruses can also acquire genes by genome rearrangements and bacteria can acquire mobile genetic elements that carry genes that may encode determinants that facilitate increased fitness in certain conditions.
Synthesis
‘For a zoonotic pathogen the risk of becoming human-to-human transmissible depends on further adaptation to the human host. For efficient fecal-oral transmission amplifiers in the transmission interface appear crucial.’
Knowledge gaps and outlook
The focus of the public health and emerging infectious disease communities on emerging viruses causing severe infections, has resulted in the discovery of several possible determinants of sustained human-to-human transmission of zoonotic viruses. However, the likelihood of sustained human-to-human fecal-oral transmission after a zoonotic event of any pathogen type is difficult to assess as these events are hardly described in current literature. One could conclude that zoonotic pathogens rarely become human-to-human transmissible through the fecal-oral route because there may be need for dual adaptation, i.e. to the harsh conditions in the environment in addition to the human host, and therefore these events are rare. However, we cannot exclude that we may be missing some of these events, because it can be difficult to distinguish between strictly human and zoonotic pathogens once the latter have established themselves in the human population or because they remain undistinguished with the use of current clinical microbiology tools. For example, recent results strongly suggest that a pig roundworm can act as an important source of human ascariasis [45, 46, 47] but this can go unnoticed as the human and pig parasite population show minor phenotypic and genotypic differences. Zoonotic pathogens that transmit via the fecal-oral route appear to cause similar clinical symptoms compared to other (related) enteric pathogens and further research is not pushed due the relative lack of (more) serious disease. We thus may neglect relevant pathogens and even if we do study these pathogens, the results may have undesired economic consequences for agriculture and food sectors. In addition, until recently we lacked tools for studying important zoonotic events; whilst the small genomes and rapid evolution of viruses allow identification of novel causative agents with limited sequencing effort, such analysis is much more complicated for bacterial and parasitic enteric pathogens which have relatively large genomes. Some experimental models for fecal-oral transmission between hosts have been described, such as for norovirus [48••], but there is a general lack of suitable animal models to study fecal-oral transmission. In fact, most research on host-pathogen interactions is focused on mechanisms of pathogenicity rather than on mechanisms of transmission, whilst the latter is crucial for the development of intervention strategies to prevent further spread and to prevent sustained human-to-human transmission of zoonotic pathogens.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The authors would like to thank all participants of the Dahlem ‘Inter human barriers’ workshop for their contributions and Ryan Kissinger (NIAID, NIH) for designing the figures. This work was supported by ANTIGONE (grant numbers 278976); EdW is supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, US National Institutes of Health; PF receives funding from the EU FP7 project PREPARE (grant number 602525); MdG is supported by the EU grant COMPARE (grant number 643476) and the Virgo Consortium, funded by the Dutch government (project number FES0908).
References
- 1.Short K.R., Richard M., Verhagen J.H., van Riel D., Schrauwen E.J., van den Brand J.M., Manz B., Bodewes R., Herfst S. One health, multiple challenges: the inter-species transmission of influenza A virus. One Health. 2015;1:1–13. doi: 10.1016/j.onehlt.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martella V., Banyai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 3.Adams N.L., Byrne L., Smith G.A., Elson R., Harris J.P., Salmon R., Smith R., O’Brien S.J., Adak G.K., Jenkins C. Shiga toxin-producing Escherichia coli O157, England and Wales, 1983–2012. Emerg Infect Dis. 2016;22:590–597. doi: 10.3201/eid2204.151485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.http://www.who.int/influenza/human_animal_interface/en/
- 6.Singh P., Teal T.K., Marsh T.L., Tiedje J.M., Mosci R., Jernigan K., Zell A., Newton D.W., Salimnia H., Lephart P. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome. 2015;3:45. doi: 10.1186/s40168-015-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stecher B., Robbiani R., Walker A.W., Westendorf A.M., Barthel M., Kremer M., Chaffron S., Macpherson A.J., Buer J., Parkhill J. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taur Y., Pamer E.G. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis. 2013;26:332–337. doi: 10.1097/QCO.0b013e3283630dd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arrieta M.C., Stiemsma L.T., Amenyogbe N., Brown E.M., Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson M.J., Jeffery I.B., Conde S., Power S.E., O’Connor E.M., Cusack S., Harris H.M., Coakley M., Lakshminarayanan B., O'Sullivan O. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 11.Makison Booth C. Vomiting Larry: a simulated vomiting system for assessing environmental contamination from projectile vomiting related to norovirus infection. J Infect Prev. 2014;15:176–180. doi: 10.1177/1757177414545390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widmer G., Lee Y., Hunt P., Martinelli A., Tolkoff M., Bodi K. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect Genet Evol. 2012;12:1213–1221. doi: 10.1016/j.meegid.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuthbert J.A. Hepatitis A: old and new. Clin Microbiol Rev. 2001;14:38–58. doi: 10.1128/CMR.14.1.38-58.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng P.K., Wong D.A., Tong L.K., Ip S.M., Lo A.C., Lau C.S., Yeung E.Y., Lim W.W. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman V.M., Albarrak A.M., Omrani A.S., Albarrak M.M., Farah M.E., Almasri M., Muth D., Sieberg A., Meyer B., Assiri A.M. Viral shedding and antibody response in 37 patients with middle east respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilantika C., Sedyaningsih E.R., Kasper M.R., Agtini M., Listiyaningsih E., Uyeki T.M., Burgess T.H., Blair P.J., Putnam S.D. Influenza virus infection among pediatric patients reporting diarrhea and influenza-like illness. BMC Infect Dis. 2010;10:3. doi: 10.1186/1471-2334-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W., Tang F., Fontanet A., Zhan L., Zhao Q.M., Zhang P.H., Wu X.M., Zuo S.Q., Baril L., Vabret A. Long-term SARS coronavirus excretion from patient cohort, China. Emerg Infect Dis. 2004;10:1841–1843. doi: 10.3201/eid1010.040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z., Liu Y., Xu L., Guan W., Zhang X., Qi T., Shi B., Song Z., Liu X., Wan Y. Extra-pulmonary viral shedding in H7N9 Avian Influenza patients. J Clin Virol. 2015;69:30–32. doi: 10.1016/j.jcv.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Gaudreau C., Rodrigues-Coutlee S., Pilon P.A., Coutlee F., Bekal S. Long-lasting outbreak of erythromycin- and ciprofloxacin-resistant Campylobacter jejuni subspecies jejuni from 2003 to 2013 in men who have sex with men, Quebec, Canada. Clin Infect Dis. 2015;61:1549–1552. doi: 10.1093/cid/civ570. [DOI] [PubMed] [Google Scholar]
- 21.de Man H., van den Berg H.H., Leenen E.J., Schijven J.F., Schets F.M., van der Vliet J.C., van Knapen F., de Roda Husman A.M. Quantitative assessment of infection risk from exposure to waterborne pathogens in urban floodwater. Water Res. 2014;48:90–99. doi: 10.1016/j.watres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Bidawid S., Farber J.M., Sattar S.A. Survival of hepatitis A virus on modified atmosphere-packaged (MAP) lettuce. Food Microbiol. 2001;18:95–102. [Google Scholar]
- 23.Karch H., Denamur E., Dobrindt U., Finlay B.B., Hengge R., Johannes L., Ron E.Z., Tonjum T., Sansonetti P.J., Vicente M. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Mol Med. 2012;4:841–848. doi: 10.1002/emmm.201201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Singletary L.A., Karlinsey J.E., Libby S.J., Mooney J.P., Lokken K.L., Tsolis R.M., Byndloss M.X., Hirao L.A., Gaulke C.A., Crawford R.W. Loss of multicellular behavior in epidemic African nontyphoidal Salmonella enterica serovar Typhimurium ST313 strain D23580. MBio. 2016;7:e02265. doi: 10.1128/mBio.02265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrated that the loss of gene functions can help pathogens to adapt to the human host.
- 25.Russell C.A., Fonville J.M., Brown A.E., Burke D.F., Smith D.L., James S.L., Herfst S., van Boheemen S., Linster M., Schrauwen E.J. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raman L., Dalton H.J. Year in review 2015: extracorporeal membrane oxygenation. Respir Care. 2016;61:986–991. doi: 10.4187/respcare.04985. [DOI] [PubMed] [Google Scholar]
- 27.Ye X., Van J.N., Munoz F.M., Revell P.A., Kozinetz C.A., Krance R.A., Atmar R.L., Estes M.K., Koo H.L. Noroviruses as a cause of diarrhea in immunocompromised pediatric hematopoietic stem cell and solid organ transplant recipients. Am J Transpl. 2015;15:1874–1881. doi: 10.1111/ajt.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siebenga J.J., Beersma M.F., Vennema H., van Biezen P., Hartwig N.J., Koopmans M. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J Infect Dis. 2008;198:994–1001. doi: 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- 29.Murk J.L., de Vries A.C., GeurtsvanKessel C.H., Aron G., Osterhaus A.D., Wolthers K.C., Fraaij P.L. Persistent spiking fever in a child with acute myeloid leukemia and disseminated infection with enterovirus. J Clin Virol. 2014;61:453–455. doi: 10.1016/j.jcv.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Greendyke W.G., Pereira M.R. Infectious complications and vaccinations in the posttransplant population. Med Clin North Am. 2016;100:587–598. doi: 10.1016/j.mcna.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 31.van der Vries E., Stittelaar K.J., van Amerongen G., Veldhuis Kroeze E.J., de Waal L., Fraaij P.L., Meesters R.J., Luider T.M., van der Nagel B., Koch B. Prolonged influenza virus shedding and emergence of antiviral resistance in immunocompromised patients and ferrets. PLoS Pathog. 2013;9:e1003343. doi: 10.1371/journal.ppat.1003343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn G., Klapsa D., Wilton T., Stone L., Minor P.D., Martin J. Twenty-eight years of poliovirus replication in an immunodeficient individual: impact on the Global Polio Eradication Initiative. PLoS Pathog. 2015;11:e1005114. doi: 10.1371/journal.ppat.1005114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawley T.D., Bouley D.M., Hoy Y.E., Gerke C., Relman D.A., Monack D.M. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Marzel A., Desai P.T., Goren A., Schorr Y.I., Nissan I., Porwollik S., Valinsky L., McClelland M., Rahav G., Gal-Mor O. Persistent infections by nontyphoidal Salmonella in humans: epidemiology and genetics. Clin Infect Dis. 2016;62:879–886. doi: 10.1093/cid/civ1221. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that NTS strains of persistently infected humans acquired antimicrobial resistance and virulence genes that possibly affect host-pathogen interactions.
- 35.DuPont H.L., Levine M.M., Hornick R.B., Formal S.B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 36.Atmar R.L., Opekun A.R., Gilger M.A., Estes M.K., Crawford S.E., Neill F.H., Graham D.Y. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalton C.B., Austin C.C., Sobel J., Hayes P.S., Bibb W.F., Graves L.M., Swaminathan B., Proctor M.E., Griffin P.M. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N Engl J Med. 1997;336:100–105. doi: 10.1056/NEJM199701093360204. [DOI] [PubMed] [Google Scholar]
- 38.Hu L., Crawford S.E., Czako R., Cortes-Penfield N.W., Smith D.F., Le Pendu J., Estes M.K., Prasad B.V. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruvoen-Clouet N., Belliot G., Le Pendu J. Noroviruses and histo-blood groups: the impact of common host genetic polymorphisms on virus transmission and evolution. Rev Med Virol. 2013;23:355–366. doi: 10.1002/rmv.1757. [DOI] [PubMed] [Google Scholar]
- 40.Purcell R.H., Emerson S.U. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Cao D., Meng X.J. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect. 2012;1:e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen H.T., Shukla P., Torian U., Faulk K., Emerson S.U. Hepatitis E virus genotype 1 infection of swine kidney cells in vitro is inhibited at multiple levels. J Virol. 2014;88:868–877. doi: 10.1128/JVI.02205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caccio S.M., Chalmers R.M. Human cryptosporidiosis in Europe. Clin Microbiol Infect. 2016;22:471–480. doi: 10.1016/j.cmi.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 44••.Speich B., Croll D., Furst T., Utzinger J., Keiser J. Effect of sanitation and water treatment on intestinal protozoa infection: a systematic review and meta-analysis. Lancet Infect Dis. 2016;16:87–99. doi: 10.1016/S1473-3099(15)00349-7. [DOI] [PubMed] [Google Scholar]; This systemic review and meta-analysis shows how lack of clean water is associated with increased risk of intestinal protozoa infection.
- 45.Zhou C., Li M., Yuan K., Deng S., Peng W. Pig Ascaris: an important source of human ascariasis in China. Infect Genet Evol. 2012;12:1172–1177. doi: 10.1016/j.meegid.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 46.Nejsum P., Betson M., Bendall R.P., Thamsborg S.M., Stothard J.R. Assessing the zoonotic potential of Ascaris suum and Trichuris suis: looking to the future from an analysis of the past. J Helminthol. 2012;86:148–155. doi: 10.1017/S0022149X12000193. [DOI] [PubMed] [Google Scholar]
- 47.Betson M., Nejsum P., Stothard J. Ascaris. Elsevier Science; Holland: 2013. From the twig tips to the deeper branches: new insights into evolutionary history and phylogeography of Ascaris; pp. 265–285. [Google Scholar]
- 48••.Rocha-Pereira J., Jochmans D., Neyts J. Prophylactic treatment with the nucleoside analogue 2′-C-methylcytidine completely prevents transmission of norovirus. J Antimicrob Chemother. 2015;70:190–197. doi: 10.1093/jac/dku363. [DOI] [PubMed] [Google Scholar]; The authors developed a murine fecal-oral transmission model.