Abstract
Bombyx mori nucleopolyhedrovirus (BmNPV) is a major pathogen causing severe economic loss. Previous studies have revealed that some proteins in silkworm digestive juice show antiviral activity. In this study, antiviral activity examination of different resistant strains showed that the digestive juice of the resistant strain (A35) had higher inhibition to virus than the susceptible strain (P50). Subsequently, the label-free quantitative proteomics was used to study the midgut digestive juice response to BmNPV infection in P50 and A35 strains. A total of 98 proteins were identified, of which 80 were differentially expressed proteins (DEPs) with 54 enzymes and 26 nonenzymatic proteins by comparing the proteomes of infected and non-infected P50 and A35 silkworms. These DEPs are mainly involved in metabolism, proteolysis, neuroactive ligand receptor interaction, starch and sucrose metabolism and glutathione metabolism. After removing the genetic background and individual immune stress response proteins, 9 DEPs were identified potentially involved in resistance to BmNPV. Further studies showed that a serine protease, an alkaline phosphatase and serine protease inhibitor 2 isoform X1 were differentially expressed in A35 compared to P50 or post BmNPV infection. Taken together, these results provide insights into the potential mechanisms for silkworm digestive juice to provide resistance to BmNPV infection.
Signifcance: Bombyx mori nucleopolyhedrovirus (BmNPV) is highly pathogenic, which has a great impact on the sericulture. BmNPV entered the midgut lumen and exposed to digestive juices after oral infection. Previous studies have revealed that some proteins in silkworm digestive juice show antiviral activity, however, current information on the digestive juice proteome of high resistant silkworm strain after BmNPV challenge compared to susceptible strain is incomprehensive. Here, we combined label-free quantification method, bioinformatics, RT-qPCR and western blot analysis and found that BmNPV infection causes some protein changes in the silkworm midgut digestive juice. The DEPs were identified in the digestive juices of different resistant strains following BmNPV infection, and screened out some proteins potentially related to resistance to BmNPV. Three important differentially expression proteins were validated by independent approaches. These findings uncover the potential role of silkworm digestive juice in providing resistance to BmNPV and supplemented the profile of the proteome of the digestive juices in B. mori.
Keywords: Bombyx mori, Label-free, Digestive juice, Bombyx mori nucleopolyhedrovirus (BmNPV)
Graphical abstract
Highlights
-
•
The digestive juice antiviral activity of resistant strain was higher than susceptible strain.
-
•
Silkworm digestive juice was analyzed by label-free quantitative proteomics.
-
•
Eighty DEPs were identified in different resistant strains following BmNPV infection.
-
•
Some DEPs were validated by RT-qPCR and western blot.
-
•
These results provide insights into the potential mechanisms for B. mori resistant to BmNPV infection.
1. Introduction
Bombyx mori is a completely domesticated insect that is important for silk production (sericulture). Improvements to sericulture could improve incomes of farmers in several developing countries, such as China, Thailand, India. It is considered to be a lepidopteron model insect for basic and applied research and is key for increasing knowledge about other insects and organisms [1,2]. Bombyx mori nucleopolyhedrovirus (BmNPV) [3] is a major viral pathogen that is currently poses a challenge to the sericulture industry, mainly because there are not effective prevention methods to limit viral infection [4].
In the insect, the larval midgut lumen is a complex bioreactor where consumed leaves encounter the first defensive barrier consisting of secreted digestive enzymes in the gut [5]. The digestive juice exists in the midgut lumen, and contains a variety of digestive enzymes and inorganic salts. These enzymes decompose organic matter into small peptides, carbohydrates, and lipids that are absorbed by epithelial cells and enter the circulatory system [6]. Previous studies have indicated that the enzymes form digestive juice not only function to digest food, but also play an important role in weakening or killing pathogens [5]. In silkworms, some digestive enzymes involved in antiviral activity have already been cloned and characterized, such as red fluorescent protein [7], lipase [8] and serine protease [9]. Alkaline trypsin was also purified from the digestive juice of B. mori larvae and showed strong antiviral activity [10]. Previous studies on insect immunity have focused on the fat body, midgut, and hemolymph. The protein composition of silkworm digestive juice and the mechanism of its antiviral proteins in the lumen after BmNPV challenge still require further study.
With the rapid development of proteomic techniques in combination with increased genome sequence information, analysis of proteomic information has become one of the most important strategies for characterizing protein expression profiles and metabolic changes in diverse biological systems [11]. Two-dimensional electrophoresis (2-DE) and mass spectrometry (MS) techniques, isobaric tags for relative and absolute quantification (iTRAQ), Tandem Mass Tags (TMT) and label-free methods are attractive proteomic analysis technologies for researching the interaction between pathogens and B. mori. Wang et al. [12] performed a comparative subcellular proteomics analysis of the silkworm larval midgut response to BmNPV infection by 2-DE combined with MS. The iTRAQ method was also used to explore the molecular mechanisms of B. mori response to B. mori cypovirus (BmCPV) [13] and BmNPV [14] challenge. Additionally, the role of Hsp90 in BmN cells response to BmNPV infection was explored by TMT method [15]. The label-free quantitative technique is gaining fast traction in the proteomics field due to its rapid, straightforward, and low-cost required for measuring protein expression levels in complex biological samples [16,17].
In recent years, some proteins were confirmed having the functions related to BmNPV resistance or infection in B. mori. B. mori C-lysozyme (BmC-LZM) and BmAtlastin-n could enhance the resistance to BmNPV by overexpression in larvae and cells [18,19]. Furthermore, a GP64-binding protein E3 ubiquitin-protein ligase SINA-like 10 (SINAL10) [20] and chaperonin containing t-complex polypeptide 1 (TCP-1) [21] could stimulate BmNPV proliferation in BmN cells. Some studies regarding the role of the B. mori larval midgut in response to BmNPV infection have been conducted in previous studies utilizing RNA-Seq [22], 2-DE combined MS [12], label free [16] and iTRAQ methods [14]. Hu et al. [6] investigated changes in the protein composition of the silkworm digestive juice between non-infected and infected samples by shotgun and MS techniques. Their research, however, only involved the P50 susceptible strain. Although there is little information available on the digestive juice proteome of different resistant strains after BmNPV challenge. Our present study, notably, firstly made sure the virus inhibition difference of different resistant strain and following BmNPV infection. Consequently, we identified DEPs between A35 and P50 using the label-free proteomic technique. Further, possible functions of identified DEPs and their potential involvement in resistance to BmNPV infection are discussed. Our study provides insights into the functions of silkworm digestive juice in antiviral activity.
2. Materials and methods
2.1. Silkworm rearing and virus preparation
The BmNPV-susceptible silkworm strain P50 (LC50 = 1.03 × 105 OBs/mL) and BmNPV-resistant silkworm strain A35 (LC50 = 5.90 × 107 OBs/mL) were maintained in the School of Life Sciences, Anhui Agricultural University, Hefei, China [22]. All larvae were reared using fresh mulberry leaves. The first to third instar larvae were reared at 27 ± 1 °C, 75 ± 5% relative humidity with 12 h day/night cycles, and the last two instar larvae were reared at 25 ± 1 °C, under the same humidity and photoperiod as above.
The BmNPV T3 strain was maintained in our laboratory and purified according to the protocol reported by Rahman et al. [23]. The third day of fifth instar larvae were selected for oral feeding experiment according to Hu et al. [6], and in each of the three independent experiments were administered 5 μL of BmNPV suspended in water (1 × 107 OBs/mL) per os and the control was treated with 5 μL of sterile water.
2.2. Digestive juice and midgut collection
Digestive juice was collected from the control groups (P50- and A35-) and infected groups (P50+ and A35+) at approximately 24 h after inoculation by electric shock (35 V). Mulberry leaf residues in obtained juice were discarded by centrifugation at 4000 rpm, for 10 min at 4 °C, the precipitate was abandoned, and supernatant was freeze-dried in powder form and stored at −80 °C for further use. Meanwhile, midgut samples were dissected and mixed together to eliminate individual genetic differences. The supernatant and midgut were then frozen in liquid nitrogen and stored at −80 °C for further use.
2.3. Incubation of BmNPV with silkworm midgut digestive juice
The non-infected digestive juices of P50 and A35 were collected and BmNPV (1 × 107 OBs/mL) was incubated with the digestive juices at a volume ratio of 4:1, for 1 h at room temperature, respectively. After incubation, the same volume of ddH2O was added to dilute BmNPV to 1 × 106 OBs/mL. Subsequently, each the first day of fifth instar P50 strain silkworm larva was fed with 5 μL incubated BmNPV. The ddH2O treated BmNPV were used as control. The midgut tissues were collected at 24 h, 48 h and 72 h post infection.
2.4. Investigation of BmNPV proliferation by real-time quantitative polymerase chain reaction
Total genomic DNA of silkworm midgut tissue was extracted according to the method reported by Maxim et al. [24] with some modifications. In brief, 100 mg of ground midgut tissue was mixed with 1.0 mL lysis buffer (10 mmol/L Tris-HCl pH 8.0, 0.1 mol/L EDTA pH 8.0, 0.5% SDS) and final concentration of 400–500 μg/mL Proteinase K (Tiangen, Beijing, China). After incubation at 50 °C for 5 h, equal volume of phenol/ chloroform/ isoamylol (25:24:1) was used for extraction and centrifuged at 12,000 rpm for 10 min at RT. The supernatant was collected and performed the extraction method again. After twice extraction, the supernatant was precipitated in absolute ethanol. The pellet was collected and wash with 70% ethanol after centrifuged at 7500 rpm for 10 min at RT. The final pellet was resuspended in 100 μL TE buffer and the A260/280 ratio and DNA concentration of all samples were detected using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, New York, NY, USA). BmNPV proliferation changes upon differently treated BmNPV infected in larvae was determined by estimating viral DNA using specific primers of the VP39 gene by RT-qPCR [21]. There were three biological sample replicates, and each biological sample replicate included three technique replicates. The statistical analysis was conducted using ANOVA and an LSD a posteriori test using SPSS (p < .05).
2.5. Sample preparation and SDS-PAGE
Digestive juice powder was dissolved in a moderate amount of SDT buffer (4% SDS w/v, 100 mM Tris-HCl, 0.1 M DTT, pH 7.6) and boiled for 15 min. Afterward, it was centrifuged at 12000 g at 4 °C for 40 min and protein in the supernatant was quantified with the BCA Protein Assay Kit (Bio-Rad, USA). The sample was then stored at −80 °C. Proteins (30 μg) for each sample were mixed with 5 × loading buffer and boiled for 5 min. The protein samples were separated on a 12% SDS-PAGE gel. Protein bands were visualized by Coomassie Blue R-250 staining.
2.6. Filter-aided sample preparation (FASP digestion)
For each protein sample (200 μg), DTT was added and the mixture was boiled for 5 min. Subsequently, the solution was allowed to cool to room temperature. The detergent, DTT, and other low-molecular-weight components were removed using UA buffer (8 M Urea, 150 mM Tris-HCl pH 8.0) and by repeated ultra-filtration (microcon units, 10 kD). Then 100 μL iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues and the samples were incubated for 30 min in darkness. The filters were washed with 100 μL UA buffer three times and with 100 μL 25 mM NH4HCO3 buffer twice. Finally, the protein suspensions were digested with 4 μg trypsin (Promega) in 40 μL 25 mM NH4HCO3 buffer overnight at 37 °C, and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Empore™ SPE Cartridges (standard density, bed I.D. 7 mm, volume 3 mL, Sigma), concentrated by vacuum centrifugation and reconstituted in 40 μL of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of the 0.1% (g/L) solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins [25].
2.7. Capillary high-performance liquid chromatography (HPLC)
Each fraction was injected for nano LC-MS/MS analysis. The peptide mixture was loaded onto a reverse phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18)connected to the C18-reverse phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min controlled by IntelliFlow technology. A two-hour gradient was used as follows:0–55% buffer B for 110 min, 55%- 100% buffer B for 5 min, hold in 100% buffer B for 5 min.
2.8. Liquid chromatography (LC)-electrospray ionization (ESI) tandem MS (MS/MS) analysis
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 120 min. The mass spectrometer was operated in positive ion mode. MS data was acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. Automatic gain control (AGC) target was set to 3e6, and maximum inject time to 10 ms. Dynamic exclusion duration was 40 s. Survey scans were acquired at a resolution of 70,000 at m/z 200 and resolution for HCD spectra was set to 17,500 at m/z 200, and isolation width was 2 m/z. Normalized collision energy was 30 eV and the underfill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
2.9. Sequence database search and data analysis
The MS data were analyzed using MaxQuant software version1.5.3.17 (Max Planck Institute of Biochemistry, Germany) [26]. The MS data were searched against the uniprot_bmnpv_sgbp. fasta. A main search was set at a precursor mass window of 6 ppm. The search was performed based on an enzymatic cleavage rule for trypsin and allowed a maximum of two missed cleavage sites and a mass tolerance of 20 ppm for fragment ions for the purpose of MaxQuant identification of the maximum number of peptides for mass and retention time calibration. Carbamidomethylation of cysteine residues was defined as a fixed modification, whereas the N-terminal acetylation and methionine oxidation of proteins were defined as variable modifications for the database search. The cutoff global false discovery rate (FDR) for peptide and protein identification was set at 0.01. Label-free quantification was carried out in MaxQuant as previously described [27]. Intensity-based, absolute quantification (iBAQ) in MaxQuant was performed on the identified peptides to quantify protein abundance [28].
2.10. Bioinformatic analysis
Functional annotation of the DEPs was performed using the Blast2GO (Version 3.3.5) [29] program against a non-redundant protein database. The corresponding gene ontology (GO) terms were extracted using the most homologous proteins with the Perl program. GO has three ontologies: molecular functions, cellular components, and biological processes. The GO annotation results were plotted using the Web Gene Ontology Annotation Plot (WEGO) (http://wego.genomics.org.cn/cgibin/wego/index.pl). The FASTA protein sequences of DEPs were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/tools/blast/) [30]. The corresponding KEGG pathways were analyzed.
2.11. RT-qPCR validation of DEPs
Total RNA from each midgut sample was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocols. RNA was quantified by measuring the absorbance at 260 nm using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, New York, NY, USA). The purity of all RNA samples was assessed at an absorbance ratio of A260/280 and A260/230, and the integrity of RNA was confirmed by 1.0% agarose gel electrophoresis. First strand cDNA was synthesized using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer's instructions. The RT-qPCR reaction was carried out in a 25 μL reaction mixture containing 12.5 μL of SYBR Premix Ex Taq (TaKaRa, Osaka, Japan). PCR amplification was performed in triplicate wells. B. mori glceraldehyde-3-phosphate dehydrogenase (BmGAPDH) was used as a reference gene. The thermal cycling profile consisted of initial denaturation at 95 °C for 30 s and 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 20 s. The reactions were performed in 96-well plates with a Multicolour Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). Relative expression levels were calculated using the 2−△△Ct method [35]. There were three biological sample replicates, and each biological sample replicate included three technique replicates. The statistical analysis was conducted using ANOVA and an LSD a posteriori test using SPSS (p < .05). The gene-specific primers (Table S1) were designed using Primer Premier 5 (Premier Biosoft International, Palo Alto, CA, USA).
2.12. Antibody preparation and western blot analysis
Western blot analysis was performed to validate the protein abundance in the P50−, P50+, A35− and A35+ samples. Four DEPs, serine protease (SP), alkaline phosphatase (AP) and serine protease inhibitor 2 isoform X1 (Spn-2) were selected to confirm the results by western blotting. The anti-AP antibody was purchased from HuaAn Biotechnology limited company (HUABIO, Hangzhou, China). Recombinant pET-28a-BmSP and pET-28a-BmSpn-2 (GenBank Accession numbers: AY945211.1 and XM_012692302.1, respectively) were constructed and transformed into BL21 (DE3) competent cell to express the recombinant BmSP and BmSpn-2 proteins. The recombinant proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Qiagen, Hilden, Germany) following the manufacturer's protocol, then, the purified recombinant BmSP and BmSpn-2 proteins were injected into New Zealand White rabbits respectively to prepare antibodies.
Total proteins were extracted from the midgut samples as above described. The protein extracts (30 μg) were separated on 12% SDS-PAGE gel and transferred to a polyvinylidenedifluoride (PVDF) membrane. The PVDF membranes were blocked with 5% non-fat milk in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM K2HPO4, pH 7.5, 0.1% v/v Tween-20) for 2 h at room temperature, washed with PBST three times, then incubated with primary antibody (1:500 dilution) for 2 h at room temperature. After washing, antigen-antibody complexes were detected with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:10000 dilution) in blocking buffer for 1 h. After another series of washes, immobilized conjugates on the membrane were visualized in HRP substrate solution (Tiangen, Beijing, China). Three biologically independent individuals were used. The densitometric intensity of protein bands was measured using ImageJ software (Bio-Rad, Hercules, CA, USA).
3. Results
3.1. Larval midgut digestive juices inhibited BmNPV proliferation in silkworm larvae
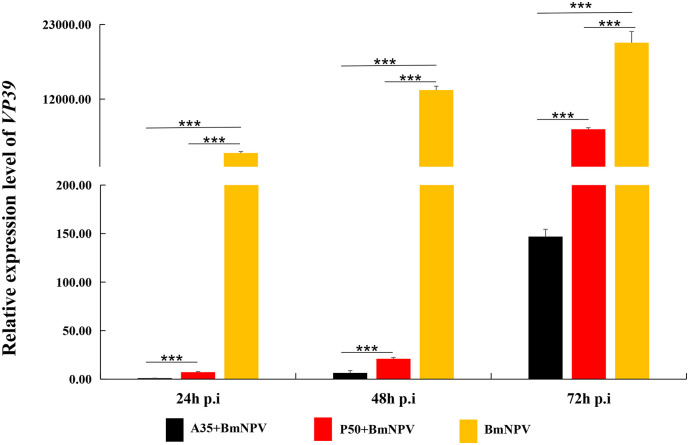
Previous studies showed that there were a lot of proteases and proteins could inhibit the infection of BmNPV in midgut digestive juice. To further validate the diversity inhibition function of digestive juices to virus infection, the dynamic proliferation of BmNPV in the midgut of P50 in different treatment groups at different time point post infection was detected using RT-qPCR. Melting curve analysis confirmed that specific amplification was achieved using a pair of primers of the BmNPV VP39 gene. The results show that BmNPV copy numbers in control were significantly higher than that of BmNPV group incubated with the P50 digestive juice (P50 + BmNPV) and that of A35 digestive juice (A35 + BmNPV) at all three selected time points (Fig. 1 ). Additionally, BmNPV proliferation in P50 + BmNPV were significantly increased than that of in A35 + BmNPV at all three selected time points. These result indicated that there are some components in the digestive juice of A35 and P50 can inhibited BmNPV activity, and the digestive juice inhibition of resistant strain (A35) was higher than that of susceptible strain (P50).
Fig. 1.
The antiviral activity of digestive juice in silkworm. Proliferation of BmNPV was assessed using RT-qPCR by analyzing relative transcription levels of VP39 at 24 h, 48 h and 72 h post infection in the midguts.“P50 + BmNPV”,“A35 + BmNPV”and “BmNPV”indicates P50- digestive juice, A35- digestive juice and ddH2O mixed with BmNPV. There were three biological sample replicates, and each biological sample replicate included three technique replicates.The statistical analysis was conducted using ANOVA and an LSD a posteriori test using SPSS (*** P < .001).
3.2. Analysis of protein band patterns derived from the digestive juice of resistant and susceptible strains following BmNPV infection
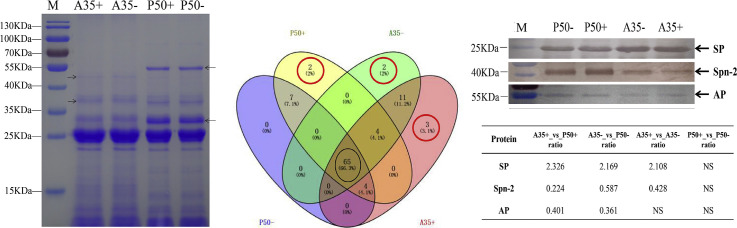
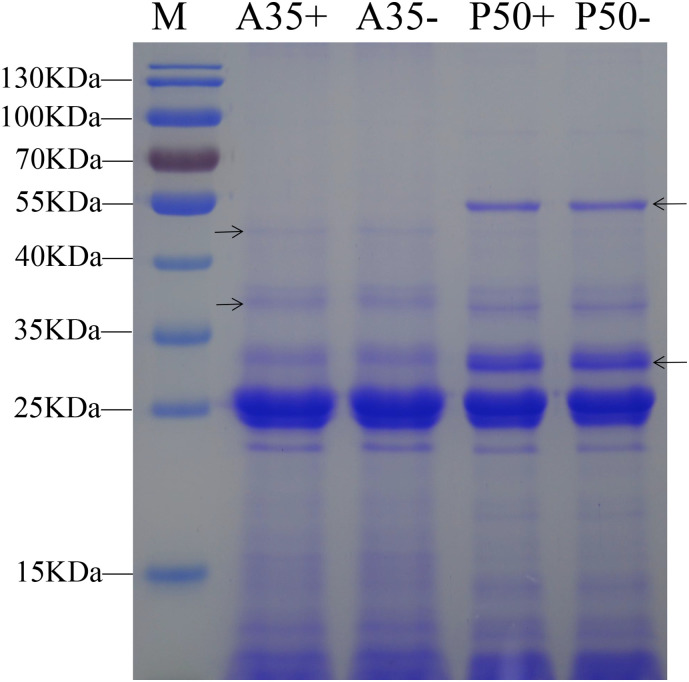
In this study, the proteins of silkworm digestive juice were extracted from strains of differing resistance. Before label-free analysis, band patterns from 30 μg of protein material was analyzed by SDS-PAGE. The protein band patterns were similar between P50- and P50+ group, and the same phenomenon was observed in the A35 strain. However, comparing strains of different resistance revealed that band patterns between the two were significantly different (Fig. 2 ). For example, 55 and 30 kDa bands specially were visible when investigating patterning for the P50 strain, whereas 45 and 37 kDa bands were only visible when using proteins derived from the A35 strain. These results show that proteins derived from the digestive juice of susceptible and resistant strains are significantly different from each other.
Fig. 2.
Proteins extracted from silkworm midgut digestive juices were separated by SDS-PAGE. M: protein marker, the plus and minus represent BmNPV-challenged group and control group, respectively. Different bands in the different strain are shown with the black arrows.
3.3. Digestive juice protein profiles following BmNPV infection
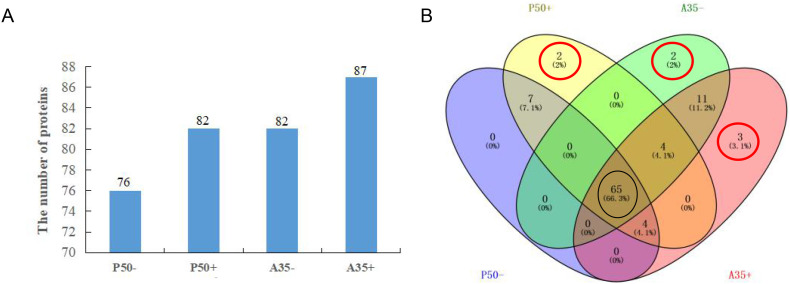
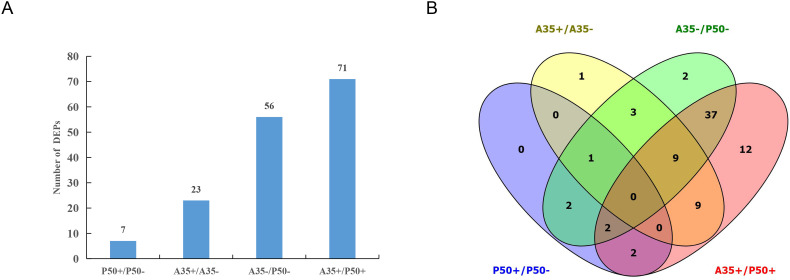
To better understand the difference of digestive juice protein components in silkworm susceptible and resistant strains, a quantitative strategy was employed to identify proteins according to mass spectrum data. A total of 98 proteins, 89 of which with least 3 unique peptides, were identified in four groups. The molecular mass of the proteins ranged from 7.9 kDa to 455.2 kDa (Table S2). In the P50 strain, 76 and 82 proteins were identified in P50- and P50+ groups, respectively. While in the A35 strain, 83 proteins and 87 were identified in A35- and A35+ group, respectively. Interestingly, the number of proteins increased after BmNPV infection in both the P50 and A35 strain (Fig. 3A), There were 65 total proteins identified from all tested groups (Fig. 3B), but there were some proteins specifically expressed in one group. We found 9 unique proteins (including 2 unique proteins in P50+ group) that appeared in the P50 strain and 16 unique proteins (including 2 and 3 unique proteins in A35- and A35+ group, respectively) in the A35 strain (Fig. 3B). 69% of the total proteins identified were enzymes and include a serine protease, lipase, carboxypeptidase and other digestive enzymes (Fig. S1, Table S2).
Fig. 3.
The distribution of total proteins. (A) Summary of total digestive juice proteins identified by a Label-free by approach in four groups. The numbers indicate the quantity of proteins that were expressed in each group. (B) Venn analysis of total digestive juice proteins in the four groups. The numbers and percentages denote the amount and ratio of proteins that were expressed in each group, respectively. Purple indicates P50-; yellow indicates P50+; green indicates A35- and pink indicates A35+. The red circle indicates the number of proteins only expressed in each unique group, and the black circle indicates the number of proteins expressed in all groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Identification of differentially expressed proteins (DEPs)
The DEPs were defined based on a 1.5-fold change threshold (with a fold change >1.5 or < 0.67, p < .05) or specifically expressed in comparisons between groups according to mass spectrum data. A total of 80 DEPs were identified and groups for comparison consisted of P50+ vs P50-, A35+ vs A35-, A35+ vs P50+ and A35- vs P50-). Of the 80 proteins identified in this experiment, there were 54 enzymes and 26 nonenzymatic DEPs (Table S3). Comparisons between infected and non-infected B. mori within the same strain revealed only seven DEPs in the P50 stain, and 23 DEPs in the A35 strain. Additionally, comparisons between A35 and P50 strains with the same treatment revealed 56 and 71 DEPs were identified in A35- vs P50- and A35+ vs P50+ groups, respectively (Fig. 4A). Based on Venn diagram analysis, only one DEP was present in both groups (P50+ vs P50- and A35+ vs A35-). While 48 DEPs were found in the A35+ vs P50+ and A35- vs P50- (Fig. 4B). These results suggested that there are more variations in proteins of digestive juices in resistant strains than in susceptible strains following BmNPV infection.
Fig. 4.
The distribution of differentially expressed proteins. (A) Summary of DEPs in four groups. The numbers denote the number of proteins that were expressed in each group. (B) Venn analysis of DEPs in the four groups. The numbers denote the number of proteins that were expressed in each group. Blue indicates P50+ vs P50-; pink indicates A35+ vs P50+; yellow indicates A35+ vs A35-; and green indicates A35- vs P50-. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. GO and KEGG pathways analysis of the DEPs
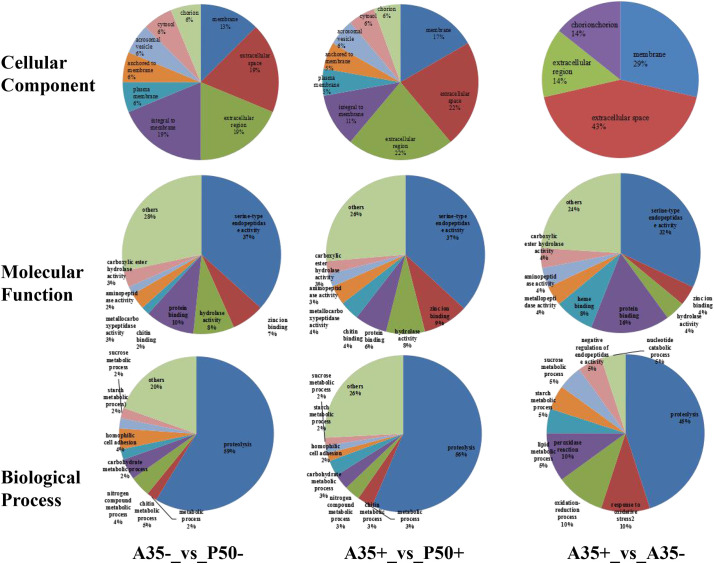
To gain insight into the functional categories of DEPs expressed in response to BmNPV, GO and KEGG analysis were conducted according to the numbers of DEPs 23, 56 and 71 in A35+ vs A35-, A35+ vs P50+ and A35- vs P50-, respectively (Fig. 4A). The GO analysis was divided into three main categories: biological processes, molecular functions and cellular components. For the cellular component category, there were more DEPs categorized as integral to membranes in A35 vs P50 were decreased from 19% group to 11% following BmNPV infection, while DEPs of the same category increased from 13% to 17%. With regards to molecular function, the largest number of DEPs were related to serine-type endopeptidases, while DEPs involved in protein binding in A35 vs P50 were decreased from 10% to 6% following BmNPV infection. For biological processes category, there were largest number of DEPs associated with proteolysis, and the DEPs related to chitin metabolic process in A35 vs P50 were decreased from 5% to 3% following NPV infection. For the resistant strain A35 group, the most predominantly DEPs were annotated on membrane, serine-type endopeptidase activity and proteolysis activity with 29%, 32% and 45% at biological processes, molecular functions and cellular components following BmNPV infection, respectively (Fig. 5 ).
Fig. 5.
Gene ontology (GO) analysis of DEPs between each group. Functional assignments if proteins their corresponding associated biological processes, molecular function and cellular components are shown. The percentage of DEPs that could be assigned to the different categories was indicated.
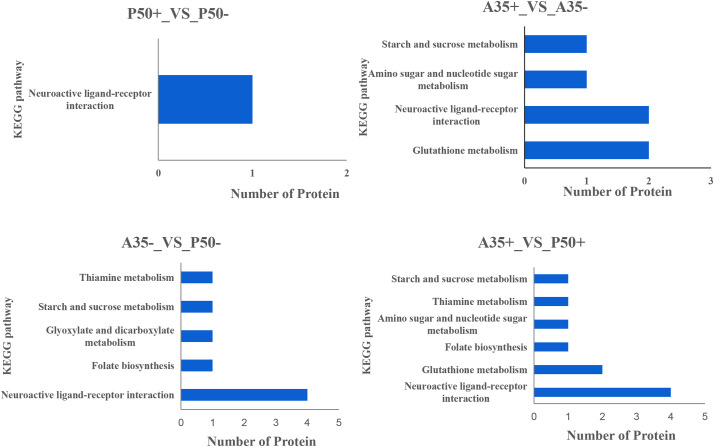
The DEPs between different groups were subjected to KEGG pathway analysis (Fig. 6 ). KEGG enrichment results show that most DEPs in A35+ vs A35-, A35- vs P50- and A35+ vs P50+ were involved in neuroactive ligand receptor interaction, while in P50+ vs P50-, only trypsin, alkaline A was associated with this pathway. Additionally, the DEPs related to starch and sucrose metabolism, folate biosynthesis, and thiamine metabolism and were enriched in A35 vs P50 group, while DEPs related to glycine were specially found in A35- vs P50-. DEPs related to amino sugar and nucleotide sugar metabolism were specially identified in A35+ vs P50+. Lastly, in the A35+ vs A35- comparable group, the identified DEPs were annotated in only four pathways (Fig. 6).
Fig. 6.
KEGG enrichment analysis of DEPs. To the left of each plot are KEGG terms. Under each plot the number proteins are shown.
3.6. Analysis of the enzymes in DEPs response between different strains and after BmNPV infection
Among the DEPs identified in this study, 54 enzymes were identified in the digestive juice of different strains post BmNPV infection, including serine protease, lipase, aminopeptidase, carboxypeptidase, among other enzymes (Table 1 ). In total, 29 of the DEPs identified in all four groups were serine proteases and the majority of these proteins were up regulated upon infection with BmNPV. In the A35 strain, seven serine protease DEPs were up regulated and another seven serine protease of DEPs specifically expressed following BmNPV infection. Compared to P50-, 11 serine protease DEPs were up regulated in A35-, and three more serine protease DEPs were up-regulated following BmNPV infection. Only two and three lipase and aminopeptidase were differentially expressed. The expression level of lipase member H-A was up-regulated in resistant strain A35 and increased in expression after BmNPV infection, while there was no difference in P50+ vs P50-. However, the pancreatic triacylglycerol lipase was only identified within P50 strain. The expression level of aminopeptidase N was down-regulated in both A35+ vs A35- and A35+ vs P50+, while aminopeptidase N-7 was specially expressed in P50 and A35+. Interestingly, six carboxypeptidase proteins were up-regulated in A35- vs P50- and A35+ vs P50+ or specifically expressed in A35 strain and P50+. Additionally, other 14 enzymes were differentially expressed between the two strains or following BmNPV infection, including AP, chitinase and peroxidase. These results indicate that expression level and the type of enzymes present were different in resistant vs non-resistant strains, or following NPV infection.
Table 1.
Differentially expressed Enzymes proteins in different resistant strain following BmNPV infection.
| Protein ID | Protein description | Accession | A35+ vs. P50+ ratio | A35- vs. P50- ratio | A35+ vs. A35- ratio | P50+ vs. P50- ratio |
|---|---|---|---|---|---|---|
| Serine protease | ||||||
| Bm_nscaf2674_066 | trypsin alkaline A-like protein, partial [Bombyx mori] | APS85762 | 780.424 | 3186.616 | NS | NS |
| Bm_scaffold644_4 | collagenase [Bombyx mori] | XP_004927691 | 11.067 | 8.279 | 1.554 | NS |
| Bm_nscaf2983_048 | trypsin, alkaline A [Bombyx mori] | XP_004923290 | 5.134 | 3.897 | NS | NS |
| Bm_nscaf2902_287 | chymotrypsin-like serine protease precursor [Bombyx mori] | NP_001040430 | 3.04 | 5.172 | NS | NS |
| Bm_scaffold769_2 | serine protease precursor [Bombyx mori] | NP_001036826 | 2.785 | 2.98 | NS | NS |
| Bm_nscaf2674_017 | LOW QUALITY PROTEIN: trypsin, alkaline C [Bombyx mori] | XP_012549322 | 2.449 | 2.367 | NS | NS |
| Bm_scaffold644_3 | serine protease [Bombyx mori] | AAX39409 | 2.326 | 2.169 | 2.108 | NS |
| Bm_nscaf2674_063 | trypsin, alkaline C [Bombyx mori] | XP_004930040 | 2.29 | NS | 2.552 | NS |
| Bm_nscaf3058_335 | collagenase-like [Papilio xuthus] | XP_013167497 | 2.128 | 1.768 | NS | NS |
| Bm_nscaf2993_279 | trypsin, alkaline C [Bombyx mori] | XP_004929525 | 2.091 | 1.674 | NS | NS |
| Bm_nscaf2902_225 | LOW QUALITY PROTEIN: transmembrane protease serine 9-like [Helicoverpa armigera] protease serine 9-like [Helicoverpa armigera] | XP_021201549 | 1.787 | NS | 1.547 | NS |
| Bm_nscaf3058_362 | brachyurin [Bombyx mori] | XP_004928997 | 1.734 | 1.572 | NS | NS |
| Bm_nscaf1962_13 | trypsin alpha-3 [Bombyx mori] | XP_004927140 | 1.658 | 1.723 | NS | NS |
| Bm_nscaf2902_226 | chymotrypsin-like protease C8 [Heliothis virescens] | ABR88238 | 1.533 | NS | NS | NS |
| Bm_nscaf2983_050 | trypsin-like serine protease [Bombyx mori] | NP_001243950 | 0.611 | NS | 2.061 | NS |
| Bm_nscaf2901_48 | ovochymase-2 [Bombyx mori] | XP_021203513 | 0.61 | 0.521 | NS | NS |
| Bm_nscaf3058_328 | 35 kDa protease precursor [Bombyx mori] | NP_001037037 | 0.58 | NS | NS | NS |
| Bm_nscaf3058_337 | serine protease 3 [Bombyx mori] | XP_004928982 | 0.431 | 0.533 | NS | NS |
| Bm_nscaf2953_114 | collagenase isoform X1 [Bombyx mori] | XP_004922848 | A35+ | A35- | NS | N |
| Bm_nscaf3058_329 | collagenase- partial [Bombyx mori] | XP_004928979 | A35+ | A35- | NS | N |
| Bm_nscaf3058_338 | collagenase [Bombyx mori] | XP_004928983 | A35+ | A35- | NS | N |
| Bm_scaffold644_6 | collagenase [Bombyx mori] | XP_004927692 | A35+ | A35- | 2.579 | N |
| Bm_nscaf2674_064 | trypsin, alkaline C [Bombyx mori] | XP_004930042 | A35+ | A35- | NS | N |
| Bm_scaffold644_5 | serine protease precursor [Bombyx mori] | NP_001036903 | A35+ | A35- | NS | N |
| Bm_nscaf2795_043 | venom serine protease [Bombyx mori] | XP_004922529 | A35+ | N | A35+ | N |
| Bm_nscaf2883_014 | collagenase [Bombyx mori] | XP_004923933 | P50+ | P50- | N | NS |
| Bm_nscaf2983_046 | trypsin, alkaline C [Bombyx mori] | XP_004923288 | P50+ | P50- | N | NS |
| Bm_nscaf2986_178 | trypsin, alkaline A [Bombyx mori] | XP_004931376 | NS | A35- | 0.654 | P50+ |
| Bm_nscaf2983_049 | trypsin-like protease precursor [Bombyx mori] | NP_001093273 | NS | NS | 1.705 | NS |
| Lipase | ||||||
| Bm_nscaf2529_051 | lipase member H-A [Bombyx mori] | XP_004932346 | 9.716 | 4.164 | 3.025 | NS |
| Bm_nscaf3032_110 | pancreatic triacylglycerol lipase [Bombyx mori] | XP_004924199 | P50+ | P50- | N | NS |
| Aminopeptidase N | ||||||
| Bm_nscaf2889_046 | aminopeptidase N [Bombyx mori] | AAC33301 | 0.373 | NS | 0.599 | NS |
| Bm_nscaf2889_049 | membrane alanyl aminopeptidase [Bombyx mori] | XP_021208930 | NS | A35- | NS | P50+ |
| Bm_nscaf2889_053 | aminopeptidase N-7 [Bombyx mori] | AFK85023 | 0.039 | P50- | A35+ | NS |
| Carboxypeptidase | ||||||
| Bm_nscaf2953_014 | carboxypeptidase B-like precursor [Bombyx mori] | NP_001296550 | 4.23 | 3.55 | NS | NS |
| Bm_nscaf2818_010 | carboxypeptidase B [Bombyx mori] | XP_004923345 | 3.481 | 3.155 | NS | NS |
| Bm_nscaf2818_078 | zinc carboxypeptidase [Bombyx mori] | XP_012553139 | A35+ | A35- | NS | N |
| Bm_nscaf3098_76 | carboxylesterase CarE-7 [Bombyx mori] | ABY57298 | A35+ | A35- | NS | N |
| Bm_nscaf2575_105 | esterase FE4 [Bombyx mori] | XP_021208942 | A35+ | A35- | NS | N |
| Bm_nscaf2953_011 | zinc carboxypeptidase [Bombyx mori] | XP_021208707 | NS | A35- | NS | P50+ |
| Other enzymes | ||||||
| Bm_nscaf1898_224 | akaline nuclease precursor [Bombyx mori] | NP_001091744 | 2.496 | NS | NS | NS |
| Bm_nscaf1681_013 | beta-1,3-glucanase [Bombyx mori] | ACU57045 | 1.567 | 1.625 | NS | NS |
| Bm_nscaf1898_444 | venom dipeptidyl peptidase 4 [Bombyx mori] | XP_021204723 | 0.639 | NS | NS | NS |
| Bm_scaffold416_08 | prostatic acid phosphatase [Bombyx mori] | XP_012546808 | 0.598 | NS | NS | NS |
| Bm_nscaf2925_05 | alkaline phosphatase [Bombyx mori] | BAG41975 | 0.401 | 0.361 | NS | NS |
| Bm_nscaf2827_48 | alpha-amylase precursor [Bombyx mori] | NP_001166624 | 0.0004 | P50- | A35+ | NS |
| Bm_nscaf3058_115 | apyrase [Bombyx mori] | XP_004928875 | A35+ | N | A35+ | N |
| Bm_nscaf2818_054 | n-acetylneuraminate lyase [Bombyx mori] | XP_021208662 | A35+ | A35- | NS | N |
| Bm_nscaf3097_36 | chitin deacetylase 5b [Helicoverpa armigera] | ADB43612 | P50+ | P50- | N | NS |
| Bm_nscaf1289_1 | chitinase isoform 3 precursor [Bombyx mori] | NP_001166832 | P50+ | N | N | P50+ |
| Bm_nscaf2838_097 | 15-hydroxyprostaglandin dehydrogenase [NAD(+)] [Bombyx mori] | XP_012543884 | P50+ | N | N | P50+ |
| Bm_nscaf3056_04 | peroxidase [Bombyx mori] | XP_012545623 | N | A35- | A35- | N |
| Bm_nscaf1681_231 | peroxidase isoform X1 [Bombyx mori] | XP_012550815 | N | A35- | A35- | N |
| Bm_nscaf2818_052 | N-acetylneuraminate lyase isoform X1 [Bombyx mori] | XP_004923368 | N | N | A35+ | N |
NS: non-significant protein in comparable group, N: no expressed protein in comparable group, P50-, P50+, A35- and A35+: the protein only expressed in P50-, P50+, A35- and A35+ group.
3.7. Analysis of DEPs associated with immune response between different strains and after BmNPV infection
Unlike mammals, insects do not produce antibodies and instead rely on innate immunity through humoral and cellular responses. DEPs related to the immune response were identified between resistant and non-resistant strains after BmNPV infection. In total, four DEPs involved in the immune response were identified belonging to either the prophenoloxidase (PPO) pathway or having pattern recognition receptor functions (Table 2 ). These included serine protease inhibitor 16 precursor (Spn-16), Spn-2, antitrypsin isoform X1 (Spn-1) and peptidoglycan recognition protein precursor (PRP). The expression level of Spn-2 and Spn-1 were down-regulated in resistant strain A35 compared with the non-resistant P50 strain, and also down-regulated following BmNPV infection. Meanwhile, DEPs identified during comparisons of the aforementioned groups were not differentially expressed when comparing between P50+ and P50- treatment groups. Additionally, the Spn-16 was only expressed in the P50 strain and no significant differences P50+ vs P50- was observed. The expression level of PRP was reduced in A35+ vs P50+, while there was no significant differences observed within the other three groups.
Table 2.
DEPs by label-free associated with immunity in silkworm in different resistant strain after BmNPV infection.
| Protein ID | Protein description | Accession | A35 + _vs_P50+ ratio | A35-_vs_P50- ratio | A35 + _vs_A35- ratio | P50 + _vs_P50- ratio |
|---|---|---|---|---|---|---|
| PPO pathway | ||||||
| Bm_nscaf2888_098 | serine protease inhibitor 2 isoform X1 [Bombyx mori] | XP_012547756 | 0.224 | 0.587 | 0.428 | NS |
| Bm_nscaf2979_2 | antitrypsin isoform X1 [Bombyx mori] | XP_021204540 | 0.293 | 0.281 | 0.579 | NS |
| Bm_nscaf2641_2 | serine protease inhibitor 16 precursor [Bombyx mori] | NP_001139708 | P50+ | P50- | N | NS |
| Pattern recognition receptor | ||||||
| Bm_nscaf3058_130 | peptidoglycan recognition protein precursor [Bombyx mori] | NP_001243949 | 0.463 | NS | NS | NS |
NS: non-significant protein in comparable group, N: no expressed protein in comparable group, P50-, P50+, A35- and A35+: the protein only expressed in P50-, P50+, A35- and A35+ group.
3.8. The analysis of selected DEPs involved in resistance to BmNPV
Utilizing Venn diagram analysis allowed for the removal of DEPs related to either genetic background (A35- vs P50-) and individual immune stress response proteins (P50+ vs P50-) (Fig. 4B). Nine DEPs were identified, including two uncharacterized proteins, four serine proteases, N-acetylneuraminate lyase isoform X1, aminopeptidase N and apyrase, potentially related to resistance to BmNPV in both groups (A35+ vs A35- and A35+ vs P50+). The expression level of the 9 DEPs involved in resistance to BmNPV were up- or down-regulated in two of the groups compared (A35+ vs A35- and A35+ vs P50+) while no significant differences in expression were observed between the other two groups. Trypsin, alkaline C and transmembrane protease serine 9-like proteins were up-regulated in both groups. Trypsin-like serine protease were up-regulated in A35+ vs A35- but down-regulated in A35+ vs P50+. Finally, the venom serine protease was specially expressed in A35+. Additionally, N-acetylneuraminate lyase isoform X1 and apyrase were specially expressed in A35+, and only aminopeptidase N was down-regulated in both A35+ vs A35- and A35+ vs P50+ groups (Table 3 ).
Table 3.
The DEPs potentially related to resistance to BmNPV.
| Protein ID | Protein description | Accession | A35 + _vs_P50+ ratio | A35-_vs_P50- ratio | A35 + _vs_A35- ratio | P50 + _vs_P50- ratio |
|---|---|---|---|---|---|---|
| Bm_scaffold1148_1 | uncharacterized protein LOC101744440 [Bombyx mori] | KPJ17799 | 8.441 | NS | 3.408 | NS |
| Bm_nscaf2655_107 | uncharacterized protein LOC101735588 [Bombyx mori] | XP_021209046 | 3.783 | NS | 2.072 | NS |
| Bm_nscaf2674_063 | trypsin, alkaline C [Bombyx mori] | XP_004930040 | 2.290 | NS | 2.552 | NS |
| Bm_nscaf2902_225 | transmembrane protease serine 9-like [Helicoverpa] armigera] | XP_021201549 | 1.787 | NS | 1.547 | NS |
| Bm_nscaf2983_050 | trypsin-like serine protease [Bombyx mori] | NP_001243950 | 0.611 | NS | 2.061 | NS |
| Bm_nscaf2889_046 | aminopeptidase N [Bombyx mori] | AAC33301 | 0.373 | NS | 0.599 | NS |
| Bm_nscaf2795_043 | venom serine protease [Bombyx mori] | XP_004922529 | A35+ | N | A35+ | N |
| Bm_nscaf2818_052 | N-acetylneuraminate lyase isoform X1 [Bombyx mori] | XP_004923368 | A35+ | N | A35+ | N |
| Bm_nscaf3058_115 | apyrase [Bombyx mori] | XP_004928875 | A35+ | N | A35+ | N |
NS: non-significant protein in comparable group, N: no expressed protein in comparable group, P50-, P50+, A35- and A35+: the protein only expressed in P50-, P50+, A35- and A35+ group.
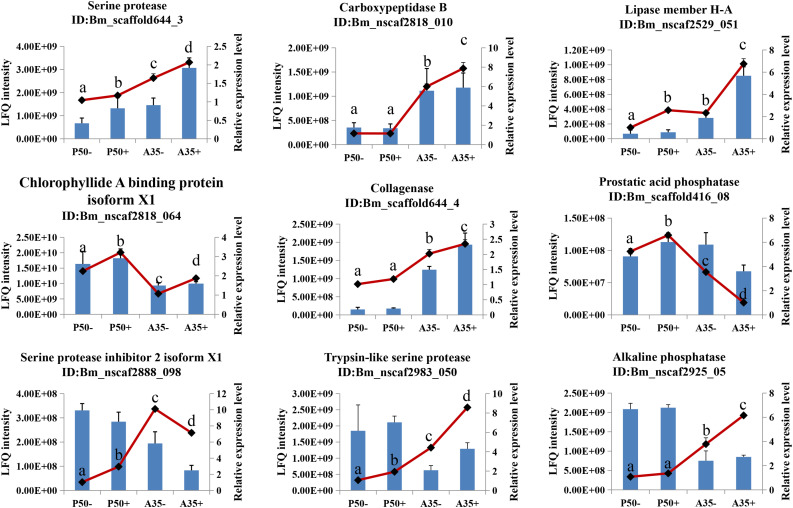
3.9. Validation of DEPs at the transcriptional level in the midgut and protein level of digestive juice
Digestive juice proteins are synthesized by midgut cells and secreted into the midgut lumen. To analyze the level of transcription of the genes corresponding to proteins in digestive juice, the cDNA of midgut RNA was used as a template for qPCR reactions according to Hu et al. [6]. To measure between protein and transcript abundance, RT-qPCR was conducted to analyze transcript levels of nine identified DEPs, selected from the 80 total DEPs identified as an accumulation of those identified in all four groups (Fig. 7 ). The results show that the transcriptional levels of six tested DEPs were consistent with the label free data, and transcript abundance of the other three DEPs (AP, Trypsin-like serine protease and Spn-2) were partially consistent with the label free data.
Fig. 7.
Differential expression levels of 9 DEPs corresponding to the co-regulated genes of midgut in both strains of BmNPV-infected and control silkworm larvae at 24 h post inoculation. The y-axis on the left side indicates the expression level of candidate proteins. The x-axis on the right side indicates the relative expression level of candidate gene mRNA transcripts. Data were normalized using BmGADPH and represented as means ± standard errors of the means from three independent experiments. Relative expression levels were calculated using the 2-△△Ct method. Statistical analysis was conducted using the SPSS software. Significant differences are indicated by different letters (p < .05).
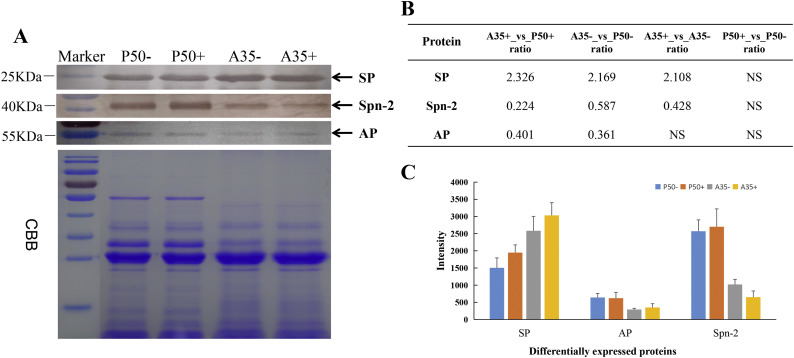
Western blotting was used to further confirm the observed changes in protein expression in the P50 and A35 strains following BmNPV infection. The SP, AP and Spn-2 proteins in digestive juice were selected for western blotting analysis. Because there exists no appropriate control for digestive juice, Coomassie Blue R-250 staining was performed according to Rao et al. [31]. The results show that SP was increased in resistant and susceptible strain following BmNPV infection. The AP protein was down-regulated in A35 compared to P50, but there was no difference following BmNPV infection. The Spn-2 was down-regulated in A35 compared to P50 and also down-regulated in A35 strain following BmNPV infection. However, the protein had similar expression levels in P50+ and P50- (Fig. 8A, B). The Western blotting results using all the 3 DEPs were consistent with the Label free data obtained in this study (Fig. 8C).
Fig. 8.
Confirmation of three differentially expressed proteins (SP, AP and Spn-2) with Western blotting. A, The protein levels of SP, AP and Spn-2 in P50−, P50+, A35− and A35+. 30 μg each of midgut digestive juice protein was analyzed by western blot with antiserum (top). Total proteins were Coomassie Brilliant Blue (CBB)-stained as the loading control (bottom). B, Label-free results from 3 DEPs in P50 and A35 strains showing BmNPV-infected and control groups. C, Densitometric intensity of the Western blot was analyzed by using Image J software. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Previous studies have indicated that some digestive juice proteins involved in antiviral activity, such as red fluorescent protein [7], lipase [8], serine protease [9] and alkaline trypsin [10] in silkworms. Here, we confirmed that the digestive juices of resistant strain (A35) had stronger anti-BmNPV activity than those of susceptible strain (P50) by RT-qPCR. Additionally, the SDS-PAGE results suggested that the proteins components of digestive juice had significantly difference between the two different resistant strains. These results implied that the content of antiviral proteins in the digestive juices of resistant strains is higher than that of susceptible strains, or that resistant strains contain antiviral proteins not found in susceptible strains.
In previous study, Xue et al. [32] found that the peak responses appeared at 24 h BmNPV post infection according to results of gene expression kinetics in Bm5 cells. Furthermore, Muhammad et al. [33] results showed that the BmNPV begins to massive replication in the midgut at this time. Additionally, the most of DEPs were identified in midgut of different resistant strains at 24 h BmNPV post infection [14]. To comprehensively understand the function of proteins present in the digestive juice of B. mori in response to BmNPV infection, the 24 h time point post BmNPV infection was selected to analysis of the digestive juice from two silkworm strains by label-free proteomic. In total, 98 unique proteins were identified, including 68 enzymes and 30 non-enzymatic proteins. Included in that total number are 72, 82, 82 and 87 proteins identified from P50-, P50+, A35- and A35+ strains, respectively. These results demonstrate that proteins derived from digestive juice mainly consist of enzymes, and are consistent with previous reports in B. mori and Helicoverpa armigera [5,6]. Compared to studies by Hu et al. [6] which investigated protein expression in the digestive juice of silkworms, >45 and 15 proteins were identified by Label-free quantitative approaches in P50- and P50+, respectively. Notably, eight specially expressed proteins were identified in our four experiment groups. Compared with other proteomics technologies, the Label-free method provides higher sequence coverage and ultimately detects a higher number of DEPs [34,35].
Eighty proteins were identified found to differentially expressed when comparing between resistant and non-resistant strains +/− challenge by BmNPV. Among the proteins identified were 54 enzymes which mainly included serine proteases, lipases and aminopeptidases. There were also 26 nonenzymatic proteins which mainly included serine protease inhibitors and uncharacterized proteins. The DEP data was validated through the quantification of transcript abundance, however, three DEPs tested (AP, Trypsin-like serine protease and Spn-2) were only partially consistant with the label free data following BmNPV infection. Digestive juice proteins were synthesized by midgut cells and secreted into the midgut lumen. However, mRNA from the midgut can be regulated at both transcriptional and translational levels before proteins accumulate in digestive juice. Examples of the regulatory mechanisms possibly affecting protein levels include translational levels, such as mRNA stability, splicing, miRNA regulation and protein stability [13,14]. Generally, only 60% - 70% of the genes have transcript and protein abundance levels that are comparable as detected by qRT-PCR and proteomic methodology [36].
4.1. Enzymes response to BmNPV infection in different resistant strain
The midgut digestive juice contains a large number of enzymes, such as lipase, serine protease and aminopeptidase. The enzymes in the digestive juice of silkworms are synthesized and secreted by epithelial cells, with their main function being the hydrolysis of macromolecular compounds in mulberry leaves, such as sugars, proteins, and lipids. As a result, these enzymes play important roles in growth and development in B. mori. Especially, as the previous study results confirmed, because that some enzymes function to protect against BmNPV.
Serine protease (SP) is a class of proteolytic enzymes with serine activity centers that play an important role in digestion, growth, development, and immune response in insects [37,38]. It is the most important type of digestive enzyme in the lepidopteran gut and accounts for about 95% of digestive activity [39]. In the present study, a total of 29 SPs were identified that were differentially expressed within the digestive juice of different resistant silkworm strains. The expression level of most SPs were up-regulated in the highly resistant A35 strain compared with susceptible P50 strain and further up-regulated in highly resistant A35 strain following NPV infection. Additionally, four SPs were identified that were potentially related to resistance to BmNPV in digestive juice by removing those DEPs related to either genetic background (A35- vs P50-) and individual immune stress response proteins (P50+ vs P50-). Nakazawa and Ponnuvel [9,10] previously reported that alkaline trypsin and SP-2 purified from the digestive juice of B. mori larvae had strong antiviral activity against BmNPV under in vitro conditions. Bm-SP142 could effectively impair the ability of the virus to infect BmN cells and silkworms, and over-expression of Bm-SP142 could inhibit viral propagation in BmN cells [40]. Additionally, Chen et al. [41] found that the some anti-BmNPV genes expressed significantly higher in the resistant strains compared to susceptible strains. These results suggested that SPs, an insect digestive enzyme, can be a potential antiviral factor against BmNPV at the initial site of viral infection. It was implied that the high expression level of SPs in digestive juice is one potential reason the resistant silkworm strain is not susceptible to NPV infection.
Lipases are ubiquitous enzymes in nature, and play a crucial role in fat metabolism by catalyzing the hydrolysis of triacylglycerol to free fatty acids and glycerol [42,43]. Ponnuvel et al. [8] purified a lipase from the digestive juice of B. mori larvae and confirmed that it had strong antiviral activity against BmNPV under in vitro conditions. In this study, two lipases (lipase member H-A and pancreatic triacylglycerol lipase) were differentially expressed. After BmNPV infection, the expression level of Lipase member H-A increased 3 times in the A35 strain, while no significant differences occurred in the P50 strain. These results suggest that the silkworm strain having higher expression level of lipase may also have stronger resistance to viral infection. Jiang et al. [44] found that in transgenic line silkworm over-expressing an endogenous antiviral gene lipase-1, mortality decreased to approximately 33% compared with the non-transgenic line. As the results suggest, we suspect that not only one lipase protein had antiviral functions in silkworm larvae digestive juice, and other proteins in the lipase family had similar functions.
Aminopeptidase N (APN), belonging to zinc-dependent metalloproteinase, is a major component in the insect gut epithelial membrane, with a primary function to cleave N-terminal amino acids. APN not only catalyzes protein proteolytic processes, but also is involved in the pathogenic process as the receptor of pathogenic toxin [45]. APN was the first molecule identified as a Cry toxin-binding protein from Manduca sexta [46,47], and it is an extensively studied putative receptor from other lepidopteran insect pests [47,48]. In the present study, three APNs (aminopeptidase N, membrane alanyl aminopeptidase and aminopeptidase N-7) were identified from four experiment groups. Interestingly, the three APNs were down-regulated in A35 compared to P50 strains and also downregulated following BmNPV. APN acts as the viral receptor of some group I coronaviruses, such as human coronavirus 229E and transmissible gastroenteritis virus (TGEV), in order to allow the virus to enter to cell [49,50]. A study by Lucas et al. [51] indicated that membrane alanyl aminopeptidase N acts as a receptor for a plant virus in the pea aphid vector in vitro. Additionally, an APN interaction between BmDNV VD1 and ORF4 was identified by Co-immunoprecipitation [52]. We hypothesized that the APNs probably act as the receptor for BmNPV, or alternatively accelerate infection by interaction with BmNPV. The relationship between BmNPV and APN need to be explored in the future.
Regarding the other 21 enzymes within the identified DEPs, two DEPs related to chitin degradation were identified in all four groups being compared, including one chitin deacetylase (CDA, chitin deacetylase 5b) and one chitinase (chitinase isoform 3 precursor). Interestingly, the two enzymes were specially expressed in the susceptible BmNPV P50 strain (Table 1). In Helicoverpa armigera, the expression level of chitin deacetylase-like proteins were down-regulated following infection with H. armigera single nucleopolyhedrovirus, which might reduce the susceptibility for baculovirus infection by decreasing its PM permeability [53]. This result suggested that the different resistance of silkworm strains might be related to expression level of chitinolytic enzymes by affecting the PM permeability after the virus infection. On the other hand, there were two differentially expressed phosphatase proteins, including prostatic acid phosphatase (PAP) and alkaline phosphatase (AP). The expression level of PAP was down-regulated in A35 strain compared with P50 strain. Although the alkaline and acid phosphatase were reported to be involved in the transport of glucose and fatty acids on the intestinal wall membrane in B. mori [54], Wang et al. [22] found that the expression levels of PAP were decreased following BmNPV infection in resistant strains, and suggested that PAP could directly or indirectly participate in repressing BmNPV replication in host cells. Lee et al. [55] also reported that the lysosomal acid phosphatase 2 (ACP2) was required for membrane fusion during influenza A virus (IAV) entry in mammal cells. Meanwhile, the expression level of AP in the P50 strain was higher than in the A35 strain (by Label free and Western blotting) (Fig. 7). These results were consistent with a previous report that showed the expression level of AP in midgut was up-regulated in P50 compared with BC9 (resistant BmNPV stain) [14]. Additionally, AP activity was decreased in the silkworm embryonic cells after BmNPV infection [56]. In summary, PAP and AP might play an important role in response to virus infection.
4.2. Non-enzymatic proteins response to BmNPV infection in midgut digestive juice
Previous studies found that the midgut digestive juice not only contains a large number of enzymes but also contain other non-enzymatic proteins [6]. In this study, thirty non-enzymatic proteins were identified in four groups, including 26 DEPs in the four groups compared here.
The larval midgut is targeted by baculovirus, which uses it as a portal of entry to later establish a systemic infection within the insect body. Thus, the immune response plays an important role in the defense against pathogens [57]. Here, four DEPs, including Spn-1, Spn-2, Spn-16 and PRP, are known to be related to the immune response were identified. Melanization is a major immune responses against large pathogens or parasites in arthropods, and phenoloxidase (PO) is a key enzyme in the melanization process [58]. In the insect immune response, Spn can bind to the active center of a serine protease during the extracellular serine protease cascade, which makes the serine protease unable to activate the next level serine protease or PPO, thereby inhibiting the cascade reaction, and finally inhibiting melanization [[59], [60], [61]]. In this study, the Spn-1 and Spn-2 proteins were found that it significantly down-regulated in A35+ vs A35- and A35- vs P50-. Interestingly, the Spn-16 specially expressed in P50 strain. Pan et al. [62] found that the expression of Spn-2 in the midgut were significantly increased in susceptible strain after infected with BmNPV, while there was no significant change in resistant strain. These results indicated that Spn-2 may be involved in the B. mori antiviral response. Additionally, serpin-5 and serpin-9 can promote baculovirus infection by inhibiting melanizationin in Helicoverpa armigera [63]. Based on these findings, we speculated that the down-regulation of expression of Spn in resistant strains or following BmNPV infection might lead to inhibition of serine proteases and enhance host melanization to suppress infection by BmNPV in resistant strains. Pathogen-associated molecular patterns are pathogen molecules recognized by cells of the innate immune system (by pattern recognition receptors [PRRs]) [64]. Peptidoglycan recognition protein precursors (PRPs) were down-regulated in A35+ compared to A35-, but expression levels showed no difference in the P50 strain after BmNPV infection. Yu et al. [14] found that the pattern-recognition receptor-related proteins showed a different response to BmNPV infection in resistant strain vs non-resistant strains. As above all, the DEPs related to the immune response might play an important role in response to BmNPV infection in digestive juice.
In this study, a chlorophyllide A binding protein isoform X1 (CHBP) protein and red fluorescence protein were found in digestive juice. Interestingly, the expression level of CHBP was up-regulated but no significantly in control compared to BmNPV-challenged samples. However, it was significantly down-regulated in A35- compared to P50- strains. The red fluorescent protein (RFP) had antiviral properties and the red fluorescence was suggested to be derived from the binding between insect midgut proteins and chlorophyllid α, the prosthetic group of chlorophyll. Moreover, it was also suggested that anti-BmNPV activity was attributed to chlorophyllid α rather than the protein [65,66]. Therefore, we speculated that the CHPB had an antiviral function in digestive juice, but is not the main contributing factor to silkworm resistance in the resistant strains. The peritrophic membrane (PM) as a first line of defense and is not absolutely responsible for the prevention of invasion of pathogenic microorganisms in insects, but the structural integrity of the PM may delay the invasion rate and delay the infection process [67,68]. Insect intestinal mucin (IIM) is an important part of the PM and plays an important role in protecting insects from microbial infection. Wang et al. [69] identified the first invertebrate intestinal mucin from Trichoplusia ni larva and showed that it can resist the degradation of protease in the midgut. Here, two IIMs (mucin-2, insect intestinal mucin precursors) were identified from digestive juice. The expression level of mucin-2 and insect intestinal mucin precursor were down-regulated in A35+ vs P50+ and A35- vs P50-, respectively. Wang et al. [70] reported that the IIM can be degraded by enhancin, a synergistic protein produced by Trichoplusia ni granulovirus (TnGV), which specifically interacts with IIM, leading to the destruction of the integrity of the PM and thereby increasing the chances of the virus entering epithelial cells and killing insects. Consequently, we speculated that the BmNPV had an enhancin homologue with a similar function as IIM that might play an important role in BmNPV infection. There are also some other non-enzymatic DEPs have been showed response to virus infection, we will further study these proteins and elucidate their roles in BmNPV infection in the future.
5. Conclusion
Previous studies have revealed that some proteins in silkworm digestive juice show antiviral activity. Here, we examined the antiviral activity of silkworm digestive juice and observed that the digestive juice of the resistant strain (A35) had stronger antiviral activity than the susceptible strain (P50). Additionally, the DEPs were identified that were likely responsible for the silkworm response to BmNPV in different resistant vs susceptible strains based on label free quantitative proteomics. In summary, 80 proteins were found to be differentially expressed, including 54 enzymes and 26 nonenzymatic proteins. DEPs identified here are mainly involved in proteolysis and associated with neuroactive ligand receptor interaction. After removing the genetic background and individual immune stress response proteins, we identified nine DEPs likely to be involved in response to BmNPV infection. These findings provide an overview of the protein content of the digestive juice of resistant and susceptible silkworm strains and their responses to BmNPV infection, They also expound on current knowledge of the mechanism of silkworm resistance to BmNPV.
The following are the supplementary data related to this article.
Classification of total protein in digestive juices of silkworm midgut.
Primers used in RT-qPCR for validation of DEPs
Summary of digestive juice proteins identified by a Label free technology
Summary of differentially expressed proteins (DEPs)
Acknowledgments
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 31973002, 31472148), International Cooperation Project of Anhui Province (No. 1804b06020345) and Anhui International Joint Research and Development Center of Sericulture Resources Utilization (No. 2017R0101).
Declaration of Competing Interest
The authors have declared no conflict of interest.
References
- 1.Goldsmith M.R., Shimada T., Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005;50:71–100. doi: 10.1146/annurev.ento.50.071803.130456. [DOI] [PubMed] [Google Scholar]
- 2.Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., Pan G., Xu J., Liu C., Lin Y., Qian J., Hou Y., Wu Z., Li G., Pan M., Li C., Shen Y., Lan X., Yuan L., Li T., Xu H., Yang G., Wan Y., Zhu Y., Yu M., Shen W., Wu D., Xiang Z., Yu J., Wang J., Li R., Shi J., Li H., Li G., Su J., Wang X., Li G., Zhang Z., Wu Q., Li J., Zhang Q., Wei N., Xu J., Sun H., Dong L., Liu D., Zhao S., Zhao X., Meng Q., Lan F., Huang X., Li Y., Fang L., Li C., Li D., Sun Y., Zhang Z., Yang Z., Huang Y., Xi Y., Qi Q., He D., Huang H., Zhang X., Wang Z., Li W., Cao Y., Yu Y., Yu H., Li J., Ye J., Chen H., Zhou Y., Liu B., Wang J., Ye J., Ji H., Li S., Ni P., Zhang J., Zhang Y., Zheng H., Mao B., Wang W., Ye C., Li S., Wang J., Wong G.K., Yang H. Biology Analysis, A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L., Xia Q.Y. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect. Biochem. Molec. 2014;48:1–7. doi: 10.1016/j.ibmb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen S., Hou C., Bi H., Wang Y., Xu J., Li M., James A.A., Huang Y., Tan A. Transgenic CRISPR/Cas9-mediated viral gene targeting for antiviral therapy of Bombyx mori nucleopolyhedrovirus (BmNPV) J. Virol. 2017;91 doi: 10.1128/JVI.02465-16. (02465-02416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pauchet Y., Muck A., Svatos A., Heckel D.G., Preiss S. Mapping the larval midgut lumen proteome of Helicoverpa armigera, a generalist herbivorous insect. J. Proteome Res. 2008;7:1629–1639. doi: 10.1021/pr7006208. [DOI] [PubMed] [Google Scholar]
- 6.Hu X., Zhu M., Wang S., Zhu L., Xue R., Cao G., Gong C. Proteomics analysis of digestive juice from silkworm during Bombyx mori nucleopolyhedrovirus infection. J. Proteome. 2015;15:2691–2700. doi: 10.1002/pmic.201400475. [DOI] [PubMed] [Google Scholar]
- 7.Sunagar S.G., Savanurmath C.J., Hinchigeri S.B. The profiles of red fluorescent proteins with antinucleopolyhedrovirus activity in races of the silkworm Bombyx mori. J. Insect Physiol. 2011;57:1707–1714. doi: 10.1016/j.jinsphys.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Ponnuvel K.M., Nakazawa H., Furukawa S., Asaoka A., Ishibashi J., Tanaka H., Yamakawa M. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J. Virol. 2003;77:10725–10729. doi: 10.1128/JVI.77.19.10725-10729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakazawa H., Tsuneishi E., Ponnuvel K.M., Furukawa S., Asaoka A., Tanaka H., Ishibashi J., Yamakawa M. Antiviral activity of a serine protease from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Virology. 2004;321:154–162. doi: 10.1016/j.virol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Ponnuvel K.M., Nithya K., Sirigineedi S., Awasthi A.K., Yamakawa M. In vitro antiviral activity of an alkaline trypsin from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Arch. Insect. Biochem. 2012;81:90–104. doi: 10.1002/arch.21046. [DOI] [PubMed] [Google Scholar]
- 11.Liu H., Chen B., Hu S., Liang X., Lu X., Shao Y. Quantitative proteomic analysis of germination of Nosema bombycis spores under extremely alkaline conditions. Front. Microbiol. 2016;7:1459. doi: 10.3389/fmicb.2016.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Yu H., Xu J., Zhang S., Yu D., Liu M., Wang L. Comparative subcellular proteomics analysis of susceptible and near-isogenic resistant Bombyx mori (Lepidoptera) larval Midgut response to BmNPV infection. Sci Rep-UK. 2017;7:45690. doi: 10.1038/srep45690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao K., Deng X.Y., Shang M.K., Qin G.X., Hou C.X., Guo X. iTRAQ-based quantitative proteomic analysis of midgut in silkworm infected with Bombyx mori cytoplasmic polyhedrosis virus. J. Proteome. 2017;152:300–311. doi: 10.1016/j.jprot.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Yu H., Wang X., Xu J., Ma Y., Zhang S., Yu D., Fei D., Muhammad A. iTRAQ-based quantitative proteomics analysis of molecular mechanisms associated with Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic strains. J. Proteome. 2017;165:35–50. doi: 10.1016/j.jprot.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Wu P., Shang Q., Huang H., Zhang S., Zhong J., Hou Q., Guo X. Quantitative proteomics analysis provides insight into the biological role of Hsp90 in BmNPV infection in Bombyx mori. J. Proteome. 2019;203:103379. doi: 10.1016/j.jprot.2019.103379. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Xia D., Zhao Q., Zhang G., Zhang Y., Qiu Z., Shen D., Lu C. Label-free proteomic analysis of silkworm midgut infected by Bombyx mori nuclear polyhedrosis virus. J. Proteome. 2019;200:40–50. doi: 10.1016/j.jprot.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Hongxia W., Sophie A., Hicks L.M. Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering. J. Proteome Res. 2012;11:487–501. doi: 10.1021/pr2008225. [DOI] [PubMed] [Google Scholar]
- 18.Chen T.T., Tan L.R., Hu N., Dong Z.Q., Hu Z.G., Jiang Y.M., Chen P., Pan M.H., Lu C. C-lysozyme contributes to antiviral immunity in Bombyx mori against nucleopolyhedrovirus infection. J. Insect Physiol. 2018;108:54–60. doi: 10.1016/j.jinsphys.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu T., Dong X., Pan C., Du G., Wu Y., Yang J., Peng C., Cheng L., Pan M. A newly discovered member of the Atlastin family, BmAtlastin-n, has an antiviral effect against BmNPV in Bombyx mori. Sci Rep-UK. 2016;6:28946. doi: 10.1038/srep28946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng M., Kong X., Zhang J., Xu W., Wu X. Identification of a novel host protein SINAL10 interacting with GP64 and its role in Bombyx mori nucleopolyhedrovirus infection. Virus Res. 2018;247:102–110. doi: 10.1016/j.virusres.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Wang X.Y., Shao Z.M., Chen Q.Y., Xu J.P., Sun X., Xu Z.P., Li M.W., Wu Y.C. Knockdown of BmTCP-1β delays BmNPV infection in vitro. Front. Microbiol. 2019;10:578. doi: 10.3389/fmicb.2019.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X.Y., Yu H.Z., Geng L., Xu J.P., Yu D., Zhang S.Z., Ma Y., Fei D.Q. Comparative Transcriptome analysis of Bombyx mori (Lepidoptera) larval Midgut response to BmNPV in susceptible and near-isogenic resistant strains. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman M.M., Gopinathan K.P. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 2004;101:109–118. doi: 10.1016/j.virusres.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Brevnov M.G., Pawar H.S., Janna M., Calandro L.M., Furtado M.R., Shewale J.G. Developmental validation of the PrepFiler forensic DNA extraction kit for extraction of genomic DNA from biological samples. J. Forensic Sci. 2010;54:599–607. doi: 10.1111/j.1556-4029.2009.01013.x. [DOI] [PubMed] [Google Scholar]
- 25.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Mrthods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 26.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Luber C.A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2013;495:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 29.Götz S., Garcíagómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talón M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang P.J., Zhan M.Y., Ye C., Yu X.Q., Rao X.J. Molecular cloning and characterization of a short peptidoglycan recognition protein from silkworm Bombyx mori. Insect Mol. Biol. 2017;26:665–676. doi: 10.1111/imb.12330. [DOI] [PubMed] [Google Scholar]
- 32.Xue J., Qiao N., Zhang W., Cheng R.L., Zhang X.Q., Bao Y.Y., Xu Y., Gu L.Z., Han J.D., Zhang C.X. Dynamic interactions between Bombyx mori nucleopolyhedrovirus and its host cells revealed by transcriptome analysis. J. Virol. 2012;86:7345–7359. doi: 10.1128/JVI.07217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhammad A., Toufeeq S., Yu H.Z., Wang J., Zhang S.Z., Li B., Li Z., Yang L.A., Hu P., Ma Y., Xu J.P. Molecular Characterization of Two Mitogen-Activated Protein Kinases: p38 MAP Kinase and Ribosomal S6 Kinase From Bombyx mori (Lepidoptera: Bombycidae), and Insight Into Their Roles in Response to BmNPV Infection. J. Insect. Sci. 2019;19 doi: 10.1093/jisesa/iey134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trinh H.V., Grossmann J., Gehrig P., Roschitzki B., Schlapbach R., Greber U.F., Hemmi S. iTRAQ-based and label-free proteomics approaches for studies of human adenovirus infections. Int. J. Proteomics. 2013;2013:1–16. doi: 10.1155/2013/581862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latosinska A., Vougas K., Makridakis M., Klein J., Mullen W., Abbas M., Stravodimos K., Katafigiotis I., Merseburger A.S., Zoidakis J. Comparative analysis of label-free and 8-Plex iTRAQ approach for quantitative tissue proteomic analysis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian Q. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Rawlings N.D., Barrett A.J. Evolutionary families of peptidases. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Z., Lopez D.L., Kanost M.R., Evans J.D., Jiang H. Comparative analysis of serine protease-related genes in the honey bee genome: possible involvement in embryonic development and innate immunity. Insect Mol. Biol. 2010;15:603–614. doi: 10.1111/j.1365-2583.2006.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan A., Giri A.P., Gupta V.S. Structural and functional diversities in lepidopteran serine proteases. Cell. Mol. Biol. Lett. 2006;11:132–154. doi: 10.2478/s11658-006-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G., Zhou Q., Qiu L., Yao Q., Chen K., Tang Q., Hu Z. Serine protease Bm-SP142 was differentially expressed in resistant and susceptible Bombyx mori strains, involving in the defence response to viral infection. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y., Wang X., Du C., Gao J., Xu J. Expression analysis of several antiviral related genes to BmNPV in different resistant strains of silkworm, Bombyx mori. J. Insect Sci. 2014;14:76. doi: 10.1093/jis/14.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaeger K.E., Reetz M.T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Li J., Zhao X., Qian C., Wei G., Zhu B., Liu C. Expression and characterization of a lipase-related protein in the malpighian tubules of the Chinese oak silkworm, Antheraea pernyi. B. Entomol. Res. 2016;106:615–623. doi: 10.1017/S0007485316000365. [DOI] [PubMed] [Google Scholar]
- 44.Jiang L., Wang G., Cheng T., Yang Q., Jin S., Lu G., Wu F., Xiao Y., Xu H., Xia Q. Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch. Virol. 2012;157:1323–1328. doi: 10.1007/s00705-012-1309-8. [DOI] [PubMed] [Google Scholar]
- 45.Budatha M., Meur G., Dutta-Gupta A. A novel aminopeptidase in the fat body of the moth Achaea janata as a receptor for Bacillus thuringiensis cry toxins and its comparison with midgut aminopeptidase. Biochem. J. 2007;405:287–297. doi: 10.1042/BJ20070054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdullah M.A.F., Valaitis A.P., Dean D.H. Identification of a Bacillus thuringiensis Cry11Ba toxin-binding aminopeptidase from the mosquito, Anopheles quadrimaculatus. BMC Biochem. 2006;7:16. doi: 10.1186/1471-2091-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopal R., Sivakumar S., Neema A., Pawan M., Bhatnagar R.K. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem. 2002;277:46849–46851. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi K., Yaoi K., Nagino Y., Hara H., Kitami M., Atsumi S., Miura N., Sato R. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella – their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002;519:215–220. doi: 10.1016/s0014-5793(02)02708-4. [DOI] [PubMed] [Google Scholar]
- 49.Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linz L.B., Sijun L., Chougule N.P., Bonning B.C. In vitro evidence supports membrane Alanyl Aminopeptidase N as a receptor for a plant virus in the Pea Aphid vector. J.Virol. 2015;89 doi: 10.1128/JVI.01479-15. (01479-01415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li G., Zhou Q., Hu Z., Wang P., Tang Q., Chen K., Yao Q. Determination of the proteins encoded by BmBDV VD1-ORF4 and their interacting proteins in BmBDV-infected Midguts. Curr. Microbiol. 2015;70:623–629. doi: 10.1007/s00284-014-0765-7. [DOI] [PubMed] [Google Scholar]
- 53.Jakubowska A.K., Silvia C., Gordon K.H., Juan F., Salvador H. Downregulation of a chitin deacetylase-like protein in response to baculovirus infection and its application for improving baculovirus infectivity. J. Virol. 2010;84:2547–2555. doi: 10.1128/JVI.01860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sridhara S., Bhat J.V. Alkaline and acid phosphatases of the silkworm, Bombyx mori. L. J. Insect. Physiol. 1963;9:693–701. [Google Scholar]
- 55.Lee J., Kim J., Son K., Min J.Y. Acid phosphatase 2 (ACP2) is required for membrane fusion during influenza virus entry. Sci Rep-UK. 2017;7:43893. doi: 10.1038/srep43893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matindoost L., Sendi J.J., Soleimanjahi H., Etebari K., Rahbarizade F. The effects of BmNPV on biochemical changes in primary cultures of Bombyx mori embryonic tissue. In. Vitro. Cell. Dev. Biol. Anim. 2008;44:121–127. doi: 10.1007/s11626-008-9083-3. [DOI] [PubMed] [Google Scholar]
- 57.Crava C.M., Jakubowska A.K., Escriche B., Herrero S., Bel Y. Dissimilar regulation of antimicrobial proteins in the midgut of Spodoptera exigua larvae challenged with Bacillus thuringiensis toxins or Baculovirus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerenius L., Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol. Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 59.Tong Y., Jiang H., Kanost M.R. Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J. Biol. Chem. 2005;280:14932–14942. doi: 10.1074/jbc.M500532200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Yu H.Z., Ye C.J., Ma Y., Li X., Fan T., Chen F.S., Xu J. Bombyx mori Serpin6 regulates prophenoloxidase activity and the expression of antimicrobial proteins. Gene. 2017;610:64–70. doi: 10.1016/j.gene.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Wang L., Liu H., Fu H., Zhang L., Guo P., Xia Q., Zhao P. Silkworm serpin32 functions as a negative-regulator in prophenoloxidase activation. Dev. Comp. Immunol. 2019;91:123–131. doi: 10.1016/j.dci.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Ye P., Chuan X.H., Peng L., Ping C.K., Qin Y., Qin C.H., Lu G., Qing H.Y., Lin W. Molecular cloning, expression and characterization of Bmserpin-2 gene from Bombyx mori. Acta Biochim. Pol. 2009;56:671–677. [PubMed] [Google Scholar]
- 63.Yuan C., Xing L., Wang M., Wang X., Yin M., Wang Q., Hu Z., Zou Z. Inhibition of melanization by serpin-5 and serpin-9 promotes baculovirus infection in cotton bollworm Helicoverpa armigera. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Wet B.J., Gordon S. 2002. Pattern Recognition Receptor; pp. 1118–1127. [Google Scholar]
- 65.Bao Y.Y., Tang X.D., Lv Z.Y., Wang X.Y., Tian C.H., Xu Y.P., Zhang C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics. 2009;94:138–145. doi: 10.1016/j.ygeno.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 66.Hayashiya K. Red fluorescent protein in the digestive juice of the silkworm larvae fed on host-plant mulberry leaves. Entomol. Ntomol. Exp. Appl. 2011;24:428–436. [Google Scholar]
- 67.Wang P., Granados R.R. Observations on the presence of the peritrophic membrane in larval Trichoplusia ni and its role in limiting baculovirus infection. J. Invertebr. Pathol. 1998;72:57–62. doi: 10.1006/jipa.1998.4759. [DOI] [PubMed] [Google Scholar]
- 68.Derksen A.C.G., Granados R.R. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virrology. 1988;167:242–250. doi: 10.1016/0042-6822(88)90074-8. [DOI] [PubMed] [Google Scholar]
- 69.Wang P., Granados R.R. Molecular cloning and sequencing of a novel invertebrate intestinal mucin cDNA. J. Biol. Chem. 1997;272:16663–16669. doi: 10.1074/jbc.272.26.16663. [DOI] [PubMed] [Google Scholar]
- 70.Wang P., Granados R.R. An intestinal mucin is the target substrate for a baculovirus enhancin. P. Natl. Acad. Sci. USA. 1997;94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Classification of total protein in digestive juices of silkworm midgut.
Primers used in RT-qPCR for validation of DEPs
Summary of digestive juice proteins identified by a Label free technology
Summary of differentially expressed proteins (DEPs)