Highlights
-
•
Influenza virus replication depends on activation by airway proteases of the host.
-
•
One prominent enzyme, TMPRSS2, also activates some other respiratory viruses.
-
•
Inhibitors of these trypsin-like proteases display anti-influenza activity.
-
•
These agents can be either peptidomimetic active-site or exosite inhibitors.
-
•
This new drug concept could be broadly used against several respiratory viruses.
Abstract
To enter into airway epithelial cells, influenza, parainfluenza- and coronaviruses rely on host cell proteases for activation of the viral protein involved in membrane fusion. One protease, transmembrane protease serine 2 (TMPRSS2) was recently proven to be crucial for hemagglutinin cleavage of some human influenza viruses. Since the catalytic sites of the diverse serine proteases linked to influenza, parainfluenza- and coronavirus activation are structurally similar, active site inhibitors of these airway proteases could have broad therapeutic applicability against multiple respiratory viruses. Alternatively, superior selectivity could be achieved with allosteric inhibitors of TMPRSS2 or another critical protease. Though still in its infancy, airway protease inhibition represents an attractive host-cell targeting approach to combat respiratory viruses such as influenza.
Current Opinion in Virology 2017, 24:16–24
This review comes from a themed issue on Antiviral strategies
Edited by Lieve Naesens and Fabien Zoulim
For a complete overview see the Issue and the Editorial
Available online 14th April 2017
http://dx.doi.org/10.1016/j.coviro.2017.03.018
1879-6257/© 2017 The Authors. Published by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
On a global scale, seasonal influenza A and B viruses cause around 3–5 million cases of severe illness each year resulting in about 250 000–500 000 deaths [1]. The existing drugs directing the influenza neuraminidase (oseltamivir, zanamivir, peramivir and laninamivir) or M2 proton channel (amantadine and rimantadine), show rather modest clinical efficacy and established or potential resistance. Besides, pandemic variants may at any time emerge due to gene reassortment between human and zoonotic influenza viruses and rapid virus spread in a globalized society. Novel medications with an inventive mode of action are urgently needed to treat or prevent severe influenza infections in vulnerable populations [2]. A recent WHO BRaVe (Research needs for the Battle against Respiratory Viruses) [3] report calls for host cell targeting antiviral approaches, since these are anticipated to exhibit a higher barrier for drug resistance. We here focus on a new paradigm to combat influenza or other respiratory virus infections, which is directed towards airway proteases with an essential role in virus activation. Complementary to our concise analysis, some detailed reviews were recently published elsewhere [4, 5•, 6, 7, 8].
The influenza hemagglutinin as antiviral target
The influenza virus hemagglutinin (HA) mediates two events in virus entry: (i) the HA globular head binds to sialylated cell surface glycans resulting in virus endocytosis; and (ii) the HA stem refolds at endosomal pH to liberate the fusion peptide, causing fusion of the viral and endosomal membranes and release of the viral genome into the host cell cytoplasm. Hence, HA targeting entry blockers may (i) interact with the HA receptor binding site to inhibit virus attachment; or (ii) bind to the HA stem region and inhibit fusion, for instance by preventing its refolding at acidic pH. Design of broad influenza HA blockers is anything but easy, given the high sequence variability among HA subtypes; antigenic drift of HA; different receptor usage of avian versus human influenza viruses; and multivalent nature of the HA-receptor interaction. For instance, small molecule influenza fusion inhibitors appear inadequate since they generally show subtype specificity and rapid emergence of resistance [9].
An alternative and indirect strategy is to prevent the proteolytic activation of HA by inhibiting the host cell proteases responsible thereof. In influenza virus-infected cells, HA is synthesized as the precursor protein HA0 (∼75 kDa), which assembles into a noncovalently linked homotrimer. To obtain fusion capacity, HA0 needs to be cleaved, by a cellular protease, into HA1 (∼50 kDa) and HA2 (∼25 kDa). This enables insertion of the fusion peptide (now located at the N-terminus of HA2) into a negatively charged cavity, priming the HA for low pH-dependent fusion [7].
Highly pathogenic avian influenza A viruses (HPAIVs) of subtypes H5 and H7 possess a multibasic cleavage site that is cleaved by ubiquitously expressed proteases, explaining the systemic infections of these viruses in avian species [7]. The multibasic cleavage site R-X-K/R-R, with Arg at position P4, is processed in the trans-Golgi network by furin and proprotein convertase (PC) 5/6 [7], calcium-dependent serine proteases of the subtilisin superfamily which have a different fold yet the same catalytic triad (Asp–His–Ser) as trypsin-like enzymes [10]. For HPAIVs possessing a K-K/R-X-R motif, the P4-Lys significantly suppresses cleavage efficiency by furin and PC 5/6. These HAs are activated by type II transmembrane serine proteases (TTSPs), more specifically mosaic serine protease large-form (MSPL) or its splice variant TMPRSS13 [11].
In contrast, the HA0 proteins of human influenza A and B viruses possess a cleavage loop with a monobasic (i.e. single arginine) cleavage site. This feature is also present in pandemic influenza A viruses, whether of H1N1 (including the viruses of 1918 and 2009), H2N2 or H3N2 subtype. Inhibition of the HA-cleaving host cell proteases induces production of non-infectious virions, thus halting further virus replication. This type of inhibitors could have two major strengths: (i) resistance is less likely to develop since a host protein is targeted instead of a highly mutable viral protein; and (ii) broad activity against diverse respiratory viruses may be envisaged since, besides influenza virus, several other respiratory viruses rely on activation by similar proteases (see below). Hence, these drugs might also be used to tackle outbreaks by respiratory viruses for which no specific antiviral drugs are available.
Host proteases involved in HA0 cleavage activation
As summarized in Table 1 , diverse trypsin-like proteases (TLPs) are able to activate human influenza viruses in vitro or in vivo. During virus propagation in chicken eggs, HA0 cleavage is performed by blood clotting factor Xa in the allantoic fluid [12], while in Madin Darby canine kidney (MDCK) cell cultures, addition of trypsin is needed for multicycle replication [13]. As for HA activation in the airways of infected persons, several TLP candidate enzymes have been identified, but the picture is still far from complete. A role is now well established for transmembrane protease serine 2 (TMPRSS2), a member of the TTSP family, which are integral membrane proteins with an extracellular C-terminal serine protease domain and an N-terminal cytoplasmic domain. TMPRSS2-knockout (KO) mice were found to survive infection with an H1N1 or H7N9 influenza virus that was lethal in wild-type (WT) mice [14••, 15, 16]. Furthermore, a genome wide association study showed that patients with higher TMPRSS2 expression levels had higher risk for severe infections by the 2009 pandemic H1N1 virus and increased susceptibility to influenza H7N9 [17••]. For the other circulating human influenza A subtype, H3N2, the protease profile is less well defined and appears to be strain-dependent. Compared to WT, TMPRSS2-KO mice proved equally [15] or slightly less vulnerable [14••] to lethal H3N2 infection. A third H3N2 strain was avirulent in TMPRSS2-KO mice but became lethal after ten passages in mice [16, 18]. Since the passaged virus carried an N-glycosylation mutation at the bottom of the HA stalk region, the loss of this glycan may alter the accessibility of the cleavage loop and provide access to an alternative host protease [18].
Table 1.
Experimental data on the role of airway proteases in the activation of influenza-, corona- and paramyxoviruses
| Protease | Virus | In vitro evidence | In vivo evidence |
|---|---|---|---|
| TMPRSS2 | Influenza A and B virus | The HA0s from H1N1, H2N2, H3N2 or influenza B are cleaved in cells overexpressing TMPRSS2 enabling trypsin-independent replication [25••, 36, 66]. Knockdown of TMPRSS2 in Calu-3 cells inhibits cleavage of H1N1 [67] | TMPRSS2-KO mice survive lethal infection with influenza H1N1 or H7N9 [15, 16]. In contrast, influenza B virus is lethal in TMPRSS2-KO mice [37] |
| SARS, MERS and 229E coronaviruses | The coronavirus S-protein is cleaved when co-expressed with TMPRSS2 [31, 62•, 64, 68] | ||
| Metapneumovirus and parainfluenzavirus | The viruses efficiently replicate in cells overexpressing TMPRSS2 and their F protein is cleaved [58, 59] | ||
| TMPRSS4 | Influenza A virus | HA0 from H1N1 is cleaved in TMPRSS4-expressing cells. Knockdown of TMPRSS2 and TMPRSS4 reduces H1N1 virus replication [19, 20, 21] | H3N2 replication is significantly reduced but not abolished, in TMPRSS2/TMPRSS4-double KO mice [21] |
| HAT | Influenza A and B virus | The HA0s from H1N1, H2N2 and H3N2 or influenza B are cleaved in cells overexpressing HAT enabling trypsin-independent replication [25••, 36, 54, 66] | This remains to be investigated in the HAT-KO mouse model [26] |
| SARS and MERS coronaviruses | The coronavirus S-protein is cleaved when co-expressed with HAT [31], hence activating the S protein for cell-cell fusion [69] | ||
| TMPRSS13/MSPL | Influenza A virus; SARS and MERS coronaviruses | The HA0s from subtypes H1, H2 or H3; and HPAIV H5 and H7 with KKKR motif (not recognized by furin), and S-proteins from SARS or MERS coronavirus, are cleaved when co-expressed with MSPL or its splice variant TMPRSS13 [11, 31] | This remains to be investigated in MSPL-KO mice, which display abnormal skin development [70] |
| Matriptase | Influenza A virus | Knockdown of matriptase in Calu-3 cells leads to reduced growth of H1N1 virus [28] Overexpressed HA0 from subtypes H1 or H9 (with RSSR/RSRR motif) is cleaved when cells are exposed to recombinant matriptase [29, 30] |
These studies are hindered by the fact that matriptase-KO mice are unviable [32] |
| DESC1 | Influenza A virus; SARS and MERS coronaviruses | The influenza HA0s subtypes H1, H2 and H3, and S-proteins from SARS or MERS coronavirus are cleaved when co-expressed with DESC1 [29] | |
| KLK5, KLK12 | Influenza A virus | Exogenous KLK5 cleaves overexpressed HA0 from subtypes H1 or H3, whereas KLK12 activates subtypes H1 and H2 [35] | |
| Plasmin | Influenza A/H1N1, strain A/WSN/33 | A/WSN/33 neuraminidase recruits plasminogen, which after conversion into plasmin leads to HA0 cleavage [34] | |
| Soluble serine proteases, for example tryptase Clara | Influenza A virus | Lungs from rats or pigs contain diverse trypsin-like proteases which, when isolated, cleave the HA0 protein of influenza H3N2. The relevance for infections of humans is uncertain [33, 71, 72, 73] | |
TMPRSS2: transmembrane protease serine 2; TMPRSS4: transmembrane protease serine 4; HAT: human airway trypsin-like protease; TMPRSS13: transmembrane protease serine 13; MSPL: mosaic serine protease large-form; DESC1: differentially expressed in squamous cell carcinoma 1; KLK: kallikrein.
A second TLP candidate is the closely related TMPRSS4 enzyme [19, 20]. When infected with an influenza H3N2 strain, TMPRSS2/TMPRSS4 double-KO mice displayed lower morbidity and mortality, though showing some body weight loss, pathology and processing of HA [21]. Thus, besides TMPRSS2 and TMPRSS4, additional proteases are able to cleave and activate the HA of H3N2 viruses. A possible explanation is provided by a conformational difference seen in available HA0 crystal structures [22, 23]. In H3 HA0, the cleavage loop extends from the protein surface, whereas in H1 HA0, the cleavage site is less exposed. Hence, H3 HA0 may be more accessible to diverse proteolytic enzymes [21]. This might allow for more efficient replication of the H3N2 virus, possibly explaining why the symptoms of H3N2 infections tend to be more severe compared to those of the H1N1 subtype [24].
A role for human airway trypsin-like protease (HAT) was proposed several years ago [25••] but still awaits in vivo verification in the existing and viable HAT-KO mouse model [26]. A systematic analysis of the HA0 cleavage pattern for 16 influenza A HA subtypes, was performed in cell assays using coexpression of HA0 plus TMPRSS2 or HAT protease [27•]. Under these testing conditions, cleavage by TMPRSS2 seemed, overall, more efficient than that by HAT, although marked subtype dependence was seen and neither of the two proteases was able of activating all 16 HA subtypes. Analysis of the cleavage site sequences of these 16 HAs did not point to any amino acid as being an absolute determinant for recognition by TMPRSS2 or HAT [27•].
Besides TMPRSS2, TMPRSS4 and HAT, other members of the TTSP family, namely matriptase [28, 29, 30], DESC1 (differentially expressed in squamous cell carcinoma gene 1) [31], MSPL (mosaic serine protease large-form) and its splice variant TMPRSS13 [11, 31] can mediate cell membrane-associated HA0 cleavage of certain HA subtypes. These investigations were performed in HA0-expressing cells which were exposed to recombinant protease or engineered to overexpress the protease under study. The effect of matriptase was further demonstrated in Calu-3 cells (a cell line derived from human bronchial epithelia), in which matriptase gene knockdown resulted in significant impairment of influenza H1N1 replication [28]. In vivo investigations are hindered by the fact that matriptase-KO mice are non-viable [32].
Additionally, secreted proteases like plasmin [33, 34] and kallikrein (KLK) types 5 and 12 [35] can perform H1 and H3 HA0 cleavage [35]. Although all these proteases are able to activate influenza virus in cell culture, there is no evidence yet confirming their role in the airways of influenza-infected individuals.
In the case of influenza B virus, the HA0 activation mechanism is largely unexplored. In cell culture, overexpression of TMPRSS2 or HAT resulted in HA0 cleavage and multicycle replication of influenza B virus [36]. This disagrees, however, with the in vivo finding that influenza B virus displays efficient HA0 cleavage and full pathogenicity in TMPRSS2-KO mice [37]. Unlike influenza A, influenza B virus does not absolutely require exogenous trypsin to replicate in certain MDCK cell lines, indicating that these cells express an as yet unidentified protease that acts on the HA of influenza B but not (or far less efficiently) on that of influenza A [38, 39].
Inhibition of airway proteases as antiviral strategy
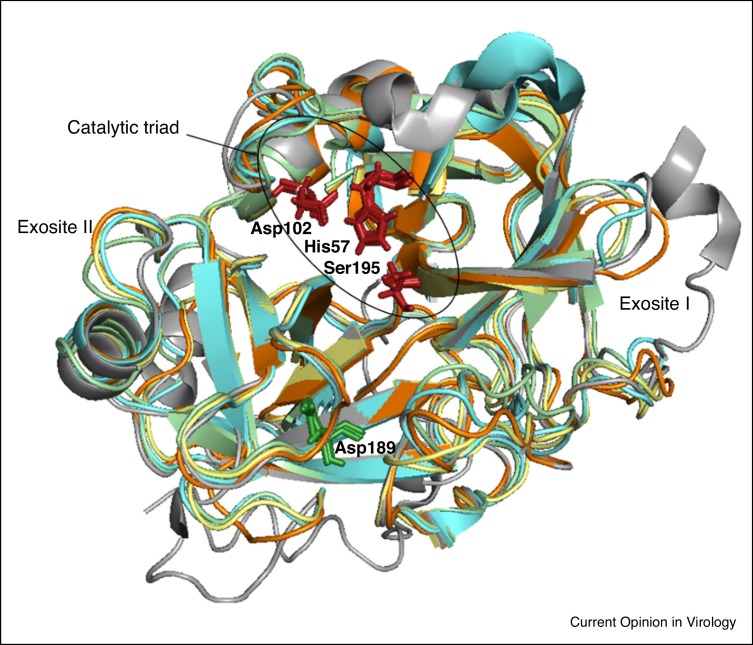
All serine proteases mentioned above are classified as trypsin-like proteases (TLP) since they cleave peptide bonds after a positively charged amino acid (arginine or lysine). This P1 specificity is linked to the presence of a negatively charged Asp residue at the bottom of the S1 specificity pocket (Figure 1 ). As of today, crystal structures are available for matriptase [40], DESC1 [41] and several kallikreins [42], but not for TMPRSS2, TMPRSS4 or HAT. The crystallographic information indicates a high degree of structural homology in the active site regions of the TLPs, which contain the catalytic triad His57, Asp102 and Ser195 (chymotrypsin numbering). Outside this catalytic domain, several exosites exist with higher structural diversity and a critical role in substrate binding and recognition [43]. Consequently, active site targeting inhibitors have a higher chance of possessing a broader activity spectrum, whereas exosite or allosteric binding is preferred to develop highly selective TLP inhibitors.
Figure 1.
Superposition of known TLP crystal structures: trypsin (pdb: 3MFJ, yellow); matriptase (pdb: 1EAX, cyan blue); DESC1 (pdb: 2OQ5, green); KLK5 (pdb: 2PSX, orange); and thrombin (pdb: 3RLW, grey). Residues shown in stick model represent the catalytic triad (in red font) or aspartic residue in the S1 specificity pocket (in green font). Performed with PyMol Molecular Graphics System™. Laporte et al. unpublished.
Proof-of-concept (pre)clinical evidence is already available, since the broad serine protease inhibitors aprotinin and camostat suppress influenza virus replication in cell culture and mouse models, through inhibition of serine proteases involved in HA0 cleavage and inflammation [44, 45]. Moreover, aerosolized aprotinin (an approved therapy in Russia) was shown to shorten symptom duration in humans infected with influenza or parainfluenza virus [44]. However, aprotinin is a protein with unfavorable pharmacokinetics whereas camostat is a covalent binder; both are non-specific protease inhibitors with potential side-effects. To achieve agents with superior potency and safety, specific inhibitors of the relevant airway proteases, particularly TMPRSS2, need to be designed.
The classical type of protease inhibitors are peptidomimetic substrate analogues. TLP inhibitors with high binding affinity are obtained by coupling a P1-Arg mimicking moiety to a peptidyl portion with optimized size and composition, to enable tight fitting in the active site. Among two series of TMPRSS2 inhibitors containing either a 4-amidinobenzylamide or a sulfonylated 3-amidinophenylalanylamide, several analogues displayed K i values below 20 nM in an enzymatic assay with TMPRSS2 protein produced in Escherichia coli [46•]. The best inhibitor (92 in Table 2 ) had a K i value of 0.9 nM and, interestingly, produced clear inhibition of HA0 cleavage and virus replication in Calu-3 cells, which endogenously express HA0-activating proteases like TMPRSS2. Inhibitor 5 (Table 2), which contains 4-amidinobenzylamide at P1, proline at P2 and d-arginine at P3, was also nicely active (K i value of 19 nM) against recombinant HAT enzyme, indicating that inhibitors with dual activity against these two relevant airway proteases can be achieved [47]. Unfortunately, compound 5 also produced strong inhibition of the related TLPs thrombin, plasmin and factor Xa, suggesting that the P2 proline substitution entails reduced selectivity [47].
Table 2.
Described inhibitors for TMPRSS2 and related TLPs
| Name and structure |
Ki (nM) in enzymatic assay |
Antiviral activity in cell culture | References | ||||
|---|---|---|---|---|---|---|---|
| TMPRSS2 | HAT | Matriptase | Thrombin | FXa | |||
| Peptidomimetic inhibitors | |||||||
Inhibitor ‘92’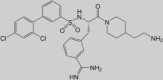
|
0.9 | 1700 | NA | 20 | 50 | Inhibits H1N1 and H3N2 replication in Calu-3 cells | [46•] |
Inhibitor ‘5′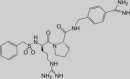
|
20 | 19 | 55 | 3.5 | 2.4 | Inhibits H1N1 and H3N2 replication in HAT-expressing MDCK cells | [47] |
Inhibitor ‘1′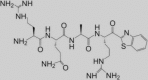
|
NA | 0.0084 | 0.000011 | 0.637 | NA | Inhibits H1N1 replication in Calu-3 cells | [28, 49] |
| Non-peptidomimetic inhibitors | |||||||
Bromhexine
|
0.8 | NA | 59 | >100 | NA | Nonea | [52] |
0591-5329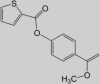
|
0.9 | NA | 75.3 | >100 | NA | Nonea | [52] |
4401-0077
|
2.7 | NA | >100 | >100 | NA | Nonea | [52] |
4554-5138
|
1.4 | NA | 78.3 | >100 | NA | Nonea | [52] |
8008-1235
|
2.6 | NA | >100 | >100 | NA | Nonea | [52] |
NA, not available.
Laporte, unpublished data.
Besides this selectivity issue, the safety of serine protease inhibitors largely depends on whether or not the binding is reversible. The classical inhibitors show covalent binding to the catalytic serine, which is in most cases (pseudo-)irreversible, for example with camostat [48]. Such irreversible protease inhibitors may pose a safety issue. However, the matriptase inhibitor 1 (Table 2) possesses a ketobenzothiazole moiety that forms a covalent yet reversible bond with the catalytic serine [49]. This compound caused a significant reduction of influenza H1N1 replication in Calu-3 cells [28]. Whether this can solely be attributed to inhibition of matriptase is unclear, since the effect of 1 on other relevant proteases like TMPRSS2 was not reported.
An entirely different approach aims at developing exosite binders or allosteric inhibitors [50, 51]. Discovery of these inhibitors requires high-throughput screening (HTS) or fragment-based approaches, unlike the peptidomimetic active site binders which can be rationally designed from known peptide substrates. A biochemical HTS with recombinant TMPRSS2 protein yielded five hit compounds, including bromhexine, which is used as a mucolytic drug [52]. These hits (Table 2) displayed selectivity for TMPRSS2 (i.e. poor to no activity against matriptase, trypsin or thrombin) which, combined with their non-peptidomimetic structure suggests that they could be exosite or allosteric inhibitors. Bromhexine was shown to suppress prostate cancer metastasis in a mouse model, in line with the role of TMPRSS2 overexpression in promoting prostate cancer metastasis [52]. As for influenza, we evaluated all five compounds in virus-infected Calu-3 cells but did not observe any inhibitory effect (Laporte, unpublished data), indicating that hit-to-lead optimization is still required.
Another factor to take into account is the compound’s ability to permeate the cell membrane. For example, it is unclear whether the influenza suppression by the polypeptidic (6 kDa) compound aprotinin occurs extracellularly or, rather, after the compound has been internalized [53]. Influenza HA0 cleavage can occur in the trans-Golgi network during transport of newly synthesized HA0 along the secretory pathway; or at the apical cell membrane when virions are being released from infected cells; or subsequently during entry into a new host cell [5•, 53]. TMPRSS2 is mainly localized in the trans-Golgi network where it cleaves newly synthesized HA0. On the other hand, since HAT mainly resides on the cell membrane with its protease domain directed towards the extracellular space, this protease can cleave HA0 of both released and incoming virus [54].
Potential for safe and broad spectrum antiviral therapy
Despite the growing interest in host cell factors as a target for antiviral intervention [55], possible toxicity is a logical concern. This seems not verified for the TMPRSS2, TMPRSS4 and HAT airway proteases since HAT, TMPRSS2 and TMPRSS4 (single or double)-knockout mice are perfectly healthy [21, 56, 57]. Matriptase however is indispensable and ubiquitously expressed, with a known role in the development of the epidermis, hair follicles, and cellular immune system [32].
One unique benefit of blocking TMPRSS2 and related airway proteases is that, besides influenza virus, several other respiratory viruses could be targeted. TMPRSS2 was shown to activate the fusion proteins of some paramyxoviruses (i.e. metapneumovirus [58], trypsin-dependent parainfluenza subtypes and Sendai virus [59]). Likewise, for some human coronaviruses including the deadly SARS and MERS variants [8, 60, 61, 62•], TMPRSS2 cleavage activates the viral spike (S)-protein at the cell surface enabling cathepsin-independent host cell entry [63, 64]. The S-proteins of the MERS and SARS coronaviruses were also cleaved when co-expressed with DESC1 or MSPL protein [31]. There is also in vivo evidence since camostat proved effective in protecting mice against a lethal infection by SARS coronavirus [65]. Since an antiviral medication for these paramyxoviruses and coronaviruses is currently lacking, broader-acting airway protease inhibitors could be well suited to fill this gap.
Finally, airway protease inhibitors could be combined with more conventional antiviral drugs to obtain a synergistic effect or reduce the risk of resistance. For instance, the combination of oseltamivir with the serine protease inhibitor BAPA (benzylsulfonyl-d-Arg-Pro-4-amidinobenzylamide) was found to suppress influenza virus replication in human airway epithelial cells at remarkably lower concentrations than treatment with each inhibitor alone [36].
Conclusion
Trypsin-like proteases of the human airways represent unique targets to suppress infections with influenza or other respiratory viruses which rely on these enzymes for their replication. The significance of TMPRSS2 is supported by recent proof, from mouse or clinical studies, that this protease is essential for replication of influenza H1N1 and H7N9 viruses. TMPRSS2 protease inhibitors may act by two different mechanisms: strong yet, preferably, reversible binding to the active site; or interference with an exosite or allosteric domain. An advantage of active site binders would be the potential to suppress not only TMPRSS2, but also other relevant airway proteases including TMPRSS4 and HAT. The design of selective inhibitors would be facilitated by a better insight into the protein structures of these proteases. More research is also needed to precisely define the protease cleavage profile of human A/H3N2 and, particularly, influenza B viruses. Whether used as mono- or combination therapy, these airway protease inhibitors could represent a unique class of host cell-targeting drugs with a broad spectrum of antiviral activity.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
The authors would like to thank all the members of their research team for dedicated assistance.
References
- 1.WHO Influenza (seasonal) fact sheet. Bull. World Health Org. 2016;211:2–5. [Google Scholar]
- 2.Naesens L., Stevaert A., Vanderlinden E. Antiviral therapies on the horizon for influenza. Curr. Opin. Pharmacol. 2016;30:106–115. doi: 10.1016/j.coph.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 3.WHO . WHO; 2013. Research Needs for the Battle against Respiratory Viruses (BRaVe) [Google Scholar]
- 4.Böttcher-Friebertshäuser E., Klenk H.-D., Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog. Dis. 2013;69:87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Garten W., Braden C., Arendt A., Peitsch C., Baron J., Lu Y., Pawletko K., Hardes K., Steinmetzer T., Böttcher-Friebertshäuser E. Influenza virus activating host proteases: identification, localization and inhibitors as potential therapeutics. Eur. J. Cell Biol. 2015;94:375–383. doi: 10.1016/j.ejcb.2015.05.013. [DOI] [PubMed] [Google Scholar]; Comprehensive review on influenza-activating host proteases, describing in detail the biochemical, cell biological as well as antiviral aspects.
- 6.Bertram S., Glowacka I., Steffen I., Kühl A., Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010;20:298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher-Friebertshäuser E., Garten W., Matrosovich M., Klenk H.D. The hemagglutinin: a determinant of pathogenicity. Curr. Top. Microbiol. Immunol. 2014:3–34. doi: 10.1007/82_2014_384. [DOI] [PubMed] [Google Scholar]
- 8.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderlinden E., Naesens L. Emerging antiviral strategies to interfere with influenza virus entry. Med. Res. Rev. 2014;34:301–339. doi: 10.1002/med.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cera E. Serine proteases. Int. Union Biochem. Mol. Biol. Life. 2009;61:510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okumura Y., Takahashi E., Yano M., Ohuchi M., Daidoji T., Nakaya T., Böttcher E., Garten W., Klenk H.-D., Kido H. Novel type II transmembrane serine proteases, MSPL and TMPRSS13, proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J. Virol. 2010;84:5089–5096. doi: 10.1128/JVI.02605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh B., Ogasawara T., Toyoda T., Inocencio N.M., Hamaguchi M., Nagai Y. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J. 1990;9:4189–4195. doi: 10.1002/j.1460-2075.1990.tb07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenk H.D., Rott R., Orlich M., Blödorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 14••.Hatesuer B., Bertram S., Mehnert N., Bahgat M.M., Nelson P.S., Pöhlman S., Schughart K. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9:e1003774. doi: 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first study to show that TMPRSS2 knock-out mice are protected against severe pathology and death after H1N1 influenza infection.
- 15.Tarnow C., Engels G., Arendt A., Schwalm F., Sediri H., Preuss A., Nelson P.S., Garten W., Klenk H.-D., Gabriel G. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J. Virol. 2014;88:4744–4751. doi: 10.1128/JVI.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai K., Ami Y., Tahara M., Kubota T., Anraku M., Abe M., Nakajima N., Sekizuka T., Shirato K., Suzaki Y. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J. Virol. 2014;88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Cheng Z., Zhou J., To K.K.-W., Chu H., Li C., Wang D., Yang D., Zheng S., Hao K., Bossé Y. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J. Infect. Dis. 2015;212:1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]; Clinical study confirming an important role for TMPRSS2 in influenza replication. A single-nucleotide polymorphism leading to higher expression levels of TMPRSS2 in the lungs, was found to predispose to more severe illness from 2009 pandemic H1N1 or H7N9 infections.
- 18.Sakai K., Sekizuka T., Ami Y., Nakajima N., Kitazawa M., Sato Y., Nakajima K., Anraku M., Kubota T., Komase K. A mutant H3N2 influenza virus uses an alternative activation mechanism in TMPRSS2 knockout mice by loss of an oligosaccharide in the hemagglutinin stalk region. J. Virol. 2015;89:5154–5158. doi: 10.1128/JVI.00124-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.-Y., Park Y., Schneider H. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaipan C., Kobasa D., Bertram S., Glowacka I., Steffen I., Tsegaye T.S., Takeda M., Bugge T.H., Kim S., Park Y. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kühn N., Bergmann S., Kasnitz N., Lambertz R.L.O., Keppner A., van den Brand J.M.A., Pöhlmann S., Weiß S., Hummler E., Hatesuer B. The proteolytic activation of A (H3N2) influenza virus hemagglutinin is facilitated by different type II transmembrane serine proteases. J. Virol. 2016;90:4298–4307. doi: 10.1128/JVI.02693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C., Hill M., Nw L. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 23.Stevens J., Corper A.L., Basler C.F., Taubenberger J.K., Palese P., Wilson I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 24.Hayward A.C., Fragaszy E.B., Bermingham A., Wang L., Copas A., Edmunds W.J., Ferguson N., Goonetilleke N., Harvey G., Kovar J. Comparative community burden and severity of seasonal and pandemic influenza: results of the Flu Watch cohort study. Lancet Respir. Med. 2014;2:445–454. doi: 10.1016/S2213-2600(14)70034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Böttcher E., Matrosovich T., Beyerle M., Klenk H.-D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report on the importance of the airway proteases TMPRSS2 and HAT for cleavage of influenza HA0. In cells expressing any of these two these enzymes, HA0 is processed and influenza undergoes multicycle replication in the absence of trypsin.
- 26.Sales K.U., Hobson J.P., Wagenaar-Miller R., Szabo R., Rasmussen A.L., Bey A., Shah M.F., Molinolo A.A., Bugge T.H. Expression and genetic loss of function analysis of the HAT/DESC cluster proteases TMPRSS11A and HAT. PLoS One. 2011;6:e23261. doi: 10.1371/journal.pone.0023261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Galloway S.E., Reed M.L., Russell C.J., Steinhauer D.A. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013;9:e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]; Broad cell-based analysis of the proteolytic activation and membrane fusion characteristics of the 16 major influenza A HA subtypes. The authors observed marked subtype-related differences in susceptibility to trypsin, TMPRSS2 and HAT.
- 28.Beaulieu A., Gravel É., Cloutier A., Marois I., Colombo É., Désilets A., Verreault C., Leduc R., Marsault É., Richter M.V. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 2013;87:4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron J., Tarnow C., Mayoli-Nüssle D., Schilling E., Meyer D., Hammami M., Schwalm F., Steinmetzer T., Guan Y., Garten W. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J. Virol. 2013;87:1811–1820. doi: 10.1128/JVI.02320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton B.S., Gludish D.W.J., Whittaker G.R. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J. Virol. 2012;86:10579–10586. doi: 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zmora P., Blazejewska P., Moldenhauer A.-S., Welsch K., Nehlmeier I., Wu Q., Schneider H., Pöhlmann S., Bertram S. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J. Virol. 2014;88:12087–12097. doi: 10.1128/JVI.01427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.List K., Haudenschild C.C., Szabo R., Chen W., Wahl S.M., Swaim W., Engelholm L.H., Behrendt N., Bugge T.H. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 2002;21:3765–3779. doi: 10.1038/sj.onc.1205502. [DOI] [PubMed] [Google Scholar]
- 33.Murakami M., Towatari T., Ohuchi M., Shiota M., Akao M., Okumura Y., Parry M.A.A., Kido H. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 2001;268:2847–2855. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- 34.Li S., Schulman J.L., Itamura S., Palese P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J. Virol. 1993;67:6667–6673. doi: 10.1128/jvi.67.11.6667-6673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton B.S., Whittaker G.R. Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J. Biol. Chem. 2013;288:17399–17407. doi: 10.1074/jbc.M112.440362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Böttcher-Friebertshäuser E., Lu Y., Meyer D., Sielaff F., Steinmetzer T., Klenk H.-D., Garten W. Hemagglutinin activating host cell proteases provide promising drug targets for the treatment of influenza A and B virus infections. Vaccine. 2012;30:7374–7380. doi: 10.1016/j.vaccine.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Sakai K., Ami Y., Nakajima N., Nakajima K., Kitazawa M., Anraku M., Takayama I., Sangsriratanakul N., Komura M., Sato Y. TMPRSS2 independency for haemagglutinin cleavage in vivo differentiates influenza B virus from influenza A virus. Sci. Rep. 2016;6:29430. doi: 10.1038/srep29430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noma K., Kiyotani K., Kouchi H., Fujii Y., Egi Y., Tanaka K., Yoshida T. Endogenous protease-dependent replication of human influenza viruses in two MDCK cell lines. Arch. Virol. 1998;143:1893–1909. doi: 10.1007/s007050050428. [DOI] [PubMed] [Google Scholar]
- 39.Lugovtsev V.Y., Melnyk D., Weir J.P. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS One. 2013;8:1–16. doi: 10.1371/journal.pone.0075014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrich R., Fuentes-Prior P., Ong E., Coombs G., Hunter M., Oehler R., Pierson D., Gonzalez R., Huber R., Bode W. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J. Biol. Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- 41.Kyrieleis O.J.P., Huber R., Ong E., Oehler R., Hunter M., Madison E.L., Jacob U. Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family. FEBS J. 2007;274:2148–2160. doi: 10.1111/j.1742-4658.2007.05756.x. [DOI] [PubMed] [Google Scholar]
- 42.Debela M., Beaufort N., Magdolen V., Schechter N.M., Craik C.S., Schmitt M., Bode W., Goettig P. Structures and specificity of the human kallikrein-related peptidases KLK 4, 5, 6, and 7. Biol. Chem. 2008;389:623–632. doi: 10.1515/BC.2008.075. [DOI] [PubMed] [Google Scholar]
- 43.Barré O., Dufour A., Eckhard U., Kappelhoff R., Béliveau F., Leduc R., Overall C.M. Cleavage specificity analysis of six type II transmembrane serine proteases (TTSPs) using PICS with proteome-derived peptide libraries. PLoS One. 2014;9:e105984. doi: 10.1371/journal.pone.0105984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhirnov O.P., Klenk H.D., Wright P.F. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011;92:27–36. doi: 10.1016/j.antiviral.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Lee M.G., Kim K.H., Park K.Y., Kim J.S. Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Arch. Virol. 1996;141:1979–1989. doi: 10.1007/BF01718208. [DOI] [PubMed] [Google Scholar]
- 46•.Meyer D., Sielaff F., Hammami M., Böttcher-Friebertshäuser E., Garten W., Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem. J. 2013;452:331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]; Synthetic inhibitors of TMPRSS2 were rationally designed using recombinant TMPRSS2 enzyme. The authors also performed a detailed enzyme kinetic analysis of TMPRSS2 and determined its optimal peptide substrate.
- 47.Sielaff F., Böttcher-Friebertshäuser E., Meyer D., Saupe S.M., Volk I.M., Garten W., Steinmetzer T. Development of substrate analogue inhibitors for the human airway trypsin-like protease HAT. Bioorg. Med. Chem. Lett. 2011;21:4860–4864. doi: 10.1016/j.bmcl.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 48.Spraggon G., Hornsby M., Shipway A., Tully D.C., Bursulaya B., Danahay H., Harris J.L., Lesley S.A. Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations. Protein Sci. 2009;18:1081–1094. doi: 10.1002/pro.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colombo E., Désilets A., Duchêne D., Chagnon F., Najmanovich R., Leduc R., Marsault E. Design and synthesis of potent, selective inhibitors of matriptase. ACS Med. Chem. Lett. 2012;3:530–534. doi: 10.1021/ml3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hauske P., Ottmann C., Meltzer M., Ehrmann M., Kaiser M. Allosteric regulation of proteases. ChemBioChem. 2008;9:2920–2928. doi: 10.1002/cbic.200800528. [DOI] [PubMed] [Google Scholar]
- 51.Gohara D.W., Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011;29:577–585. doi: 10.1016/j.tibtech.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., Morrissey C., Corey E., Montgomery B., Mostaghel E. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhirnov O.P., Ikizler M.R., Wright P.F. Cleavage of influenza A virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J. Virol. 2002;76:8682–8689. doi: 10.1128/JVI.76.17.8682-8689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Böttcher-Friebertshäuser E., Freuer C., Sielaff F., Schmidt S., Eickmann M., Uhlendorff J., Steinmetzer T., Klenk H.-D., Garten W. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 2010;84:5605–5614. doi: 10.1128/JVI.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T., Watanabe S., Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sales K.U., Hobson J.P., Wagenaar-Miller R., Szabo R., Rasmussen A.L., Bey A., Shah M.F., Molinolo A.A., Bugge T.H. Expression and genetic loss of function analysis of the HAT/DESC cluster proteases TMPRSS11A and HAT. PLoS One. 2011;6:e23261. doi: 10.1371/journal.pone.0023261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirogane Y., Takeda M., Iwasaki M., Ishiguro N., Takeuchi H., Nakatsu Y., Tahara M., Kikuta H., Yanagi Y. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 2008;82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe M., Tahara M., Sakai K., Yamaguchi H., Kanou K., Shirato K., Kawase M., Noda M., Kimura H., Matsuyama S. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J. Virol. 2013;87:11930–11935. doi: 10.1128/JVI.01490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the SARS-coronavirus spike-protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62•.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that TMPRSS2 is involved in the activation of SARS coronavirus.
- 63.Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2016;91:e01387–e01416. doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Böttcher E., Freuer C., Steinmetzer T., Klenk H.-D., Garten W. MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine. 2009;27:6324–6329. doi: 10.1016/j.vaccine.2009.03.029. [DOI] [PubMed] [Google Scholar]
- 67.Böttcher-Friebertshäuser E., Stein D.A., Klenk H.-D., Garten W. Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J. Virol. 2011;85:1554–1562. doi: 10.1128/JVI.01294-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pöhlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Madsen D.H., Szabo R., Molinolo A.A., Bugge T.H. TMPRSS13 deficiency impairs stratum corneum formation and epidermal barrier acquisition. Biochem. J. 2014;461:487–495. doi: 10.1042/BJ20140337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kido H., Yokogoshi Y., Sakai K., Tashiro M., Kishino Y., Fukutomi A., Katunuma N. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 1992;267:13573–13579. [PubMed] [Google Scholar]
- 72.Towatari T., Ide M., Ohba K., Chiba Y., Murakami M., Shiota M., Kawachi M., Yamada H., Kido H. Identification of ectopic anionic trypsin I in rat lungs potentiating pneumotropic virus infectivity and increased enzyme level after virus infection. Eur. J. Biochem. 2002;269:2613–2621. doi: 10.1046/j.1432-1033.2002.02937.x. [DOI] [PubMed] [Google Scholar]
- 73.Chen Y., Shiota M., Ohuchi M., Towatari T., Tashiro J., Murakami M., Yano M., Yang B., Kido H. Mast cell tryptase from pig lungs triggers infection by pneumotropic Sendai and influenza A viruses. Purification and characterization. Eur. J. Biochem. 2000;267:3189–3197. doi: 10.1046/j.1432-1327.2000.01346.x. [DOI] [PubMed] [Google Scholar]