Abstract
Interferon-induced transmembrane protein (IFITM) 1, 2 and 3 genes encode a family of interferon (IFN)-induced transmembrane proteins that block entry of a broad spectrum of pathogens. However, the transcriptional regulation of these genes, especially whether there exist any enhancers and their roles during the IFN induction process remain elusive. Here, through public data mining, episomal luciferase reporter assay and in vivo CRISPR-Cas9 genome editing, we identified an IFN-responsive enhancer located 35 kb upstream of IFITM3 gene promoter upregulating the IFN-induced expression of IFITM1, 2 and 3 genes. Chromatin immunoprecipitation (ChIP), electrophoretic mobility shift assay (EMSA) and luciferase reporter assay demonstrated that signal transducers and activators of transcription (STAT) 1 bound to the enhancer with the treatment of IFN and was indispensable for the enhancer activity. Furthermore, using chromosome conformation capture technique, we revealed that the IFITM1, 2 and 3 genes physically clustered together and constitutively looped to the distal enhancer through long-range interactions in both HEK293 and A549 cells, providing structural basis for coordinated regulation of IFITM1, 2 and 3 by the enhancer. Finally, we showed that in vivo truncation of the enhancer impaired IFN-induced resistance to influenza A virus (IAV) infection. These findings expand our understanding of the mechanisms underlying the transcriptional regulation of IFITM1, 2 and 3 expression and its ability to mediate IFN signaling.
Keywords: IFN, IFITM, Enhancer, Long-range interactions, Influenza A virus
Highlights
-
•
This is the first report of an enhancer coordinately upregulating IFN-induced expression of IFITM genes
-
•
The enhancer functions through IFN-induced STAT1 binding and constitutive long-range interactions
-
•
The enhancer contributes to the IFN-induced resistance to IAV infection
1. Introduction
Interferons (IFNs) are of vital importance for the host to defense on a variety of viruses [1]. A number of interferon-stimulated genes (ISGs) activated by IFN mediate virus-inhibiting function at all stages of virus life cycle [2]. Among these, interferon-induced transmembrane (IFITM) 1, 2 and 3, a cluster of genes encoding membrane proteins, inhibit the fusion of viral envelope with endosomal membrane [3], and are therefore responsible for limiting infection by many kinds of viruses including influenza A virus (IAV), West Nile virus, Dengue virus and severe acute respiratory syndrome coronavirus [4], [5], [6], [7]. Additionally, polymorphisms within IFITM genes might lead to differences in host susceptibility to viruses. Specifically, several laboratories demonstrated that the IFITM3 coding region genetic variant rs12252-C which is supposed to alter a splice acceptor site is associated with higher susceptibility to IAV [8], [9], [10], [11].
Human IFITM1, 2 and 3 genes reside on chromosome 11 as a cluster. They are expressed basally in primary tissues and cell lines [12] and robustly induced by either type I or type II IFNs [13]. Both Type I and type II IFNs activate ISGs through Janus kinase (JAK)-STAT pathway. The difference is type I IFNs, such as IFNα and IFNβ, induce gene expression by the combinatorial binding of transcription factor interferon regulatory factor 9 (IRF9), phosphorylated STAT1 and 2 to IFN-stimulated response elements (ISREs) within the target gene promoter, while type II IFN, IFNγ, exerts its regulation through the induced binding of phosphorylated STAT1 homodimers to the gamma-activated sequence (GAS) [14]. Chromatin-remodeling factor BRG1 is also essential for IFITM1, 2 and 3 gene expressions [15], [16], as it constitutively binds to the promoters of IFITM1, 2 and 3 genes and remodels the chromatin to make the promoters accessible to STAT1/STAT1/IRF9 transcription factor complex and transcription machinery [17]. Sp1 and TEF-1 are reported to be involved in recruiting BRG1 to the promoter of IFITM3 gene [16], [18].
Enhancers are the key cis-regulatory DNA elements that increase the transcription of target genes and thus play a fundamental role in development, immunity, and disease [19], [20], [21], [22]. However, as to IFITM1, 2 and 3 genes, there are no any reported enhancers. The role and mechanism of enhancers on the expression of IFITM1, 2 and 3 remain unknown.
In this study, we surveyed the genomic context around IFITM locus and identified an IFN-responsive enhancer located 35 kb upstream of IFITM3 gene promoter. The enhancer drove the coordinate expression of IFITM1, 2 and 3 through constitutive long-range interactions and IFN-induced STAT1 binding. As a consequence, truncation of this enhancer decreased IFN-induced resistance to IAV infection.
2. Materials and methods
2.1. Plasmids
Genomic fragments of E1-1, E1-2, E1-3, E2-1, E2-2, E2-3 and R were amplified from human genome by PCR. Truncated E2-3 was amplified from the genome of HEK293 cells in which E2-3 was truncated. As STAT1 binding motif was in the primer binding region of E2-3, mutant E2-3 with three mutations in STAT1 binding motif and negative control mutant E2-3 with three mutations outside STAT1 binding motif were directly amplified using corresponding mutant primers. All the fragments were subcloned into pGL3-promoter reporter constructs respectively (Promega, USA). Table S1 contained the positions of E1-1, E1-2, E1-3, E2-1, E2-2, E2-3 and R fragments in the human genome (hg19). Primers used for PCR were listed in Table S2.
2.2. Cell lines, cytokines and virus
A549 (human lung epithelial, ATCC CCL-185) was cultured in DMEM/F12 supplemented with 10% fetal bovine serum. HEK293 (human embryonic kidney, ATCC CRL-1573) was cultured in α-MEM supplemented with 10% fetal bovine serum. The two cell lines were authenticated using short tandem repeat (STR) genotyping analysis by Beijing Microread Gene Technology Company (China).
Human interferon β-1a and interferon α-A/D were purchased from PBL Assay Science (USA) and used at indicated concentrations.
Influenza A virus A/Puerto Rico/8/1934 (H1N1) (PR8, ATCC VR-1469) was used in this study and virus titers were determined based on the 50% tissue infectious dose (TCID50) assay using Madin-Darby canine kidney (MDCK) cells according to the Reed-Muench method [23].
2.3. Luciferase reporter assay
Luciferase reporter assays were performed as described [24]. Briefly, the pGL3-promoter Firefly luciferase reporter constructs were co-transfected using lipofectamine 2000 reagent (Life technology, USA) with the vector phRG-TK (Promega, USA) which expresses synthetic Renilla luciferase to normalize the transfection efficiency. Luciferase activities were measured using the Dual-Luciferase Reporter Assay reagent (Promega, USA) on a LB 960 Centro XS3 luminometer (Berthold Technologies, Germany). Relative luciferase activities were expressed as ratios between Firefly and Renilla luciferase activities.
2.4. CRISPR
To truncate E2-3, we designed two flanking guide RNAs (5′-GGAAAACATTCCCGAACCGA-3′ and 5′-CCGAAGTGACGACCGCACCT-3′) and subcloned into pGK2.1 plasmid (Genloci Biotechnologies, China) expressing Cas9. HEK293 cells were transfected with the plasmid and selected by puromycin. Resistant cells were then cloned by limited dilution in 96-well plates and screened for E2-3-truncated clones using a PCR-based approach. Primers are as follows: 5′-CCTCCCAGGTTCACCCCATT-3′ and 5′-AAACGGAGAAAGCGCCTGG-3′. Truncated clones were validated by Sanger sequencing.
2.5. Real-time PCR for mRNA analysis
Total RNA was isolated with Trizol reagent (Life technology, USA), and reverse transcription was performed using with TransScript® All-in-one First-Strand cDNA Synthesis SuperMix for qPCR (Transgen Biotech, China). The real-time PCR primers used were listed in Table S2. Real-time PCR was performed using the IQ5 system (Bio-Rad, USA) with TransStart Tip Green qPCR SuperMix (Transgen Biotech, China), and expression was quantified relative to a housekeeping gene ACTB. All experiments were repeated at least three times.
2.6. Western blot
Western blots were carried out using antibodies against IFITM1 (#262939, BBl Life Science, China), IFITM2 (#RLT5532, Ruiying Bio, China), IFITM3 (#3776-1, Epitomics, USA), STAT1 (#sc-345, Santa Cruz, USA), phospo-STAT1 (#RLP00249, Ruiying Bio, China), NP (#ab128193, Abcam, China) and β-actin (#M1210-2, HuaAn Biotechnology, China).
Briefly, 30 μg protein samples were separated by 15% SDS-PAGE and transferred onto PVDF membranes (Millipore, USA). Membranes were blocked with 5% non-fat milk in TBS containing 0.1% Tween at room temperature for 1 h. They were next incubated with indicated primary antibodies at 4 °C overnight followed by incubation with horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (#CW0102S, CWBIO, China) or anti-rabbit antibody (#CW0103S, CWBIO, China) at room temperature for 1 h. Bound antibodies were detected by pro-light HRP chemiluminescent detection reagent (Tiangen, China), following the manufacturer's directions.
2.7. Chromatin immunoprecipitation (ChIP)
Cells were cross-linked in 1% formaldehyde and chromatin was sheared to 350–1000 bp fragments by sonication. Immune complexes containing STAT1 were enriched using antibody against STAT1 (#sc-345, Santa Cruz, USA) and protein-G magnetic beads, washed under stringent conditions, and eluted in TE buffer with 1% SDS. After reversal of cross-linking and purification, immunoprecipitated DNA was analyzed by qPCR. Primers used were listed in Table S2. Enrichment was quantified relative to input. IgG (#2729, Cell Signaling Technology, USA) immunoprecipitation was used as a negative control.
2.8. Electrophoretic mobility shift assay (EMSA)
Nuclear extract of HEK293 cells with 1000 U/mL IFNβ treatment for 4 h was prepared using Nuclear and Cytoplasmic Extraction Kit (CWBIO, China). The protein content of the nuclear extract was determined using BCA Protein Assay Kit (Tiangen, China). 5′ biotin-labeled or unlabeled oligonucleotide probes encompassing the STAT1 binding motif from enhancer E2-3 (5′-GTCGTCACTTCGGGGAAACGGAGAAA-3′) and correspondent unlabeled mutant oligonucleotide probe (5′-GTCGTCACAAGGGCCTTACGGAGAAA-3′) were synthesized by BBl Life Science (China). Briefly, 6 μg of nuclear extract was incubated in a binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT; pH 7.5) with 0.1 pmol biotin-labeled probe and 0.5 μg of poly(dI-dC) for 20 min at room temperature. For competition assays, 50-fold excess of unlabeled oligonucleotide probe was added to the binding buffer before adding the biotin-labeled probe. To perform a supershift EMSA, 9 μg antibody against STAT1 (#sc-345 X, Santa Cruz, USA) was added to the binding buffer 20 min after adding the biotin-labeled probe and incubated for another 20 min. The DNA-protein complexes were separated by electrophoresis on 5% polyacrylamide gels and transferred to a positively charged nylon membrane (GE healthcare, USA), followed by UV cross-linking. The biotinylated signals of the DNA-protein binding complex were detected using LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, USA) following the manufacturer's instructions.
2.9. Chromosome conformation capture (3C)
3C experiments were performed as described [25] with some modifications. Briefly, cells were cross-linked by 1% formaldehyde. Then nuclei were isolated and digested with Pst I restriction enzyme (New England Biolabs, USA) prior to proximity ligation with T4 DNA ligase (Thermo Fisher Scientific, USA). After the reversal of cross-links, DNA was purified and ligation frequencies of restriction fragments were analyzed by PCR, using primers specific to the restriction fragments of interest (Table S2). BAC clone CTD2344F1, covering the entire region of interest, was used to create an artificial library of ligation product to control for PCR efficiency. The ACTB locus was used to control for cross-linking efficiency between different experiments. PCR products were run on agarose gels and quantified with Image J software. All data were normalized to the BAC ligation products and the ACTB control.
2.10. Statistical analysis
All data are presented as means ± SD. We made three biological replicates in all the experiments and we performed unpaired (two tailed) Student's t-tests. Results were considered significant with P < 0.05.
3. Results
3.1. E2-3 is an IFNβ-responsive enhancer
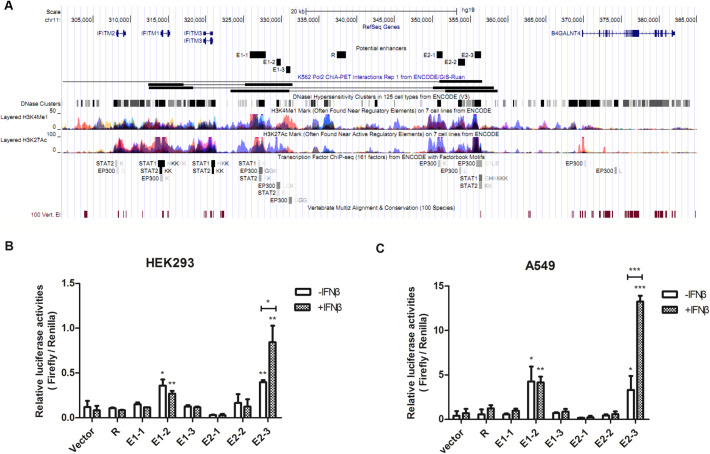
Enhancers, especially long-range enhancers usually communicate with regulated genes through physical interactions [26]. In order to identify putative enhancer elements that could regulate the expression of IFITM gene cluster, we first did the data mining using ENCODE datasets and found two regions interacting with IFITM locus supported by RNA polymerase II ChIA-PET data from K562 cells [27]. Furthermore, these two fragments were enriched with DNaseI hypersensitive sites (DHS), histone modifications of H3K4me1 and H3K27ac and binding sites of histone acetylase p300, which were all hallmarks of enhancers [28], [29]. Notably, STAT1 and STAT2, key components of type I IFN signaling pathway, were bound to the two fragments in several ChIP-seq datasets [30] (Fig. 1A).
Fig. 1.
E2-3 is an IFNβ-responsive enhancer. (A) UCSC Genome Browser output (hg19) of the IFITM genomic region, illustrating the location of the putative enhancer regions. In addition, the ENCODE integrated regulation track containing K562 Pol II ChIA-PET interactions, DNaseI hypersensitivity clusters, H3K4Me1 marks, H3K27Ac marks, transcription factor ChIP-seq data of P300, STAT1 and STAT2, and conservation are displayed. (B) Six fragments, all contained in the two regions interacting with IFITM locus, was used to enhance expression of an SV40 promoter-driven Firefly luciferase reporter gene vector (pGL3-promoter). R fragment, outside the two interacting region, was used as a negative control. Following transient co-transfection, HEK293 cells were treated with PBS or 1000 U/mL IFNβ for 24 h before collected. Results are displayed as ratios of Firefly to SV40 promoter-driven Renilla luciferase activities from three independent experiments. Statistical analyses were performed with unpaired Student's t-tests (*P < 0.05; **P < 0.01; ***P < 0.001). (C) Same as (B) except A549 cells was transfected.
To investigate the potential enhancer activity of these two fragments, we selected six loci which were overlapped with DNaseI hypersensitive sites, enriched with H3K4me1 and H3k27ac and bound by p300 within these two fragments and named them “E1-1”, “E1-2”, “E1-3”, “E2-1”, “E2-2” and “E2-3” respectively (Fig. 1A). These six loci and a negative control locus named as “R” outside these fragments were amplified and cloned upstream the luciferase promoter in pGL3-promoter constructs respectively. Then each construct was transfected into HEK293 cells, and cells were treated with 1000 U/mL IFNβ or PBS for 24 h before collection. We found that both E1-2 and E2-3 displayed enhancer activity (Fig. 1B). However, upon treatment of IFNβ, the enhancer activity of E1-2 did not change whereas the enhancer activity of E2-3 was greatly elevated (Fig. 1B), suggesting E1-2 element was a constitutive enhancer while E2-3 element was an IFNβ-responsive enhancer. Similar results were also confirmed in A549 cells (Fig. 1C). In addition, among all six loci, only E2-3 harbored a region conserved in 100 vertebrate species (Fig. 1A), thus we focused on studying E2-3 in the following research.
3.2. IFITM1, 2 and 3 are the targets of enhancer E2-3
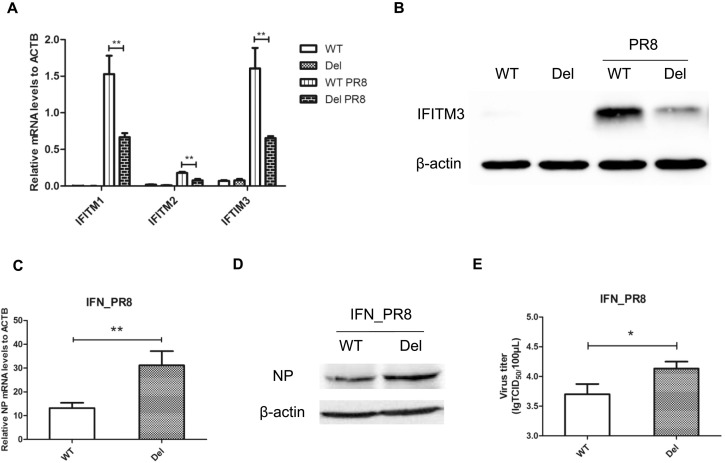
To further validate the enhancer activity of E2-3 and identify its bona fide target genes, we in vivo truncated E2-3 in HEK293 cell line using CRISPR-Cas9 technique (Fig. 2A). Homozygous clone harboring truncated E2-3 was screened and confirmed by PCR and Sanger sequencing (Fig. 2A). Luciferase reporter assay validated that the enhancer activity of the truncated E2-3 was greatly disrupted compared to that of wild type (Fig. 2B). Then we measured mRNA expression levels of IFITM1, IFITM2 and IFITM3 in the mutant and corresponding wild-type control HEK293 cells treated with IFNβ or PBS. Expectedly, we found that IFNβ-induction of IFITM1, 2 and 3 was significantly impaired in the mutant cells (Fig. 2C). This was further validated at the protein level as assessed by western blot (Figs. 2D and S1). By contrast, for the E2-3′s nearest gene B4GALNT4 gene, neither its basal expression nor induction by IFNβ could be detected in our experiments (Fig. 2C). Similar results were also observed when cells were treated with IFNα (Fig. S2), suggesting that IFITM1, 2 and 3, but not B4GALNT4 are bona fide targets of enhancer E2-3. In addition, whether IFNβ treatment or not, no difference was observed in the levels of STAT1 and phosphorylated STAT1 (P-STAT1) between the mutant and corresponding wild-type control HEK293 cells (Fig. S3), ruling out the possibility that impaired induction of IFITM1, 2 and 3 by IFN in mutant HEK293 cells was due to disruption of JAK-STAT pathway by CRISPR off-target effect.
Fig. 2.
IFITM1, 2 and 3 genes are the targets of enhancer E2-3. (A) Scheme depicting CRISPR design to truncate E2-3 in HEK293 cells (top). PCR analysis of representative HEK293 clones (bottom left) and Sanger sequencing validation of truncation (bottom right). (B) Truncated E2-3 was used to enhance expression of an SV40 promoter-driven firefly luciferase reporter gene vector. Wild-type E2-3 was used as positive control. Treatment with IFNβ and determination of reporter gene activity was performed as described in Fig. 1B. (C) Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by quantification of IFITM1, IFITM2, IFITM3 and B4GALNT4 mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests (*P < 0.05; **P < 0.01). (D) Proteins from HEK293 cultures treated as in (C) were analyzed for IFITM3 and β-actin levels by Western blots.
3.3. STAT1 binds to E2-3 with the treatment of IFNβ
To investigate the mechanism of enhancer E2-3 regulating expression of IFITM genes, we analyzed the transcription factor binding on E2-3. In accordance with E2-3 being an IFN-responsive enhancer, ENCODE ChIP-seq data showed that STAT1 bound to E2-3 in GM12878, IFN-γ-treated HeLa-S3 and IFN-α/γ-treated K562 cells while STAT2 bound to E2-3 in IFN-α-treated K562 cells [30] (Fig. 1A). Using ChIP-qPCR, we validated that STAT1 bound on E2-3 in both HEK293 and A549 cells when treated with IFNβ (Fig. 3A and B). No enrichment was found using IgG antibody (data not shown). In addition, we performed STAT1 ChIP-qPCR in HEK293 cells treated with 0, 10, 100, 1000 U/mL IFNβ respectively, and found that STAT1 had a tendency of more enrichment on E2-3 with the increasing concentration of IFNβ while it did not have such a tendency on negative control locus (Fig. 3C), indicating the specificity of STAT1 binding to E2-3 and the critical role of IFNβ in the process.
Fig. 3.
STAT1 binds to E2-3 with the treatment of IFNβ. (A) ChIP-qPCR using antibody against STAT1 in HEK293 cells treated with 1000 U/mL IFNβ or PBS for 4 h. The promoter of IFITM1 served as positive control. A locus near R fragment served as negative control. Statistical analyses were performed with unpaired Student's t-tests (*P < 0.05; **P < 0.01; ***P < 0.001). (B) Same as (A) except performed in A549 cells. (C) Same as (A) except performed in HEK293 cells treated with 10, 100, 1000 U/mL IFNβ or PBS respectively. (D–E) EMSA using biotin-labeled probe encompassing the STAT1 binding motif of enhancer E2-3 incubated with nuclear extract from IFNβ-treated HEK293. (D) For competition assays, 50-fold excess of corresponding unlabeled wild-type or mutant probe was added. (E) For supershift assay, antibody against STAT1 was added. * indicated free probe, ** indicated DNA-protein complex shift, *** indicated DNA-protein-antibody complex supershift. (F) Scheme depicting the design to mutate STAT1 binding motif of E2-3. (G) Mutant E2-3 was used to enhance expression of an SV40 promoter-driven Firefly luciferase reporter gene vector. Control mutant E2-3 with three mutations outside STAT1 binding motif was used as negative control. Treatment with IFNβ and determination of reporter gene activity were performed as described in Fig. 1B.
To determine whether STAT1 directly bound to E2-3, we performed EMSA experiment. Incubating 26-bp probe from E2-3 region containing STAT1 binding motif with nuclear extracts from IFNβ-treated HEK293 cells, we observed a shift band, which could be potently competed by unlabeled DNA with identical sequence (Fig. 3D). We further proved that this interaction was specific, as STAT1 motif mutant DNA could not compete the binding probe to nuclear proteins (Fig. 3D). Moreover, when antibody against STAT1 was added into the nuclear extract, the shift band was remarkably reduced and a weak supershift band appeared above it (Fig. 3E). These results indicated that STAT1 directly bound to E2-3 at least in vitro.
To further characterize the importance of STAT1 binding for the functionality of E2-3, we mutated three positions of STAT1 binding motif in E2-3-pGL3-promoter (Fig. 3F). Luciferase reporter assay indicated that these mutations greatly decreased the enhancer activity and enhancer activity of mutant E2-3 could no longer be induced by IFNβ, while the control mutant E2-3 which was mutated in positions other than STAT1 binding motif showed no significant difference with wild-type E2-3 (Fig. 3G), suggesting that E2-3 might exert its enhancer activity through recruiting STAT1.
3.4. E2-3 constitutively forms long-range interactions with IFITM1, 2 and 3
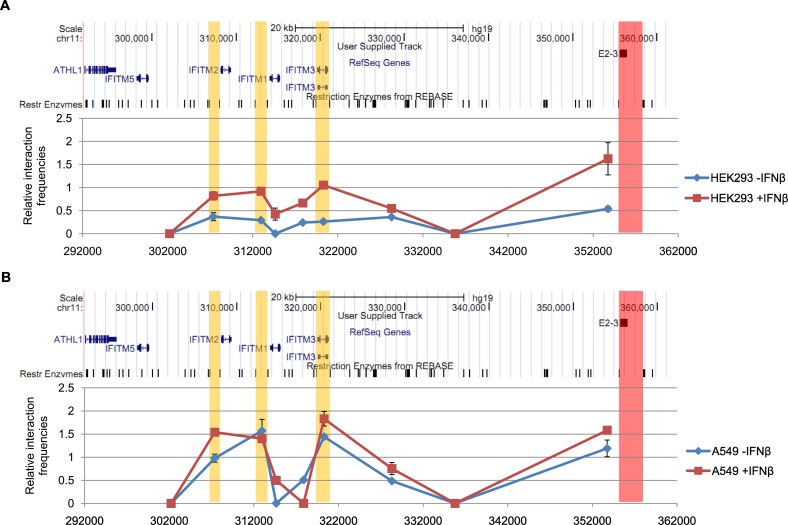
Enhancer E2-3 is about 35 kb away from the IFITM locus, so how does it overcome such a long distance to regulate the expression of IFITM1, 2 and 3 genes? Recent reports have showed that distal enhancers could form long-range interactions with the promoters of the targeted genes to exert its function [26]. To investigate whether enhancer E2-3 also behaved like this, we performed chromosome conformation capture (3C) assay in HEK293 cells [25]. Using enhancer E2-3 as anchor, we observed E2-3 looping to the promoters of IFITM1, 2 and 3. When cells were treated with IFNβ, the interaction frequency increased, in accordance with the elevated expression of IFITM1, 2 and 3 (Fig. 4A).To further validate this result, we also performed 3C assay using IFITM1, IFITM2 or IFITM3 locus as anchor respectively, and as expected, all of 3C data showed that E2-3 could interact with IFITM1, IFITM2 or IFITM3 especially when treated with IFNβ (Fig. S3, Fig. S4, Fig. S5). In addition, we observed the interactions among the promoters of IFITM1, 2 and 3 (Fig. S4, Fig. S5, Fig. S6), suggesting IFITM1, 2 and 3 genes physically clustered together and were coordinated regulated by enhancer E2-3. Similar results were also confirmed in A549 cells (Figs. 4B and Fig. S4, Fig. S5, Fig. S6). Notably, unlike in HEK293, these interaction frequencies in A549 cells seemed not to change with the treatment of IFNβ (Figs. 4B and Fig. S4, Fig. S5, Fig. S6). The mechanism underlining this phenomenon was currently not clear. While cell line authentication of HEK293 and A549 cells using cell line STR genotyping ruled out that these cell lines were misused or contaminated (Fig. S7), one possible explanation for this phenomenon might be due to cell type difference.
Fig. 4.
Enhancer E2-3 forms long-range interactions with IFITM1, 2, 3 genes. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing enhancer E2-3, the yellow bars represented the location of fragments containing the promoters of IFITM1, IFITM2 and IFITM3, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
We also measured the interactions between the truncated enhancer E2-3 and the promoters of IFITM1, 2 and 3 in mutant HEK293 cells. It seemed that the constitutive interactions between the restriction fragment containing E2-3 and restriction fragments containing IFITM1, 2 and 3 were basically unaffected by truncation of E2-3 (Fig. S8), suggesting these interactions might be mediated by factors residing outside E2-3, which were not disrupted by truncation of E2-3.
3.5. Truncation of E2-3 decreased IFNβ-induced resistance to IAV infection
As IFITM1, 2 and 3 are very important for host to resist infection of IAV [4], [8], we infected wild-type and E2-3-truncated HEK293 cells with IAV Puerto Rico/8/1934 (PR8) at a MOI of 0.1 for 24 h. Although IFITM1, 2 and 3 were induced by PR8 virus in both wild-type and E2-3-truncated HEK293 cells, the inductions were greatly impaired upon truncation of E2-3 (Fig. 5A and B), further confirming the regulation of IFITM1, 2 and 3 expression by the enhancer E2-3. Surprisingly, we did not observe a significant effect on the replication of IAV when enhancer E2-3 was truncated, as revealed by the mRNA and protein levels of virus NP and progeny virus titers determined by TCID50 assay (Fig. S9). Nonetheless, when we treated wild-type and E2-3-truncated HEK293 cells with IFNβ respectively for 24 h before infection of PR8, NP mRNA and protein levels were found to be much higher in mutant HEK293 cells (Fig. 5C and D). Moreover, consistent with virus NP expression, the viral titers were also increased in E2-3-truncated cells (Fig. 5E), indicating that enhancer E2-3 was responsible for IFNβ-induced resistance to IAV infection.
Fig. 5.
Truncation of E2-3 decreased IFNβ-induced resistance to IAV. (A) Wild-type (WT) and mutant (Del) HEK293 cells were infected with IAV PR8 at a MOI of 0.1 for 24 h followed by quantification of IFITM1, IFITM2 and IFITM3 mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests (*P < 0.05; **P < 0.01). (B) Proteins from HEK293 cultures treated as in (A) were analyzed for IFITM3 and β-actin levels by Western blots. (C) Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ for 24 h before being infected with IAV PR8 at a MOI of 0.1 for 24 h followed by quantification of virus NP mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests (**P < 0.01). (D) Proteins from HEK293 cultures treated as in (C) were analyzed for virus NP and β-actin levels by Western blots. (E) Progeny virus titers from HEK293 cultures treated as in (C) were determined with the TCID50 assay. Statistical analyses were performed with unpaired Student's t-tests (*P < 0.05).
4. Discussion
IFITM1, 2 and 3 are essential host effector molecules of interferon response against multiple pathogens [4], [5], [6], [7], thus their expression should be tightly controlled. However, previous studies on transcriptional regulation of IFITM1, 2 and 3 genes were limited to the promoter elements [15], [16], [17], [18], [31]. The role of enhancers in the transcriptional regulation of IFITM1, 2 and 3 genes remains elusive. Combining public ENCODE data mining, luciferase reporter assay and genome editing by CRISPR-Cas9, we identified an IFN-responsive enhancer located 35 kb upstream of IFITM3 gene promoter regulating the expression of IFITM1, 2 and 3 genes, thus providing the first evidence that enhancers play an important role in transcriptional regulation of these IFITM genes.
Notably, this single enhancer E2-3 could simultaneously enhance the expression of three clustered genes in the IFITM locus, suggesting it might act like a locus control region (LCR), as in the α-globin and β-globin loci [32], [33]. We also noticed that the induction of IFITM1, 2 and 3 was not completely disrupted by truncation of enhancer E2-3. Two possible explanations may be proposed: firstly, enhancer E2-3 was only truncated, not completely deleted. The residual E2-3 might still have an effect. Indeed, luciferase reporter assay indicated that the truncated E2-3 still had a minor enhancer activity under IFN treatment conditions. Secondly, given the nature of complicated chromatin interaction and regulation network in eukaryotic genomes, many studies have shown that genes could be regulated by multiple enhancers [34], [35], [36]. Therefore, it's possible that there are other redundant enhancers like E1-2, which was also identified as an enhancer in our luciferase reporter assay screening, controlling the expression of IFITM genes. After all, truncation of enhancer E2-3 decreased IFNβ-induced resistance to IAV infection, highlighting its biological importance.
STAT1 binds to the promoters of ISGs to activate gene expression in both type I and type II IFN induction [14]. However, previous ChIP-chip or ChIP-seq data indicated that only a small fraction (< 30%) of STAT1 binding sites were within 1 kb of transcriptional start sites (TSSs) [37], [38], [39]. The function of the large numbers of distal STAT1 binding sites remains unknown. Here we showed that STAT1 binding to the enhancer E2-3, which was located > 35 kb away from its target genes, was indispensable for its IFN-responsive enhancer activity. We therefore speculate that a similar enhancer function may also exist in other distal STAT1 binding sites. In addition to STAT1, STAT2 might also be involved in the regulation of IFITMs by enhancer E2-3, because STAT2 is also suggested to bind to E2-3 in IFNα-treated K562 cells, as inferred from ENCODE STAT2 ChIP-seq data. It's quite interesting that the promoters of IFITM genes already contain the binding sites for STAT1 or STAT2 while there still exists an enhancer mediating IFN/STAT signaling. A reasonable explanation might be that the distal enhancer could fine tune the expression of these important genes and thus add robust and plastic response to virus invasion and IFN stimulation. Besides the IFITM gene cluster, it has been previously reported that many other genes such as IFNβ gene [40], [41] and STAT1 gene [42] also behave like this, suggesting a general transcriptional regulation on eukaryotic gene expression.
Circumstantial evidence indicates that distal enhancers regulate gene expression through formation of physical chromatin loops [26]. Consistent with transcriptional regulation of IFITM1, 2 and 3 by the remote enhancer E2-3, we did observe long-range interactions between enhancer E2-3 and promoters of IFITM1, 2 and 3 using 3C assays in HEK293 and A549 cells. Notably, the promoters of IFITM1, 2 and 3 also clustered together physically in three dimensions to form some kind of active chromatin hub [43], which might facilitate coordinated regulation of enhancer E2-3. Moreover, these interactions or active chromatin hub already existed before treatment of IFN. This may be beneficial to rapid induction of gene expression, thus timely restricting the infection of viruses. Such pre-formed promoter-enhancer interactions have been widely reported in many biological processes like TNFα response, development and hypoxia [35], [44], [45]. Specifically, upon truncation of E2-3, the truncated E2-3 still interacted with the promoters of IFITM1, 2 and 3. A recent paper about a distal enhancer regulating PAG1 gene also reported a similar phenomenon [46]. It's possible that the driving force looping E2-3 to IFITM gene cluster might not be disrupted upon truncation of E2-3, suggesting roles of architectural factors residing outside E2-3, possibly CTCF and cohesin [47]. At last, considering the growing reports that one enhancer could contact and regulate multiple related genes, some of which are even on different chromosomes [34], [35], [45], [48], [49], we are open that E2-3 might also be able to associate with and regulate other ISGs in addition to IFITMs. These possibilities of course call for extensive efforts to examine in the future.
Collectively, our study reported the first enhancer regulating the expression of IFITM genes. This remote enhancer coordinately upregulated IFN-induced expression of IFITM1, 2 and 3 genes through constitutive long-range interactions and IFN-induced recruitment of STAT1, thus contributing to IFN-induced resistance to IAV infection. These findings expanded our insights into the roles and mechanisms of enhancers in transcriptional regulation of IFITM genes and IFN-induced resistance to virus infection, which might contribute to our understanding of the complicated mechanisms underlying innate immunity, and shed new light on the characterization of novel disease susceptibility loci.
The following are the supplementary data related to this article.
Truncation of E2-3 decreases IFNβ-induced IFITM1 and IFITM2 protein levels. Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by analysis of IFITM1 (A) and IFITM2 (B) protein levels by Western blots. β-actin was analyzed as control.
Truncation of E2-3 decreases IFNα-induced IFITM1, IFITM2 and IFITM3 mRNA and protein levels. (A). Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNα or PBS for 24 h, followed by quantification of IFITM1, IFITM2, IFITM3 and B4GALNT4 mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests (***P < 0.001). (B–C) Proteins from HEK293 cultures treated as in (A) were analyzed for IFITM1, IFITM2, IFITM3 and β-actin levels by Western blots.
Truncation of E2-3 does not affect STAT1 and P-STAT1 protein levels. Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by analysis of STAT1 (A) and P-STAT1 (B) protein levels by Western blots. β-Actin was analyzed as control.
3C analysis using fragment containing the promoter of IFITM1 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM1, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM2 and IFITM3, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
3C analysis using fragment containing the promoter of IFITM2 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM2, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM1 and IFITM2, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
3C analysis using fragment containing the promoter of IFITM3 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM3, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM1 and IFITM2, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
STR loci genotyping of HEK293 (A) and A549 (B) cell lines. The STR genotyping results were 100% match with that of corresponding cell lines in ATCC. No loci were triallelic, so the cell lines were not cross-contaminated by other cell lines.
Truncation of enhancer E2-3 did not abolish its interactions with the promoters of IFITM1, 2 and 3. (A) Mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and truncated enhancer E2-3. The red bar represented the location of the anchor fragment containing the truncated enhancer E2-3, the yellow bars represented the location of fragments containing the promoters of IFITM1, 2 and IFITM3, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except using the fragment containing the promoter of IFITM1 as anchor. (C) Same as (A) except using the fragment containing the promoter of IFITM2 as anchor. (D) Same as (A) except using the fragment containing the promoter of IFITM3 as anchor.
Truncation of enhancer E2-3 did not affect the infection of IAV. (A) Wild-type (WT) and mutant (Del) HEK293 cells were infected with IAV PR8 at a MOI of 0.1 for 24 h followed by quantification of virus NP mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests. “n.s.” meant “not significant”. (B) Proteins from HEK293 cultures treated as in (A) were analyzed for virus NP and β-actin levels by Western blots. (C) Progeny virus titers from HEK293 cultures treated as in (A) were determined with the TCID50 assay. Statistical analyses were performed with unpaired Student's t-tests. “n.s.” meant “not significant”.
Positions of clones in the genome.
Primers for cloning, RT-qPCR, ChIP-qPCR and 3C.
Transparency document
Transparency document.
Acknowledgements
We thank Cheng-Cai Lai for excellent technical assistance and Bing-Yu Ye for helpful advice. This work was supported by the National Basic Research Program of China [2013CB966802]; National Natural Science Foundation of China [31370762, 81372218] and National Key Technology R&D Program of China [2015BAI08B03]. Funding for open access charge: National Natural Science Foundation of China.
Footnotes
The Transparency document associated with this article can be found, in the online version.
Contributor Information
Yan Zhang, Email: zany1983@163.com.
Zhi-Hu Zhao, Email: zhaozh@bmi.ac.cn.
References
- 1.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey C.C., Zhong G., Huang I.C., Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu. Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., Wise H.M., Kane L., Goulding D., Digard P., Anttila V., Baillie J.K., Walsh T.S., Hume D.A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S.B., Gen I.I., Investigators M., Smyth R.L., Openshaw P.J., Dougan G., Brass A.L., Kellam P. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H., Yan H.P., Wu H., Liu J.H., Liu N., Wang D.Y., Shu Y.L., Ho L.P., Kellam P., McMichael A., Dong T. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M., Li L., Zhou M., Yuan S., Sun J., Zhu Z., Chen L., Li Q., Zhang Z., Zhang X., Lu S., Doherty P.C., Kedzierska K., Xu J. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills T.C., Rautanen A., Elliott K.S., Parks T., Naranbhai V., Ieven M.M., Butler C.C., Little P., Verheij T., Garrard C.S., Hinds C., Goossens H., Chapman S., Hill A.V. IFITM3 and susceptibility to respiratory viral infections in the community. J. Infect. Dis. 2014;209:1028–1031. doi: 10.1093/infdis/jit468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka S.S., Yamaguchi Y.L., Tsoi B., Lickert H., Tam P.P. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Sanda C., Weitzel P., Tsukahara T., Schaley J., Edenberg H.J., Stephens M.A., McClintick J.N., Blatt L.M., Li L., Brodsky L., Taylor M.W. Differential gene induction by type I and type II interferons and their combination. J. Interf. Cytokine Res. 2006;26:462–472. doi: 10.1089/jir.2006.26.462. [DOI] [PubMed] [Google Scholar]
- 14.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 15.Huang M., Qian F., Hu Y., Ang C., Li Z., Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- 16.Liu H., Kang H., Liu R., Chen X., Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 2002;22:6471–6479. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui K., Tailor P., Liu H., Chen X., Ozato K., Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell. Biol. 2004;24:4476–4486. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuddapah S., Cui K., Zhao K. Transcriptional enhancer factor 1 (TEF-1/TEAD1) mediates activation of IFITM3 gene by BRGl. FEBS Lett. 2008;582:391–397. doi: 10.1016/j.febslet.2007.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laat W.D., Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 20.Lee G.R., Fields P.E., Griffin T.J., Flavell R.A. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 21.Thanos D., Maniatis T. Virus induction of human IFN-β gene assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 22.Murakawa Y., Yoshihara M., Kawaji H., Nishikawa M., Zayed H., Suzuki H., Fantom C., Hayashizaki Y. Enhanced identification of transcriptional enhancers provides mechanistic insights into diseases. Trends Genet. 2016;32:76–88. doi: 10.1016/j.tig.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938;27 [Google Scholar]
- 24.Zhang Y., Shen W.L., Shi M.L., Zhang L.Z., Zhang Z., Li P., Xing L.Y., Luo F.Y., Sun Q., Zheng X.F., Yang X., Zhao Z.H. Involvement of aberrant miR-139/Jun feedback loop in human gastric cancer. Biochim. Biophys. Acta. 2015;1853:481–488. doi: 10.1016/j.bbamcr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 26.Visel A., Rubin E.M., Pennacchio L.A. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J., Sim H.S., Peh S.Q., Mulawadi F.H., Ong C.T., Orlov Y.L., Hong S., Zhang Z., Landt S., Raha D., Euskirchen G., Wei C.L., Ge W., Wang H., Davis C., Fisher-Aylor K.I., Mortazavi A., Gerstein M., Gingeras T., Wold B., Sun Y., Fullwood M.J., Cheung E., Liu E., Sung W.K., Snyder M., Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maston G.A., Landt S.G., Snyder M., Green M.R. Characterization of enhancer function from genome-wide analyses. Genomics Hum. Genet. 2012;13:29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- 29.Plank J.L., Dean A. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell. 2014;55:5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewin A.R., Reid L.E., Mcmahon M., Stark G.R., Kerr I.M. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhou G.L., Xin L., Song W., Di L.J., Liu G., Wu X.S., Liu D.P., Liang C.C. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol. Cell. Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., Laat W.D. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 34.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., Splinter E., de Laat W., Spitz F., Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 37.Stephen P.B., Hartman E., Nath Anjali K., Royce Thomas E., Gerstein Mark, Weissman Sherman, Snyder Michael. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhinge A.A., Kim J., Euskirchen G.M., Snyder M., Iyer V.R. Mapping the chromosomal targets of STAT1 by Sequence Tag Analysis of Genomic Enrichment (STAGE) Genome Res. 2007;17:910–916. doi: 10.1101/gr.5574907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 40.Apostolou E., Thanos D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 41.Banerjee A.R., Kim Y.J., Kim T.H. A novel virus-inducible enhancer of the interferon-beta gene with tightly linked promoter and enhancer activities. Nucleic Acids Res. 2014;42:12537–12554. doi: 10.1093/nar/gku1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuasa K., Hijikata T. Distal regulatory element of the STAT1 gene potentially mediates positive feedback control of STAT1 expression. Genes Cells. 2016;21:25–40. doi: 10.1111/gtc.12316. [DOI] [PubMed] [Google Scholar]
- 43.De L.W., Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosom. Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 44.Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 45.Platt J.L., Salama R., Smythies J., Choudhry H., Davies J.O., Hughes J.R., Ratcliffe P.J., Mole D.R. Capture-C reveals preformed chromatin interactions between HIF-binding sites and distant promoters. EMBO Rep. 2016;17:1410–1421. doi: 10.15252/embr.201642198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schorg A., Santambrogio S., Platt J.L., Schodel J., Lindenmeyer M.T., Cohen C.D., Schrodter K., Mole D.R., Wenger R.H., Hoogewijs D. Destruction of a distal hypoxia response element abolishes trans-activation of the PAG1 gene mediated by HIF-independent chromatin looping. Nucleic Acids Res. 2015;43:5810–5823. doi: 10.1093/nar/gkv506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S., Ong C.T., Hookway T.A., Guo C., Sun Y., Bland M.J., Wagstaff W., Dalton S., McDevitt T.C., Sen R., Dekker J., Taylor J., Corces V.G. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markenscoff-Papadimitriou E., Allen W.E., Colquitt B.M., Goh T., Murphy K.K., Monahan K., Mosley C.P., Ahituv N., Lomvardas S. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y., Guo W., Li P., Zhang Y., Zhao M., Fan Z., Zhao Z., Yan J. Long-range chromosome interactions mediated by cohesin shape circadian gene expression. PLoS Genet. 2016;12:e1005992. doi: 10.1371/journal.pgen.1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Truncation of E2-3 decreases IFNβ-induced IFITM1 and IFITM2 protein levels. Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by analysis of IFITM1 (A) and IFITM2 (B) protein levels by Western blots. β-actin was analyzed as control.
Truncation of E2-3 decreases IFNα-induced IFITM1, IFITM2 and IFITM3 mRNA and protein levels. (A). Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNα or PBS for 24 h, followed by quantification of IFITM1, IFITM2, IFITM3 and B4GALNT4 mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests (***P < 0.001). (B–C) Proteins from HEK293 cultures treated as in (A) were analyzed for IFITM1, IFITM2, IFITM3 and β-actin levels by Western blots.
Truncation of E2-3 does not affect STAT1 and P-STAT1 protein levels. Wild-type (WT) and mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by analysis of STAT1 (A) and P-STAT1 (B) protein levels by Western blots. β-Actin was analyzed as control.
3C analysis using fragment containing the promoter of IFITM1 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM1, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM2 and IFITM3, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
3C analysis using fragment containing the promoter of IFITM2 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM2, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM1 and IFITM2, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
3C analysis using fragment containing the promoter of IFITM3 as the anchor. (A) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and enhancer E2-3. The red bar represented the location of the anchor fragment containing the promoter of IFITM3, the yellow bars represented the location of fragments containing the enhancer E2-3 and the promoters of IFITM1 and IFITM2, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except performed in A549 cells.
STR loci genotyping of HEK293 (A) and A549 (B) cell lines. The STR genotyping results were 100% match with that of corresponding cell lines in ATCC. No loci were triallelic, so the cell lines were not cross-contaminated by other cell lines.
Truncation of enhancer E2-3 did not abolish its interactions with the promoters of IFITM1, 2 and 3. (A) Mutant (Del) HEK293 cells were treated with 1000 U/mL IFNβ or PBS for 24 h, followed by 3C assays of a 60 kb region containing IFITM locus and truncated enhancer E2-3. The red bar represented the location of the anchor fragment containing the truncated enhancer E2-3, the yellow bars represented the location of fragments containing the promoters of IFITM1, 2 and IFITM3, Pst I restriction sites were indicated as black vertical lines. (B) Same as (A) except using the fragment containing the promoter of IFITM1 as anchor. (C) Same as (A) except using the fragment containing the promoter of IFITM2 as anchor. (D) Same as (A) except using the fragment containing the promoter of IFITM3 as anchor.
Truncation of enhancer E2-3 did not affect the infection of IAV. (A) Wild-type (WT) and mutant (Del) HEK293 cells were infected with IAV PR8 at a MOI of 0.1 for 24 h followed by quantification of virus NP mRNA levels. Results of three independent experiments are shown relative to the ACTB mRNA levels. Statistical analyses were performed with unpaired Student's t-tests. “n.s.” meant “not significant”. (B) Proteins from HEK293 cultures treated as in (A) were analyzed for virus NP and β-actin levels by Western blots. (C) Progeny virus titers from HEK293 cultures treated as in (A) were determined with the TCID50 assay. Statistical analyses were performed with unpaired Student's t-tests. “n.s.” meant “not significant”.
Positions of clones in the genome.
Primers for cloning, RT-qPCR, ChIP-qPCR and 3C.
Transparency document.