Highlights
► (+)RNA viruses use conserved mechanisms to build membranous replication complexes. ► Virtually all (+)RNA viruses actively modify lipid synthesis. ► Secretory pathway components contribute to replication of many (+)RNA viruses. ► Hijacked host factors are integrated into networks not found in uninfected cells. ► Membrane composition of replication organelles differs from pre-existing membranes.
Abstract
Positive-strand RNA [(+)RNA] viruses show a significant degree of conservation of their mechanisms of replication. The universal requirement of (+)RNA viruses for cellular membranes for genome replication, and the formation of membranous replication organelles with similar architecture, suggest that they target essential control mechanisms of membrane metabolism conserved among eukaryotes. Recently, significant progress has been made in understanding the role of key host factors and pathways that are hijacked for the development of replication organelles. In addition, electron tomography studies have shed new light on their ultrastructure. Collectively, these studies reveal an unexpected complexity of the spatial organization of the replication membranes and suggest that (+)RNA viruses actively change cellular membrane composition to build their replication organelles.
Current Opinion in Virology 2012, 2:740–747
This review comes from a themed issue on Virus replication in animals and plants
Edited by Peter Nagy and Christopher Richardson
For a complete overview see the Issue and the Editorial
Available online 1st October 2012
1879-6257/$ – see front matter, © 2012 Elsevier B.V. All rights reserved.
Introduction
(+)RNA viruses infect almost all eukaryotic organisms, often inducing devastating diseases in humans, animals and plants. Despite adaptation to diverse hosts, the replication mechanisms of (+)RNA viruses display remarkable conservation. These viruses universally require cellular membranes for assembly of their replication complexes – containing viral proteins, RNA, and host factors – to amplify their genome. To convert cellular membranes into replication sites, (+)RNA viruses must have evolved efficient ways of manipulating the highly regulated cellular membrane metabolism. The small genome size of these viruses and thus limited repertoire of available resources suggests that they rely on only a few evolutionarily successful strategies shared among different viruses.
Recently, major advances have been made in understanding of virus–cell interaction of several groups of (+)RNA viruses. The pioneering research of picornavirus-induced membrane remodeling has been complemented lately by detailed examinations of the biochemistry and morphology of replication organelles of flaviviruses, alphaviruses and nidoviruses. Many important insights into virus-induced membrane modifications were gained from studies on Flock House virus and Brome Mosaic virus (BMV), which can replicate in yeast, allowing application of yeast genetic tools to elucidate the role of specific host factors. These studies revealed important similarities in the principles of remodeling cellular membranes and organizing replication sites among diverse (+)RNA viruses. The development and functioning of these structures require rewiring of cellular pathways into new configurations that are induced and regulated by viral proteins.
In this review, we will describe new data on the organization and development of replication structures and focus on the emerging concept that viral membranous replication complexes are bona fide new organelles with unique lipid and protein compositions.
Morphology of membranous replication organelles
Recently, 3D electron tomography (ET) was applied to resolve the complex spatial organization of replication membranes of picorna, flavi, corona, arteri and nodaviruses [1•, 2•, 3•, 4•, 5•, 6•]. Conventional EM and ET studies collectively show that different (+)RNA viruses initiate development of their replication structures on different cellular organelles and that the degree of the remodeling of the original cellular membrane architecture varies greatly. However, it appears that there are only a few basic configurations of replication-associated membranes: invaginations, tubular–vesicular networks, double or multimembrane vesicles, and convoluted membranes without identifiable topological units.
Distinct membranous structures may support different steps in the viral life cycle
Different combinations of membranous structures can often be simultaneously observed in one infected cell, or they can develop in a defined succession during the course of infection. It is not always clear whether these structures contribute equally to RNA replication or have other specialized functions in the viral life cycle. For enveloped viruses, distinct membrane structures may be expected to be associated with genome replication and virion maturation. Indeed, genome replication of HCV is associated with a membranous web believed to originate from ER membranes [7], while RNA encapsidation is linked to the surface of lipid droplets [8, 9]. Polyprotein processing, RNA replication and virion maturation each appear to be confined to different membrane compartments in cells infected with arthropod-borne flaviviruses [3•, 10, 11]. Time-dependent evolution of membrane structures, implicated to reflect distinct stages of virus propagation, has been described for coronaviruses [1•, 12], and recently for enteroviruses (i.e. poliovirus and coxsackievirus). Early in infection enteroviruses replicate their RNA on single membrane convoluted tubular clusters that later in infection are converted into double-membrane vesicles (DMV) and complex multimembrane structures [4•, 5•]. Although DMV may contribute to enterovirus RNA replication, they have also been implicated in non-lytic release of newly formed virions [13].
Morphology of replication structures is conserved among large taxa of viruses
(+)RNA viruses can be divided into three superfamilies: picorna-like, alpha-like and flavi-like viruses [14, 15, 16]. Interestingly, flavi-and alpha-like viruses induce formation of discrete membrane invaginations with negative membrane curvature [3•, 17•, 18, 19], while picorna-like viruses rely on positively curved convoluted tubular–vesicular membrane networks [1•, 4•, 5•, 20, 21, 22, 23]. In the invaginations, the replication machinery is located on the inner membrane surface and the replication compartment is connected to the cytoplasm with an opening wide enough to provide a supply of nucleotides and to export synthesized RNA [2•, 3•]. In tubulo-vesicular replication structures, the viral replication proteins are localized on the external membranous surface facing the cytoplasm [24]. This division may reflect an evolutionary divergence between (+)RNA virus groups and suggest that the mechanisms of membrane remodeling may be significantly different between picorna-like and other (+)RNA viruses (Figure 1 ).
Figure 1.
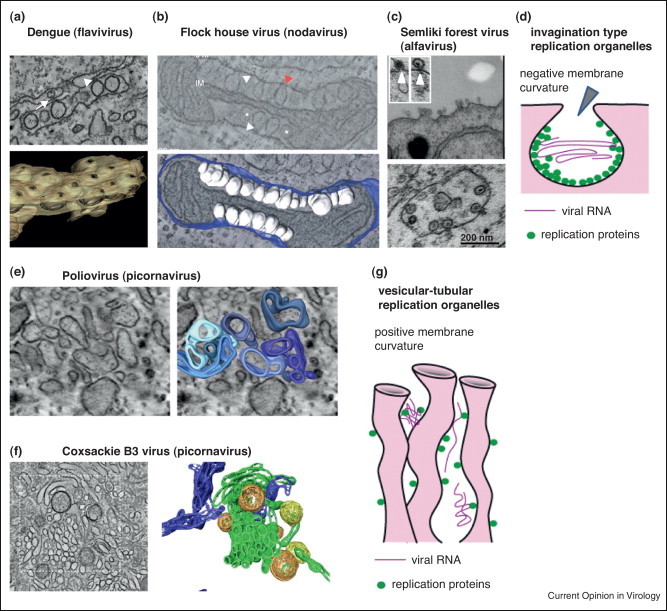
Basic principles of viral replication organelle organization. (a–d) Invagination type of replication organelles with negative membrane curvature typical for flavi-like and alpha-like viruses. Arrowheads show connection of the inner compartment with the cytoplasm. (a) EM image and tomography reconstruction of the replication structures of Dengue virus induced on ER membranes (modified from [3]). (b) EM image and tomography reconstruction of the replication structures of Flock house virus on outer mitochondria membrane (modified from [2]). (c) Spherules induced on the plasma membrane at the early stage of Semliki Forest virus infection are later translocated inside the cytoplasm (modified from [17]). (d) Schematic representation of the invagination–spherule replication organelle organization. (e–g) Vesicular–tubular replication organelles with positive membrane curvature characteristic of picorna-like viruses. (e) EM image and tomography reconstruction of the early replication structures of poliovirus (modified from [4]). (f) EM image and tomography reconstruction of the early replication structures of Coxsackie B3 virus (modified from [5]). (g) Schematic representation of the vesicular–tubular replication organelle organization.
(+)RNA viruses rewire cellular pathways to generate replication organelles
In general, remodeling of cellular membranes requires coordinated lipid sorting and/or actions of specific membrane shaping protein complexes [25, 26]. Here, we will discuss the roles of autophagy, the secretory pathway, and lipid biosynthesis in the development and/or functioning of replication organelles. For the role of protein-dependent membrane-shaping mechanisms, the reader is referred to a recent review [27].
Autophagy
Activation of at least initial steps of the autophagy pathway is well documented in cells infected with diverse (+)RNA viruses. Since a double membrane is a distinctive feature of autophagosomes, it was initially suggested that autophagy-like processes may be responsible for the generation of double membrane replication structures. However conflicting data on the importance of autophagy for replication of coronaviruses [1•, 28, 29, 30, 31] and enteroviruses [32, 33, 34] suggest that DMV are either not essential for replication, or that they bear only superficial resemblance to autophagosomes. Recently, an autophagy-independent role of LC3 in tuning the ER-associated degradation (ERAD) pathway was suggested to be important for coronavirus replication, indicating that viruses may selectively utilize autophagy/ERAD pathway components to generate replication membranes [35, 36]. Although an intact autophagy pathway may not be required for bona fide genome replication of (+)RNA viruses, autophagy may be induced upon infection as it plays an important regulatory role in the type I interferon response [37]. Indeed, activation of autophagy by HCV and Dengue virus has recently been shown to suppress the cellular capacity to activate this antiviral innate response [38, 39].
Secretory pathway
Replication of most (+)RNA viruses is intimately associated with cellular secretory pathway(-derived) membranes (Figure 2 ). Below the roles of some common membrane-remodeling host factors identified to be required for (+)RNA virus replication are discussed.
Figure 2.
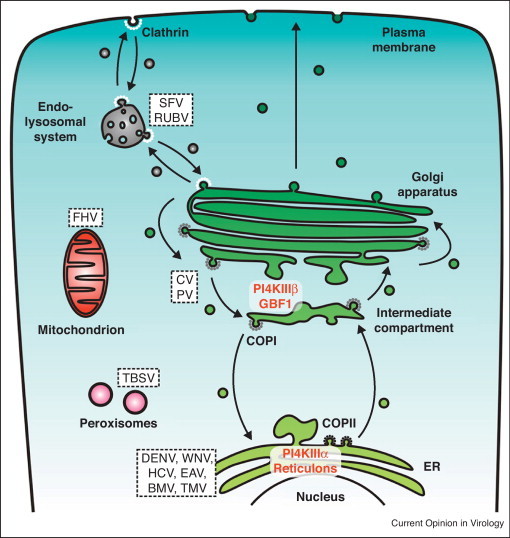
Schematic overview of the secretory pathway and the sites targeted and/or altered by (+)RNA viruses for formation of their membranous replication organelles. For reasons of simplicity, the endocytic pathway and the lysosomes are combined. Secretory pathway transport depends on specific coat complexes that contribute by mediating the bending, deformation and detachment of membranes carriers. Anterograde transport of proteins out of the ER takes place via a COP-II dependent pathway, whereas intra-Golgi transport as well as retrograde transport from the Golgi and intermediated compartment relies on COP-I-coated carries. Endocytosis takes place via clathrin-coated vesicles. The putative sites that are targeted and/or altered by different (+)RNA viruses (Family and Genera names of the viruses are given here below between parenthesis) to form their replication organelles are indicated by their position. Essential host factors and their localization are also indicated. CV, coxsackievirus (Picornaviridae, Enterovirus). DENV, dengue virus (Flaviviridae, Flavivirus). EAV, equine arteritis virus (Arteriviridae, Arterivirus). FHV, flock house virus (Nodaviridae, Alphanodavirus). HCV, hepatitis C virus (Flaviviridae, Hepacivirus). MHV, mouse hepatitis virus (Coronaviridae, Coronavirus). PV, poliovirus (Picornaviridae, Enterovirus). RUBV, rubella virus (Togaviridae, Rubivirus). SARS-CoV, severe acute respiratory syndrome coronavirus (Coronaviridae, Coronavirus). SFV, Semliki Forest virus (Togaviridae, Alphavirus). TBSV, tomato bushy stunt virus (Tombusviridae, Tombusvirus). TMV, tobacco mosaic virus (Virgaviridae, Tobamovirus). WNV, west Nile virus (Flaviviridae, Flavivirus).
GBF1
GBF1, a guanine nucleotide exchange factor for the small GTPase Arf1 and a target of brefeldin A (BFA), coordinates the formation and fusion of transport vesicles in trafficking between ER and early Golgi [40]. GBF1 was recognized as an essential factor for replication of enteroviruses [41, 42, 43], but not other picornaviruses such as cardioviruses and aphthoviruses [44, 45]. GBF1 has also been implicated in replication of HCV and mouse hepatitis coronavirus [46, 47]. Several lines of evidence suggest that the role of GBF1 in viral replication is unrelated to its normal function in transport vesicle formation and Arf1 activation. Expression of poliovirus and HCV proteins in the presence of BFA resulted in the appearance of new membrane structures that are indistinguishable from those observed without the drug, indicating that GBF1 is more probably involved in the function of replication organelles than in their formation [41, 45]. Moreover, while activated Arf1 accumulates on enterovirus replication organelles, BFA-resistant poliovirus mutants can efficiently replicate without active Arf1, and siRNA knockdown of Arf1 does not affect coxsackievirus replication [42, 48•]. Lastly, expression of truncated GBF1 deficient in supporting cellular metabolism rescued poliovirus replication in the presence of BFA [48•].
Phosphatidylinositol-4-kinase (PI4K)
Replication membranes of enteroviruses were shown to become enriched in PI4KIIIβ, but depleted of coat proteins, yielding unique uncoated phosphatidylinositol-4-phosphate (PI4P) lipid-enriched organelles [49•]. It was proposed that recruitment of PI4KIIIβ to replication organelles depends on the interaction of viral protein 3A with GBF1 as well as on the Arf1 activation function of GBF1 [45, 49•]. However, the observations that Arf1 as well as strong 3A–GBF1 interaction may be dispensable for enterovirus replication [42] (G. Belov, F. van Kuppeveld, unpublished data) argue that this model may be an oversimplification of the actual processes (Figure 3 ). Another PI4K, PI4KIIIα, is recruited to replication sites of HCV by viral protein NS5A [50, 51•]. Inhibition of PI4K activity potently blocks replication of enteroviruses and HCV, showing that PI4P is an important component of their replication membranes [49•, 52, 53]. As is the case for GBF1, PI4Ks are not required for the replication of all (+)RNA viruses and even viruses from one family may demonstrate different requirements for PI4Ks [51•, 54] (F. van Kuppeveld, unpublished data). The role of PI4P, the product of PI4K activity, is likely to mediate recruitment and/or activation of viral proteins and host factors on replication organelles (e.g. PI4P-binding cellular proteins such as oxysterol binding proteins and ceramide transport protein), where they may support RNA replication directly or indirectly by remodeling membranes (Figure 3). Poliovirus polymerase 3D has been shown to preferentially bind PI4P in a biochemical assay, but the relevance of this observation for replication in vivo remains to be elaborated [49•]. For HCV, the absence of PI4KIIIα activity induced a dramatic change in the ultrastructural morphology of the membranous replication complex, suggesting a role of PI4P in recruiting specific membrane-shaping proteins [51•].
Figure 3.
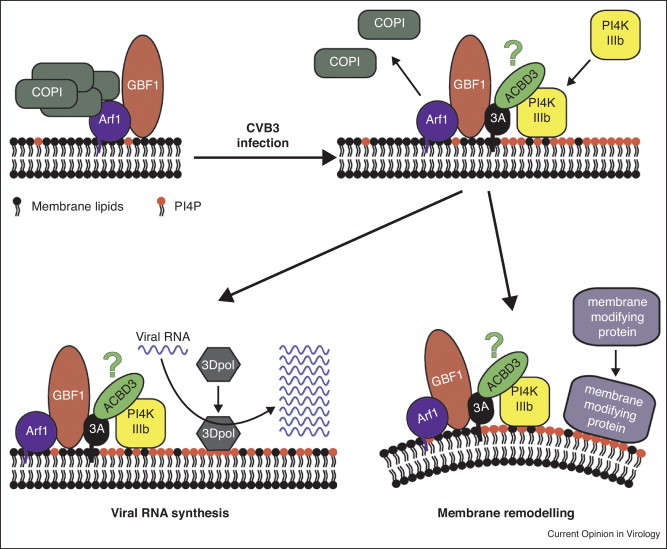
Possible functions of GBF1 and PI4KIIIβ/PI4P in formation and/or activity of enterovirus replication organelles. Enterovirus replication starts at Golgi membranes where the viral 3A protein interacts with GBF1, a GEF for Arf1 that in uninfected cells is involved in recruiting COP-I coats to membranes (left). The interaction of 3A with GBF1 interferes with COP-I recruitment, resulting in uncoated membranes that can no longer function in secretory pathway trafficking. At the same time, 3A causes an increased recruitment of PI4KIIIβ to membranes. Although this lipid kinase is an effector of Arf1 in uninfected cells, growing evidence suggests that the interaction of 3A with GBF1/Arf1 is not essential for the massive recruitment of PI4KIIIβ observed in infected cells. 3A may directly interact with PI4KIIIβ or this interaction may occur via another cellular protein (e.g. ACBD3, which has recently been reported to bind 3A as well as PI4KIIIβ [68, 69]). Enhanced recruitment of PI4KIIIβ results in PI4P enriched membranes, which may serve to activate and/or recruit viral and/or cellular proteins that are directly involved in replication of the viral RNA. One candidate protein is the viral RNA-dependent RNA polymerase, 3D, which has been shown to bind PI4P lipids in vitro. Additionally, PI4P-rich membranes may serve as docking sites for other cellular proteins that exert functions in membrane remodeling that contribute to the formation of the membranous environment suitable for RNA replication.
Taken together, these data suggest that (+)RNA viruses do not rely on a functional secretory pathway, but rather dismantle it and stitch together separate elements in new configurations that serve to support viral genome replication. Despite the critical role of GBF1 and PI4K in replication, enteroviruses can become resistant to inhibitors targeting these factors, suggesting built-in redundancy of essential virus–cell interactions [55, 56]. Other studies have shown that replication of some (+)RNA viruses can be redirected to alternative intracellular membranes, suggesting a remarkable plasticity of the viral replication machinery regarding the availability of at least some cellular factors [57, 58, 59•]. The multiple compensatory mechanisms may be important for replication in different cell types, where availability of cellular components may vary. This may serve as an essential source of evolutionary elasticity, and allow adaptation to changing conditions, including chemotherapeutic interventions.
Lipid synthesis pathways
The remarkable conservation of the overall design of the membranous replication complexes among distant (+)RNA viruses suggests shared basic mechanisms of their formation. Currently known elements of the autophagy and/or secretory pathways cannot fit the bill, since even related viruses display significant variability in requirements for these factors. The only universal feature of the (+)RNA virus infection that emerges over the years of research seems to be the obligatory modulation of cellular lipid biosynthesis pathways. Picornavirus (picorna-like superfamily) infection is long known to significantly enhance cellular phospholipid synthesis [60, 61, 62]. Investigation of BMV (alpha-like superfamily) replication in yeast system showed that formation of replication structures strongly depends on the metabolism of fatty acids and other lipids [63, 64]. Recently, new high-throughput lipidomics techniques allowed resolution of profound changes in the lipid spectrum upon Dengue virus (flavi-like superfamily) infection [65•]. Replication of many viruses has been proposed to depend on the activity of cellular fatty acid synthase (FAS). Expression of FAS is activated in HCV-infected cells [66] and the enzyme is recruited to the replication membranes of Dengue virus [67]. The apparently universal up-regulation of lipid metabolism by (+)RNA viruses raises interesting questions: Are the local changes in structural lipid composition the major driving force for formation of the membranous replication sites? Are these sites further stabilized by interaction with viral and/or cellular proteins? Do (+)RNA viruses remodel pre-existing cellular membranes, as we generally assume, or do they actually build replication membranes de novo and thus can they be truly called “novel replication organelles”?
Concluding remarks
We are only beginning to understand the complex details of biogenesis of viral replication organelles and their role in infection of (+)RNA viruses, and many questions still need to be resolved. The shared morphology of replication structures and the apparently universal requirement for active lipid synthesis strongly suggest that key steps of their formation are shared among distantly related viruses. (+)RNA viruses actively change cellular membranes, rather than rely on pre-existing membranes. They relocate key cellular regulators of membrane metabolism, and modulate their activities to create new pathways of membrane remodeling. Redundant networks of crucial virus–cell interactions that allow the generation of a favorable replication microenvironment may exist, thereby providing viruses the opportunity to adapt to and evolve under changing conditions. Understanding how viral proteins hijack regulatory mechanisms of membrane metabolism will undoubtedly bring new perspective to many areas of cell biology and will be indispensable for the development of a new generation of anti-viral control strategies.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
Acknowledgements
The authors thank Ellie Ehrenfeld for critically reading the manuscript, and Qian Feng and Jeroen Strating for help with preparation of the illustrations. Work in the lab of GB is supported by the start-up funds of the University of Maryland. Work in the lab of FvK is supported by research grants from the Netherlands Organization for Scientific Research (NWO-VICI-91812628, NWO-ECHO-700.57.001, NWO-ALW-820.02.018) and the European Union 7th Framework (EUVIRNA Marie Curie Initial Training Network, grant agreement number 264286, and SILVER Large Scale Collaborative Project, grant agreement number 260644).
Contributor Information
George A Belov, Email: gbelov@umd.edu.
Frank JM van Kuppeveld, Email: f.j.m.vankuppeveld@uu.nl.
References
- 1•.Knoops K., Kikkert M., van den Worm S.H.E., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biology. 2008;6:1957–1974. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2•.Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biology. 2007;5:2022–2034. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K.E., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host & Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Belov G.A., Nair V., Hansen B.T., Hoyt F.H., Fischer E.R., Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. Journal of Virology. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Limpens R.W., van der Schaar H.M., Kumar D., Koster A.J., Snijder E.J., van Kuppeveld F.J., Barcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. MBio. 2011;2:e00166-11. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Knoops K., Barcena M., Limpens R.W., Koster A.J., Mommaas A.M., Snijder E.J. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. Journal of Virology. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; In Refs. [1•, 2•, 3•, 4•, 5•, 6•], first insights into the three-dimensional structure of the replication sites of different (+)RNA viruses was obtained by electron tomography reconstruction studies.
- 7.Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. Journal of Virology. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biology. 2007;9:961–969. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 9.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. Journal of Biological Chemistry. 2007;282:37158–37169. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 10.Westaway E.G., MacKenzie J.M., Kenney M.T., Jones M.K., Khromykh A.A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. Journal of Virology. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie L.K., Hoenen A., Morgan G., Mackenzie J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. Journal of Virology. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulasli M., Verheije M.H., de Haan C.A.M., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cellular Microbiology. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkegaard K., Jackson W.T. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy. 2005;1:182–184. doi: 10.4161/auto.1.3.2065. [DOI] [PubMed] [Google Scholar]
- 14.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiological Sciences. 1987;4:197–202. [PubMed] [Google Scholar]
- 15.Koonin E.V., Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Critical Reviews in Biochemistry and Molecular Biology. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 16.Goldbach R., Wellink J. Evolution of plus-strand RNA viruses. Intervirology. 1988;29:260–267. doi: 10.1159/000150054. [DOI] [PubMed] [Google Scholar]
- 17•.Spuul P., Balistreri G., Kaariainen L., Ahola T. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest Virus replication complexes from the plasma membrane to modified lysosomes. Journal of Virology. 2010;84:7543–7557. doi: 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Different imaging and biochemical techniques were used to reveal the dynamic development of this alphavirus replication spherules first massively appearing on the outer cellular membrane and later transcocated into cellular interior by the endocytosis-like process.
- 18.Schwartz M., Chen J.B., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Molecular Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 19.Prod’homme D., Le Panse S., Drugeon G., Jupin I. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology. 2001;281:88–101. doi: 10.1006/viro.2000.0769. [DOI] [PubMed] [Google Scholar]
- 20.Dales S., Eggers H.J., Tamm I., Palade G.E. Electron microscopic study of the formation of poliovirus. Virology. 1965;26:379–389. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- 21.Skinner M.S., Halperen S., Harkin J.C. Cytoplasmic membrane-bound vesicles in echovirus 12-infected cells. Virology. 1968;36:241–253. doi: 10.1016/0042-6822(68)90141-4. [DOI] [PubMed] [Google Scholar]
- 22.Amako K., Dales S. Cytopathology of mengovirus infection. II. Proliferation of membranous cisternae. Virology. 1967;32:201–215. doi: 10.1016/0042-6822(67)90270-x. [DOI] [PubMed] [Google Scholar]
- 23.Monaghan P., Cook H., Jackson T., Ryan M., Wileman T. The ultrastructure of the developing replication site in foot-and-mouth disease virus-infected BHK-38 cells. Journal of General Virology. 2004;85:933–946. doi: 10.1099/vir.0.19408-0. [DOI] [PubMed] [Google Scholar]
- 24.Bienz K., Egger D., Pfister T., Troxler M. Structural and functional-characterization of the poliovirus replication complex. Journal of Virology. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley J.H., Boura E., Carlson L.A., Rozycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 27.Diaz A., Ahlquist P. Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Current Opinion in Microbiology. 2012 doi: 10.1016/j.mib.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. Journal of Biological Chemistry. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentice E., McAuliffe J., Lu X.T., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. Journal of Virology. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. Journal of Virology. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z.J., Thackrayl L.B., Miller B.C., Lynnl L.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 32.Brabec-Zaruba M., Berka U., Blaas D., Fuchs R. Induction of autophagy does not affect human rhinovirus type 2 production. Journal of Virology. 2007;81:10815–10817. doi: 10.1128/JVI.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson W.T., Giddings T.H., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biology. 2005;3:861–871. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein K.A., Jackson W.T. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. Journal of Virology. 2011;85:9651–9654. doi: 10.1128/JVI.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reggiori F., Monastyrska I., Verheije M.H., Cali T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernasconi R., Galli C., Noack J., Bianchi S., de Haan C.A., Reggiori F., Molinari M. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Molecular Cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 37.Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke P.Y., Chen S.S. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. Journal of Clinical Investigation. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrivastava S., Raychoudhuri A., Steele R., Ray R., Ray R.B. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bui Q.T., Golinelli-Cohen M.P., Jackson C.L. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Molecular Genetics and Genomics. 2009;282:329–350. doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belov G.A., Feng Q., Nikovics K., Jackson C.L., Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathogens. 2008;4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanke K.H.W., van der Schaar H.M., Belov G.A., Feng Q., Duijsings D., Jackson C.L., Ehrenfeld E., van Kuppeveld F.J.M. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. Journal of Virology. 2009;83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wessels E., Duijsings D., Niu T.K., Neumann S., Oorschot V.M., de Lange F., Lanke K.H., Klumperman J., Henke A., Jackson C.L. A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Developmental Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Gazina E.V., Mackenzie J.M., Gorrell R.J., Anderson D.A. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. Journal of Virology. 2002;76:11113–11122. doi: 10.1128/JVI.76.21.11113-11122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donnell V.K., Pacheco J.M., Henry T.M., Mason P.W. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology. 2001;287:151–162. doi: 10.1006/viro.2001.1035. [DOI] [PubMed] [Google Scholar]
- 46.Verheije M.H., Raaben M., Mari M., Lintelo E.G.T., Reggiori F., van Kuppeveld F.J.M., Rottier P.J.M., de Haan C.A.M. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathogens. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goueslain L., Alsaleh K., Horellou P., Roingeard P., Descamps V., Duverlie G., Ciczora Y., Wychowski C., Dubuisson J., Rouille Y. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. Journal of Virology. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Belov G.A., Kovtunovych G., Jackson C.L., Ehrenfeld E. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cellular Microbiology. 2010;12:1463–1479. doi: 10.1111/j.1462-5822.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; By using different GBF1 mutants the authors demonstrated that the role of GBF1 protein in poliovirus replication is different from its contribution into normal functioning of the cellular secretory pathway.
- 49•.Hsu N.Y., Ilnytska O., Belov G., Santiana M., Chen Y.H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identified PI4KIIIbeta as an important cellular factor recruited to the replication complexes of enteroviruses and proposed a model of conversion of normal cellular secretory pathway-derived membranes into viral replication organelles based on the elevated accumulation of PI4P lipid.
- 50.Berger K.L., Cooper J.D., Heaton N.S., Yoon R., Oakland T.E., Jordan T.X., Mateu G., Grakoui A., Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.S. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host & Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors showed that recruitment of PI4PIII alfa by the viral protein NS5A is important for the proper development of the hepatitis C virus replication organelles.
- 52.Arita M., Kojima H., Nagano T., Okabe T., Wakita T., Shimizu H. Phosphatidylinositol-4 kinase III beta is a target of enviroxime-like compounds for anti-poliovirus activity. Journal of Virology. 2010;85:2364–2372. doi: 10.1128/JVI.02249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger K.L., Kelly S.M., Jordan T.X., Tartell M.A., Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. Journal of Virology. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin-Acebes M.A., Blazquez A.B., de Oya N.J., Escribano-Romero E., Saiz J.C. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS ONE. 2011;6:e24970. doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crotty S., Saleh M.C., Gitlin L., Beske O., Andino R. The poliovirus replication machinery can escape inhibition by an antiviral drug that targets a host cell protein. Journal of Virology. 2004;78:3378–3386. doi: 10.1128/JVI.78.7.3378-3386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Schaar H.M., van der Linden L., Lanke K.H.W., Strating J.R., Pürstinger G., de Vries E., de Haan C.A.M., Neyts J., van Kuppeveld F.J.M. Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication. Cell Research. 2012 doi: 10.1038/cr.2012.129. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz M., Chen J., Lee W.M., Janda M., Ahlquist P. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11263–11268. doi: 10.1073/pnas.0404157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jonczyk M., Pathak K.B., Sharma M., Nagy P.D. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362:320–330. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 59•.Miller D.J., Schwartz M.D., Dye B.T., Ahlquist P. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. Journal of Virology. 2003;77:12193–12202. doi: 10.1128/JVI.77.22.12193-12202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; First report that showed that genome replication of a (+)RNA virus does not absolutely depend on a specific source of membranes but can be retargeted to another intracellullar membrane.
- 60.Mosser A.G., Caliguir L.A., Tamm I. Incorporation of lipid precursors into cytoplasmic membranes of poliovirus-infected Hela-cells. Virology. 1972;47:39. doi: 10.1016/0042-6822(72)90236-x. [DOI] [PubMed] [Google Scholar]
- 61.Schimmel H., Traub P. The effect of mengovirus infection on lipid-synthesis in cultured Ehrlich ascites tumor-cells. Lipids. 1987;22:95–103. doi: 10.1007/BF02534860. [DOI] [PubMed] [Google Scholar]
- 62.Plageman P.G., Clevelan P.H., Shea M.A. Effect of mengovirus replication on choline metabolism and membrane formation in novikoff hepatoma cells. Journal of Virology. 1970;6:800. doi: 10.1128/jvi.6.6.800-812.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Diaz A., Hao L., Gancarz B., den Boon J.A., Ahlquist P. Intersection of the multivesicular body pathway and lipid homeostasis in RNA replication by a positive-strand RNA virus. Journal of Virology. 2011;85:5494–5503. doi: 10.1128/JVI.02031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee W.M., Ishikawa M., Ahlquist P. Mutation of host Delta 9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. Journal of Virology. 2001;75:2097–2106. doi: 10.1128/JVI.75.5.2097-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65•.Perera R., Riley C., Isaac G., Hopf-Jannasch A.S., Moore R.J., Weitz K.W., Pasa-Tolic L., Metz T.O., Adamec J., Kuhn R.J. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathogens. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using modern lipidomics techniques the authors demonstrated profound changes in the lipid species accumulated in Dengue virus-infected cells.
- 66.Yang W., Hood B.L., Chadwick S.L., Liu S.F., Watkins S.C., Luo G.X., Conrads T.P., Wang T.Y. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396–1403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heaton N.S., Perera R., Berger K.L., Khadka S., Lacount D.J., Kuhn R.J., Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasaki J., Ishikawa K., Arita M., Taniguchi K. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO Journal. 2012;31:754–766. doi: 10.1038/emboj.2011.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greninger A.L., Knudsen G.M., Betegon M., Burlingame A.L., Derisi J.L. The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. Journal of Virology. 2012;86:3605–3616. doi: 10.1128/JVI.06778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


