Highlights
-
•
Broadly neutralizing antibodies against influenza A and B represents a safe and affordable alternative to convalescent plasma.
-
•
The large majority of broadly neutralizing antibodies target the stem region of HA.
-
•
Antiviral protection by anti-HA antibodies comprises both Fab- and Fc-dependent mechanisms.
-
•
Several broadly neutralizing antibodies are in development for the therapy of severe influenza infections.
Abstract
Monoclonal antibodies have revolutionized the treatment of several human diseases, including cancer, autoimmunity and inflammatory conditions and represent a new frontier for the treatment of infectious diseases. In the last decade, new methods have allowed the efficient interrogation of the human antibody repertoire from influenza immune individuals and the isolation of several monoclonal antibodies capable of dealing with the high variability of influenza viruses. Here, we will provide a comprehensive overview of the specificity, antiviral and immunological mechanisms of action and development into the clinic of broadly reactive monoclonal antibodies against influenza A and B viruses.
Current Opinion in Virology 2017, 24:60–69
This review comes from a themed issue on Antiviral strategies
Edited by Lieve Naesens and Fabien Zoulim
For a complete overview see the Issue and the Editorial
Available online 18th May 2017
http://dx.doi.org/10.1016/j.coviro.2017.03.002
1879-6257/© 2017 Elsevier B.V. All rights reserved.
Introduction
Influenza viruses are responsible for annual epidemics entailing significant morbidity and mortality, particularly in the elderly and in immune-compromised individuals [1, 2, 3].
The hemagglutinin glycoprotein (HA) is the main target of influenza A and B neutralizing antibody response to infection or vaccination. Each monomer of the trimeric HA is composed of two polypeptides generated by proteolytic cleavage of the HA0 precursor. The globular head of HA binds to sialic acid residues on target cells, while the stem region mediates the low pH-triggered fusion of viral and cellular membranes in endosomes. Sixteen subtypes of HA and two HA analogs identified in bats (H17 and H18) cluster in two groups: group 1 comprising H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17 and H18 and group 2 comprising H3, H4, H7, H10, H14 and H15. Currently circulating human viruses belong to the group 1 subtype H1N1 (derived from the 1918 and 2009 pandemics) and to the group 2 subtype H3N2 (derived from the 1968 pandemic). Other subtypes such as H2N2 (endemic in humans in 1957–1968) [4] could re-emerge and others have caused episodes of zoonotic infections with no sustained human-to-human transmission, such as the group 1 H5 [5], H9 [6] and H6 [7], and the group 2 H7 [8] and H10 subtypes [9]. Influenza B viruses exist as a single type and are represented by two co-circulating antigenically distinct lineages defined by the prototype viruses B/Victoria/1987 and B/Yamagata/1988 [10].
The second viral glycoprotein is the neuraminidase (NA) that is a mushroom-shaped tetramer that acts as a receptor-destroying enzyme, removing sialic acid residues from the surface of infected cells, thereby allowing the release and spread of budding virions. There are nine subtypes of NA clustered into two groups: group 1 N1, N4, N5 and N8 and group 2 N2, N3, N6, N7 and N9. The NA enzymatic site of influenza A and B viruses is the target of four approved anti-influenza drugs: oseltamivir, peramivir, laninamivir and zanamivir.
The M2 protein (and its influenza B orthologue BM2) are homotetramers and function as proton channels at the low pH of endosomes to trigger the uncoating of viral ribonucleoprotein (RNP) complexes [11]. M2 is poorly expressed in virions, while it is abundantly displayed on the surface of infected cells [12]. The specific M2-channel-activity inhibitors amantadine and rimantandine block infection by preventing RNP uncoating and release into the cytoplasm. However, clinical use of these drugs is currently not recommended due to widespread resistance.
Current standards of care and vaccination strategies are suboptimal to treat and prevent severe influenza A and B virus infection. Indeed, trivalent and tetravalent influenza vaccines are only partially effective in the elderly and immunocompromised individuals and in some cases the selected strains do not match with those circulating. In addition, antivirals such as NA inhibitors and M2 blockers have limited efficacy in severe cases of influenza infection if not administered within 48 hours from symptoms onset and may select for resistance.
Clinical studies in patients with severe viral pneumonia caused by viral SARS-CoV [13], 1918 and 2009 H1N1 pandemic viruses [14, 15] and H5N1 zoonotic influenza A virus [16] have shown a therapeutic benefit from the use of convalescent plasma, especially when administered early after symptom onset [17]. However, the poor supply of convalescent plasma and the low antibody titers hampered the utility of this approach. The identification during the last decade of several broadly neutralizing antibodies against influenza A and B viruses, isolated from plasma cells or memory B cells of influenza-infected or influenza-vaccinated individuals, represents a safe and affordable alternative to the use of patient-derived convalescent plasma. Indeed, recent data suggest that passive immunization using broadly neutralizing monoclonal antibodies might represent a viable approach for prophylaxis and therapy that might complement or substitute current vaccines and antivirals. We will review the current literature on broadly neutralizing antibodies against influenza viruses and discuss their mechanisms of action and clinical development.
Overview on broadly neutralizing antibodies against influenza viruses
Because of the extensive variability, the globular head of HA is targeted by antibodies that have limited breadth. These antibodies recognize antigenically similar strains within a single HA subtype, and are prone to selecting escape mutants [18]. Nonetheless, monoclonal antibodies targeting several isolates within the same subtype of influenza A (e.g., H1, H3, H5 and H7) have been described [19, 20, 21]. A similar class of anti-head antibodies with multi-lineage neutralization of influenza B viruses has also been reported and includes CR8033, targeting an epitope overlapping with the receptor binding site on HA, CR8071, 5A7 and 46B8, directed to epitopes at the base of the head (Swem L et al., US 2015/0274812) [22, 23, 24].
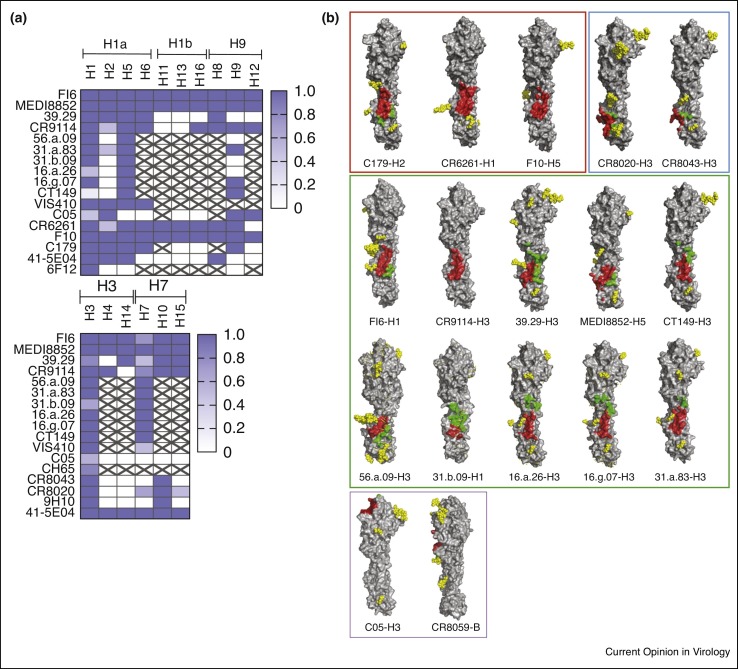
The sialic acid binding pocket in HA represents a conserved site and rare antibodies that precisely target this site have been described such as C05 [25], S139/1 [26] and F045-092 [27]. However, neutralizing activity of these antibodies is scattered against multiple group 1 and group 2 viruses and typically only a fraction of strains within each subtype are neutralized (Figure 1 a) (S139/1: H1, H2, H3, H13, H16) (F045-092: H1, H2, H3, H13).
Figure 1.
Epitope specificity and breadth of influenza A and B heterosubtypic neutralizing antibodies. (a) Breadth of reactivity of influenza A heterosubtypic neutralizing antibodies against group 1 (upper) and group 2 (lower) subtypes as derived from published data (see Table 1 for references). Color gradient used in the table indicates the fraction of strains recognized within each subtype. Strikethrough cells, not tested. (b) Footprints of antibodies C179 (4hlz), CR6261 (3gbn), F10 (3fku), CR8020 (3sdy), CR8043 (4nm8), FI6 (3ztn), CR9114 (4fqy), 39.29 (4kvn), MEDI8852 (5jw4), CT149 (4r8w), 56.a.09 (59k9), 31.b.09 (5k9o), 16.a.26 (5k9q), 16.g.07 (5kan), 31.a.83 (5kaq), C05 (4fqr) and CR8059 (4fqk, stabilized variant of CR8071) on their cognate HA ligands. Structures are grouped and boxed according to antibodies breadth towards group 1 (red box), group 2 (blue box), group 1 and 2 (green box) and anti-head antibodies (purple box). The heavy- and light-chain contact residues are depicted in red and green, respectively. HA monomer molecules are shown with the same orientation. Glycan residues are depicted with yellow spheres. Figure was made with PyMOL.
The HA stem represents a relatively conserved region that is involved in the process of viral fusion, and consequently antibodies directed to the stem region of HA are often capable of heterosubtypic neutralization across one or both influenza A groups (Table 1 ). In general, antibodies targeting the stem region of HA are rare as compared to those targeting variable regions in the globular head. The most frequent anti-stem heterosubtypic antibodies are those against group 1 viruses that were found to be elicited by both vaccination and infection by H1N1 viruses [28, 29]. X-ray structural analysis of three of such antibodies (CR6261, F10 and C179) revealed that they target a very similar epitope in the HA stem, contacting conserved residues in the Trp-21 hydrophobic pocket and Helix A regions [30, 31, 32]. Group 2 neutralizing antibodies are less frequent, possibly due to the shielding of HA stem epitopes by the conserved N38-bound glycan. Indeed, the solved structures of the Group 2 antibodies CR8043 and CR8020, and the negative-stain electron microscopy (EM) reconstruction of the 9H10 antibody, indicated that their binding region is located in the membrane-proximal base of the HA stem [33, 34, 35]. Interestingly, in group 1 viruses this region is masked by a highly conserved N21-bound glycan.
Table 1.
Characteristics of broadly neutralizing antibodies against influenza A and B viruses
| Antibody | Specificity | Breadth | VH | HCDR3 aa seq | PDB (year; Ref) | Manufacturer/developer | Trial ID | Phase | Subjects | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| F10 | HA (stem) | Group 1 | VH1-69 | ARSPSYICSGGTCVFDH | 3FKU (2009; [31]) | |||||
| MAb 6F12 | HA (stem) | Group 1 | mouse | N/A (2012; [86]) | ||||||
| C179 | HA (stem) | Group 1 | mouse | ARPKGYFPYAMDY | 4HLZ (1993, [32]) | |||||
| CR6261 | HA (stem) | Group 1 | VH1-69 | AKHMGYQVRETMDV | 3GBN, 3GBM (2008; [30, 56]) | Crucell (J&J) | NCT01406418 | Ph1 | Healthy | Complete (2013) |
| NCT02371668 | Ph2a | Exp. infected | Recruting (2017) | |||||||
| CR8020 | HA (stem) | Group 2 | VH1-18 | AREPPLFYSSWSLDN | 3SDY (2011; [34, 42•]) | Crucell (J&J) | NCT01756950 | Ph1 | Healthy | Complete (2013) |
| NCT01938352 | Ph2a | Exp. infected | Complete (2014) | |||||||
| CR8043 | HA (stem) | Group 2 | VH1-3 | ARGASWDARGWSGY | 4NM4, 4NM8 (2014; [33]) | |||||
| MAb 9H10 | HA (stem) | Group 2 | mouse | N/A (2014; [35]) | ||||||
| CT-P27** | HA (stem) | Mix Group1 and Group2 | N/A | N/A | Celltrion | KCT0001179 + KCT0001617 | Ph1 | Healthy | Complete (2013) + Unknown (submitted 2015) | |
| NCT02071914 | Ph2a | Exp. Infected | Complete (2014) | |||||||
| CR6261 + CR8020 | HA (stem) | Mix Group 1 and Group 2 | Crucell (J&J) | NCT01992276 | Ph2b | Hospitalized pts. | Withdrawn before enrollment | |||
| FI6 | HA (stem) | Group 1+2 | VH3-30 | AKDSQLRSLLYFEWLSQGYFDP | 3ZTJ, 3ZTN (2011; [36, 60]) | |||||
| VIS410 | HA (stem) | Group 1+2 | VH3-30 | AKDSRLRSLLYFEWLSQGYFNP | N/A (2015; [82, 87]) | Visterra | NCT02045472 | Ph1 | Healthy | Complete (2015) |
| NCT02468115 | Ph2a | Exp. Infected | Complete (2016) | |||||||
| 39.29* | HA (stem) | Group 1+2 | VH3-30 | AVPGPVFGIFPPWSYFDN | 4KVN (2013; [37]) | Genentech (Roche) | NCT01877785 + NCT02284607 | Ph1 | Healthy | Complete (2013 + 2015) |
| NCT01980966 | Ph2a | Exp. Infected | Complete (2014) | |||||||
| NCT02623322 | Ph2 | Non-hospitalized pts. | Recruiting (2017) | |||||||
| NCT02293863 | Ph2b | Hospitalized pts. | Recruiting (2017) | |||||||
| CT149 | HA (stem) | Group 1+2 | VH1-18 | ARDKVQGRVEVGSGGRHDY | 4R8W (2015; [38]) | |||||
| MEDI8852 | HA (stem) | Group 1+2 | VH6-1 | ARSGHITVFGVNVDAFDM | 5JW5, 5JW4, 5JW3 (2016; [39•]) | MedImmune (AstraZeneca) | NCT02350751 | Ph1 | Healthy | Complete (2015) |
| NCT02603952 | Ph1b/2a | Non-hospitalized pts. | Complete (2016) | |||||||
| 31.a.83 | HA (stem) | Group 1+2 | VH3-23 | AKDESPPIYNLMPGYYSTYYYMDV | 5KAQ (2016; [40••]) | |||||
| 56.a.09 | HA (stem) | Group 1+2 | VH6-1 | ARGSAMIFGIVIILES | 5K9K (2016; [40••]) | |||||
| 31.b.09 | HA (stem) | Group 1+2 | VH1-18 | ARDRPHILTGFDFDY | 5K9O (2016; [40••]) | |||||
| 16.a.26 | HA (stem) | Group 1+2 | VH1-18 | ARDKKQGEVVLPAASFRWFAP | 5K9Q (2016; [40••]) | |||||
| 16.g.07 | HA (stem) | Group 1+2 | VH1-18 | VRNRVQMEVSPATQSTWYMDL | 5KAN (2016; [40••]) | |||||
| 41-5E04 | HA (stem) | Group 1+2 | VH3-53 | ARDFLRGPIHDYFFYMDV | N/A (2016; [44•]) | |||||
| C05 | HA (head) | Group 1+2 | VH3-23 | AKHMSMQQLVSAGWERADLVGDAFDV | 4FNL, 4FP8, 4FQR (2012; [25]) | |||||
| CR9114 | HA (stem) | Group 1+2+B | VH1-69 | ARHGNYYYYSGMDV | 4FQH, 4FQI, 4FQV, 4FQY (2012; [23]) | |||||
| 5A7 | HA (head) | FluB | VH3-33 | ARDLQPPHSPYGMDV | N/A (2013; [22]) | |||||
| CR8033 | HA (head) | FluB | VH3-9 | AKDRLESSAMDILEGGTFDI | 4FQL (2012; [23]) | |||||
| CR8071 | HA (head) | FluB | VH1-18 | ARDVQYSGSYLGAYYFDY | 4FQJ, 4FQK (2012; [23]) | |||||
| 46B8*** | HA (head) | FluB | VH5-51 | ASGPGYSGYHYGWFDT | N/A (2015; [24]) | Genentech (Roche) | NCT02528903 | Ph1 | Healthy | Complete (2016) |
| TCN32 | M2e | FluA | VH4-59 | ARASCSGGYCILDY | N/A (?; [53, 83]) | Theraclone | NCT01390025 | Ph1 | Healthy | Complete (2012) |
| NCT01719874 | Ph2a | Exp. Infected | Complete (2012) | |||||||
MHAA4549A.
Navivumab + Firivumab.
MHAB5553A.
Intensive discovery campaigns have led to the identification of rare group 1 and 2 heterosubtypic neutralizing antibodies. The ten available X-ray structures (Figure 1b) of FI6, CR9114, 39.29, MEDI8852, CT149, 56.a.09, 31.b.09, 16.a.26 and 16.g.07, 31.a.83, [23, 36, 37, 38, 39•, 40••] revealed that these antibodies can adopt multiple binding modalities to cope with high amino acid diversity and with the structural constraints posed by the presence of conserved Group-1 and Group-2-specific glycans. Despite the restricted propensity of the stem region to mutate, escape mutants have been isolated also for anti-stem antibodies [41, 42•], and their existence is also supported by the incomplete breadth of certain anti-stem antibodies, for example, the lack of binding to human H2N2 viruses by CR6261 [30]. Finally, a rare antibody, CR9114, was shown to bind to several influenza A and B viruses tested, although neutralizing activity was limited to influenza A [23]. The breadth of CR9114 appeared to be the result of a high plasticity of its binding site that is able to accommodate the recognition of various conformations of the conserved tryptophan 21 in the presence of group-2 and influenza B-specific glycans.
Several studies identified structural and genetic requirements for the development of broadly neutralizing anti-HA influenza antibodies such as the preferential heavy variable (VH) gene usage and the presence of key somatic mutations and sequence signatures in the antibody CDRs [36, 43, 44•, 45•, 46, 47]. Of note, polyreactivity of some group 1 anti-stem antibodies has been proposed to result in B cell counter-selection in vivo [43]. This knowledge is valuable to guide the design of universal influenza vaccines [48].
Besides HA, the viral envelope proteins NA and M2, have been proposed as targets for broadly neutralizing antibodies. In animal models it was shown that NA-directed antibodies correlate with reduced severity of the disease and epidemiological studies in humans have suggested a role for NA immunity in reducing the lasting of viral shedding and symptoms [49, 50]. Recently, a universal epitope in NA conserved among all influenza A and B strains was identified and a rabbit monoclonal antibody raised against this region provided group 1 and 2 heterosubtypic protection in challenged mice [51, 52]. The highly conserved extracellular domain of M2 has been an attractive target for vaccine design [53] and antibody therapeutics, such as TCN32 [54] and Z3G1 [55].
Multiple mechanisms of action of broadly neutralizing antibodies
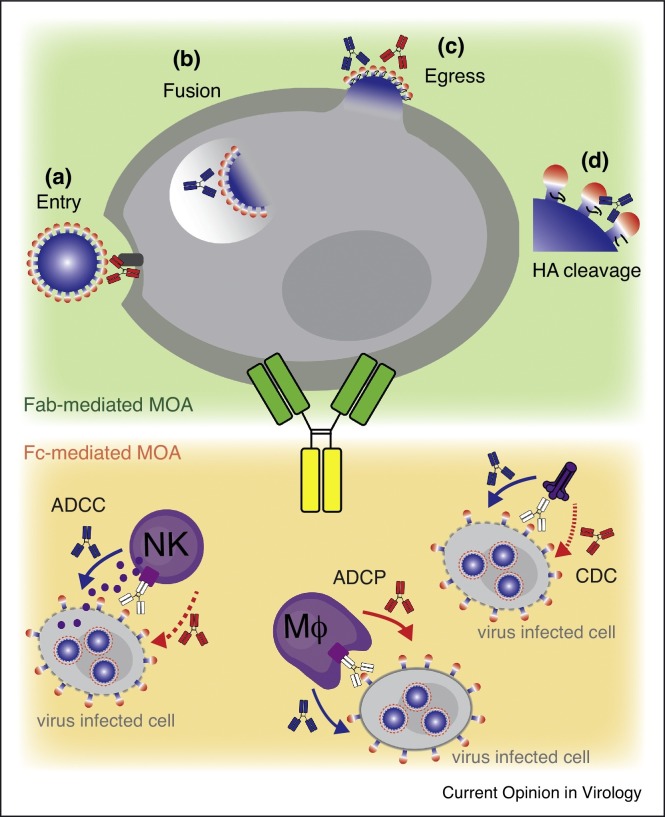
Antibodies to influenza HA can directly neutralize influenza virus at the level of entry, fusion and proteolytic cleavage, namely: (a) by blocking binding to sialic acid residues on the surface of the target cell; (b) by inhibiting endosomal viral fusion; (c) by preventing release of progeny virions from infected cells; (d) by inhibiting extracellular proteolytic cleavage of HA. The mode of viral neutralization depends on the site recognized on the trimeric HA. Thus, while anti-head antibodies work primarily by inhibiting viral entry (they are detected by the classical hemagglutination inhibition assay, HAI), anti-stem antibodies do not block binding to sialic acid (they are inactive in HAI), but instead prevent endosomal viral fusion, viral budding or HA cleavage activation (Figure 2 ) [28, 56, 57].
Figure 2.
Mechanisms of antiviral protection by anti-HA antibodies. Direct mechanisms dependent on Fab engagement [85] include: (a) Anti-head antibodies (red) block viral entry by targeting the receptor binding site on HA or indirectly by projecting the Fc over the receptor binding site [20, 23]. (b) Anti-stem antibodies (blue) interfere with the pH-driven HA rearrangement blocking the process of viral fusion in endosomes [41, 56, 57]. (c) Anti-head and anti-stem antibodies can inhibit the release of budding virions from infected cells [35, 85]. (d) Anti-stem antibodies can impede HA0 cleavage by extracellular proteases, thereby blocking proteolytic cleavage of HA into its fusogenic form [34, 39•]. Indirect mechanisms dependent of Fc engagement include: antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP) and complement-dependent cytotoxicity (CDC). Antibodies performing stronger Fc-dependent mechanism are depicted with full line arrows, while antibodies performing weaker are depicted with dotted line arows.
Additional mechanisms of action beyond viral neutralization can contribute to in vivo protection. Indeed, some broadly reactive antibodies directed to either HA head or stem and lacking in vitro neutralizing activity, showed in vivo efficacy under prophylactic settings [23, 58], Multiple studies in the last decade have further illustrated the complexity of antibody-dependent in vivo protection, which was shown to involve, in addition to the direct mechanisms described above, also indirect (i.e., Fc-dependent) mechanisms of action that are mediated by the interaction of the Fc portion with FcγRs on immune cells or with complement [59]. Importantly, Fc-dependent effector functions can be recruited by most influenza-specific antibodies, including anti-HA, anti-NA and anti-M2e. FcγR-dependent mechanisms can be assessed in vitro by measuring antibody-dependent cytotoxicity mediated by NK cells (ADCC, antibody-dependent cellular cytotoxicity) or complement (CDC, complement-dependent cytotoxicity) as well as antibody-dependent cellular phagocytosis (ADCP) [39•, 42•, 44•, 60, 61••, 62, 63, 64].
The contributions of Fc-FcγR interactions in vivo were demonstrated by using different Fc formats of antibodies in influenza virus infected wild type mice [36], or in transgenic mice expressing the full array of human FcγRs [60] or lacking individual mouse mFcγRs [65]. These studies revealed that Fc-mediated effector functions through activating FcγRs contributes to in vivo prophylactic efficacy of anti-stem antibodies at low antibody doses, while at high antibody doses protection is primarily mediated by virus neutralization [36, 60]. While there is a general agreement that anti-stem antibodies are potent inducers of ADCC in vitro and require FcγRs interaction for optimal in vivo protection, for anti-head antibodies the role of Fc-mediated effector function is still controversial [44•, 60, 66]. The role of Fc-dependent effector mechanisms in mediating antibody-dependent in vivo protection has important practical implications given the possibility to enhance these functions by glyco-engineering or mutating the Fc portion or even by a combination of the two [67, 68].
While there is a consensus on the fact that anti-stem antibodies are more efficient in engaging FcRs as compared to anti-head, the structural basis for this difference is not clear, although it may be related to a more membrane-proximal engagement of FcRs. Interestingly, recent studies suggest that ADCC of influenza-infected cells can be enhanced by the accessory interaction between HA on the infected cell and sialic acid motifs on NK cells that may stabilize the immunological synapse, thus increasing the avidity between target and NK cells [61••, 62]. These findings are consistent with the observation that the only anti-head antibodies that induce ADCC are those that do not interfere with binding of sialic acid (therefore HAI-negative) [61], Conversely, HAI positive anti-head antibodies by blocking binding to the sialic acid are not or less effective in promoting ADCC [62, 69••]. Interestingly, the interaction between HA and NK cells was found to be dependent on sialylation of the NK cell receptor NKp46 [70]. NA-mediated removal of sialic acid from the NKp46 was shown to represent an immune-evasion mechanism of influenza viruses. Reciprocally, inhibition of NA activity by oseltamivir was shown to boost NKp46 recognition and killing of infected cells [71] a finding that might explain the synergistic or additive effect observed in animals treated with anti-stem antibodies in the presence of NA-inhibitors [37]. The antibody-dependent recruitment of C1q on virions can contribute directly to antibody-mediated virus neutralization through a steric hindrance effect [72, 73], whereas C1q recruitment on infected cells activates the complement cascade leading to their killing (CDC). Interestingly, stem-specific antibodies are particularly effective in inducing CDC, possibly due to the recruitment of the complement cascade in proximity of the cell membrane [74].
Opsonophagocytosis represents another important Fc-dependent mechanism for antibody-mediated clearance of virions and infected cells (ADCP), mainly by alveolar macrophages [75]. A recent study showed that antibodies unable to induce ADCC and CDC were highly effective in ADCP, suggesting that for some antibodies phagocytosis could be the major mechanism of protection [44•]. The influence of epitope specificity in ADCP remains to be established.
The importance of Fc-mediated effector functions in vivo is well illustrated by efficacy studies using anti-M2e antibodies. These antibodies do not neutralize virus infection in vitro but show significant therapeutic efficacy in vivo by binding to the M2 protein expressed on the surface of virus-infected cells [55, 76]. The importance of phagocytosis through alveolar macrophages for in vivo immune protection by anti-M2e antibodies has been demonstrated [65], while, the role of ADCC and CDC remain controversial [55, 64, 65, 74, 76].
Because of the simultaneous occurrence of the different antiviral mechanisms listed above, there is not a precise correlation between in vitro activities and in vivo protection. In addition, cross-talk among serum antibodies of varying specificities could determine the magnitude of direct viral neutralization and Fc receptor-mediated effector functions [69••]. Other studies have also suggested a role of antibodies in enhancing disease severity. In young pigs polyclonal non-neutralizing antibodies induced enhanced respiratory disease (defined as vaccine-associated enhanced respiratory disease, VAERD) [77, 78]. In another study, immune complex formation with pre-existing non-neutralizing antibodies and consequent C4d deposition was linked to tissue pathology in severely ill individuals infected with the pandemic H1N1 virus in 2009 and with H2N2 virus in 1957 [79]. Despite the difficulties to draw conclusions from studies that analyzed the effect of the polyclonal antibody response to influenza infection, enhanced respiratory disease remains a potential concern for development of antiviral antibodies as therapeutics and is currently under investigation in multiple clinical studies.
Clinical development of broadly neutralizing antibodies
Despite advances in vaccines and small-molecule anti-viral therapeutics, there are no FDA approved drugs for use in individuals with severe influenza requiring hospitalization. Although the NA inhibitors have been used off-label in this population, their effectiveness is limited, highlighting the unmet medical need for more effective treatment of influenza in populations at high risk for morbidity and mortality. However, their effectiveness is still debated, highlighting the unmet medical need for more effective treatment of influenza in populations at high risk for morbidity and mortality [80]. Antibody-based strategies to prevent and treat influenza could be a new alternative countermeasure for influenza. Preclinical studies using broadly neutralizing antibodies showed superior efficacy and longer therapeutic windows compared to neuraminidase inhibitors in mice and ferrets [37, 39•] suggesting that antibody therapy may provide significant clinical benefit to this population.
Several anti-influenza monoclonal antibodies are being tested in various stages of clinical development (Table 1). Most of these antibody candidates are directed at the HA protein of the virus (only one against the M2 protein). Over the last 4–5 years, these antibodies have been tested in small proof-of-concept Ph2a human influenza challenge studies, in which healthy volunteers are experimentally infected with a less virulent strain of virus, and therefore cause only mild upper respiratory symptoms [81]. In these studies, administration of anti-HA antibodies, MHAA4549A and VIS410 and an anti-M2e mAb TCN-032 exhibited significant reductions in viral titer when given to subjects 24–36 hours post infection (Sloan S et al., ISIRV Options IX for the control of influenza, 2016; Trevejo JM et al., Fifth ESWI influenza conference, 2014) [82, 83]. These outcomes provide confidence in the antiviral effect of these anti-HA antibodies and anti-M2 antibodies, however ‘human viral challenge’ studies cannot be used to demonstrate efficacy for licensure according to the US FDA guidance (FDA, Influenza: Developing Drugs for Treatment and/or Prophylaxis. Guidance for Industry 2011).
The most advanced programs are primarily focused on anti-HA antibodies that can cross-react to both group 1 and 2 influenza A viruses either alone or as a cocktail for treating hospitalized patients with severe influenza. Visterra (VIS410) and Celltrion (CT-P27 (antibody cocktail)) have recently completed Ph2a studies on experimentally infected subjects, while MedImmune’s MEDI8852 and Genentech’s MHAA4549A have progressed to trials on naturally infected subjects. MedImmune has now completed a Ph2a study in non-hospitalized naturally infected patients and Genentech is currently recruiting two different phase 2 trials, one in naturally infected uncomplicated subjects, and one in naturally infected hospitalized subjects (ex-US). With several promising antibody candidates progressing to later stages of development in hospitalized subjects with severe influenza, it is worth noting that this indication presents significant challenges to clinical development. Recent failure of neuraminidase inhibitor peramivir in this indication [84], has highlighted some of the challenges that antibodies will also face, such as the lengthy recruitment timelines due to the patient population, the seasonality of the virus and the uncertainty of what pivotal trial endpoints will show enhanced efficacy over the standard of care.
Outlook
Despite over the last 30 year’s effort, designing a universal flu vaccine has been difficult due to the lack of detailed information about which epitopes within the HA stem region are key to driving broad immunity. Identification of broadly neutralizing antibodies to all subtypes of influenza viruses not only provides a basis for the design of more protective vaccine strategies but also opens the door to the alternative passive immunization approach to preventing influenza infection. New advances in antibody manufacturing capabilities, half-life extension technologies, and alternative delivery mechanisms using DNA, RNA or virus vectors may pave the way for more cost-effective immunoprophylaxis using broadly neutralizing antibodies.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Cooper B.S., Kotirum S., Kulpeng W., Praditsitthikorn N., Chittaganpitch M., Limmathurotsakul D., Day N.P.J., Coker R., Teerawattananon Y., Meeyai A. Mortality attributable to seasonal influenza A and B infections in Thailand, 2005–2009: a longitudinal study. Am. J. Epidemiol. 2015;181:898–907. doi: 10.1093/aje/kwu360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein E., Viboud C., Charu V., Lipsitch M. Improving the estimation of influenza-related mortality over a seasonal baseline. Epidemiology. 2012;23:829–838. doi: 10.1097/EDE.0b013e31826c2dda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nabel G.J., Wei C.-J., Ledgerwood J.E. Vaccinate for the next H2N2 pandemic now. Nature. 2011;471:157–158. doi: 10.1038/471157a. [DOI] [PubMed] [Google Scholar]
- 5.Webster R., Govorkova E. H5N1 influenza—continuing evolution and spread. N. Engl. J. Med. 2006;355:2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 6.Peiris M., Yuen K.Y., Leung C.W., Chan K.H., Ip P.L., Lai R.W., Orr W.K., Shortridge K.F. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J., Zhang L., Kan X., Jiang L., Yang J., Guo Z., Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin. Infect. Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 8.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 9.Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 10.Rota P.A., Wallis T.R., Harmon M.W., Rota J.S., Kendal A.P., Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 11.Schnell J.R., Chou J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zebedee S.L., Richardson C.D., Lamb R.A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Y., Wong R., Soo Y.O.Y., Wong W.S., Lee C.K., Ng M.H.L., Chan P., Wong K.C., Leung C.B., Cheng G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luke T.C., Kilbane E.M., Jackson J.L., Hoffman S.L. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann. Intern. Med. 2006;145:599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- 15.Hung I.F., To K.K., Lee C.-K., Lee K.-L., Chan K., Yan W.-W., Liu R., Watt C.-L., Chan W.-M., Lai K.-Y. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin. Infect. Dis. 2011;52:447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou B., Zhong N., Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N. Engl. J. Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- 17.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.-M., Lim W.S., Makki S., Rooney K.D., Beck C.R., Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caton A., Brockman-Schneider C.J., Yewdell J., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 19.Benjamin E., Wang W., McAuliffe J.M., Palmer-Hill F.J., Kallewaard N.L., Chen Z., Suzich J.A., Blair W.S., Jin H., Zhu Q. A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 hemagglutinin globular head. J. Virol. 2014;88:6743–6750. doi: 10.1128/JVI.03562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong X., Corti D., Liu J., Pinna D., Foglierini M., Calder L.J., Martin S.R., Lin Y.P., Walker P.A., Collins P.J. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc. Natl. Acad. Sci. 2015;112:9430–9435. doi: 10.1073/pnas.1510816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He F., Kumar S.R., Syed Khader S.M., Tan Y., Prabakaran M., Kwang J. Effective intranasal therapeutics and prophylactics with monoclonal antibody against lethal infection of H7N7 influenza virus. Antivir. Res. 2013;100:207–214. doi: 10.1016/j.antiviral.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Yasugi M., Kubota-Koketsu R., Yamashita A., Kawashita N., Du A., Sasaki T., Nishimura M., Misaki R., Kuhara M., Boonsathorn N. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog. 2013;9:e1003150–12. doi: 10.1371/journal.ppat.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai N., Swem L.R., Park S., Nakamura G., Chiang N., Estevez A., Fong R., Kamen L., Kho E., Reichelt M. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat. Commun. 2017;8:14234. doi: 10.1038/ncomms14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekiert D.C., Kashyap A.K., Steel J., Rubrum A., Bhabha G., Khayat R., Lee J.H., Dillon M.A., O'Neil R.E., Faynboym A.M. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee P.S., Yoshida R., Ekiert D.C., Sakai N., Suzuki Y., Takada A., Wilson I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. 2012;109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee P.S., Ohshima N., Stanfield R.L., Yu W., Iba Y., Okuno Y., Kurosawa Y., Wilson I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corti D., Suguitan A.L., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Investig. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrammert J., Koutsonanos D., Li G.-M., Edupuganti S., Sui J., Morrissey M., McCausland M., Skountzou I., Hornig M., Lipkin W.I. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekiert D.C., Bhabha G., Elsliger M.-A., Friesen R.H.E., Jongeneelen M., Throsby M., Goudsmit J., Wilson I.A. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.-M., Santelli E., Stec B., Cadwell G., Ali M. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyfus C., Ekiert D.C., Wilson I.A. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 hemagglutinin. J. Virol. 2013;87:7149–7154. doi: 10.1128/JVI.02975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesen R.H.E., Lee P.S., Stoop E.J.M., Hoffman R.M.B., Ekiert D.C., Bhabha G., Yu W., Juraszek J., Koudstaal W., Jongeneelen M. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekiert D.C., Friesen R.H.E., Bhabha G., Kwaks T., Jongeneelen M., Yu W., Ophorst C., Cox F., Korse H.J.W.M., Brandenburg B. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan G.S., Lee P.S., Hoffman R.M.B., Mazel-Sanchez B., Krammer F., Leon P.E., Ward A.B., Wilson I.A., Palese P. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J. Virol. 2014;88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura G., Chai N., Park S., Chiang N., Lin Z., Chiu H., Fong R., Yan D., Kim J., Zhang J. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza a antibodies. Cell Host Microbe. 2013;14:93–103. doi: 10.1016/j.chom.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y., Cho M., Shore D., Song M., Choi J., Jiang T., Deng Y.-Q., Bourgeois M., Almli L., Yang H. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 2015;6:7708. doi: 10.1038/ncomms8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Kallewaard N.L., Corti D., Collins P.J., Neu U., McAuliffe J.M., Benjamin E., Wachter-Rosati L., Palmer-Hill F.J., Yuan A.Q., Walker P.A. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell. 2016;166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides the structural and functional characterization of a new broadly neutralizing antibody that utilizes the rare VH6-1 gene carries a low level of somatic mutations and has higher potency and breadth when compared to other anti-stem antibodies. The epitope recognized by this antibody is novel and encompasses the fusion peptide and the hydrophobic groove.
- 40••.Joyce M.G., Wheatley A.K., Thomas P.V., Chuang G.-Y., Soto C., Bailer R.T., Druz A., Georgiev I.S., Gillespie R.A., Kanekiyo M. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell. 2016;166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Joyce and co-authors isolated novel group 1 and group 2 influenza A-neutralizing antibodies from H5N1 vaccinees. The sequence and structural analysis of these and other published antibodies with similar breadth revealed the existence of three classes of group 1 and group 2 broadly neutralizing antibodies directed to the HA stem.
- 41.Okuno Y., Isegawa Y., Sasao F., Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Tharakaraman K., Subramanian V., Cain D., Sasisekharan V., Sasisekharan R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe. 2014;15:644–651. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper described that the group 2 anti-stem antibody CR8020 targets HA residues that are prone to antigenic drift and host selection pressure.
- 43.Andrews S.F., Huang Y., Kaur K., Popova L.I., Ho I.Y., Pauli N.T., Henry Dunand C.J., Taylor W.M., Lim S., Huang M. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Henry Dunand C.J., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y., Tan G.S., Cruz J., Hirsh A., Zheng N.-Y. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dunand and co-authors described the isolation of neutralizing and non-neutralizing protective antibodies after H7N9 vaccination. Of note, the broadly cross-reactive non-neutralizing antibodies generated were protective through Fc-mediated effector cell recruitment.
- 45•.Pappas L., Foglierini M., Piccoli L., Kallewaard N.L., Turrini F., Silacci C., Fernandez-Rodriguez B., Agatic G., Giacchetto-Sasselli I., Pellicciotta G. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]; This paper provides an extensive analysis of the genetic and maturation requirements for the generation of group 1 VH1-69 broadly neutralizing antibodies, revealing an unexpected redundancy in the affinity maturation process.
- 46.Avnir Y., Watson C.T., Glanville J., Peterson E.C., Tallarico A.S., Bennett A.S., Qin K., Fu Y., Huang C.-Y., Beigel J.H. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires: correlates with IGHV utilization shifts and varies by ethnicity. Sci. Rep. 2016;6:20842. doi: 10.1038/srep20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lingwood D., McTamney P.M., Yassine H.M., Whittle J.R.R., Guo X., Boyington J.C., Wei C.-J., Nabel G.J. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yewdell J.W. To dream the impossible dream: universal influenza vaccination. Curr. Opin. Virol. 2013;3:316–321. doi: 10.1016/j.coviro.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rott R., Becht H., Orlich M. The significance of influenza virus neuraminidase in immunity. J. Gen. Virol. 1974;22:35–41. doi: 10.1099/0022-1317-22-1-35. [DOI] [PubMed] [Google Scholar]
- 50.Wohlbold T.J., Krammer F. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses. 2014;6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doyle T.M., Li C., Bucher D.J., Hashem A.M., Van Domselaar G., Wang J., Farnsworth A., She Y.-M., Cyr T., He R. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem. Biophys. Res. Commun. 2013;441:226–229. doi: 10.1016/j.bbrc.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 52.Doyle T.M., Hashem A.M., Li C., Van Domselaar G., Larocque L., Wang J., Smith D., Cyr T., Farnsworth A., He R. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013;100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 53.Neirynck S., Deroo T., Saelens X., Vanlandschoot P., Jou W.M., Fiers W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 54.Grandea A.G., Olsen O.A., Cox T.C., Renshaw M., Hammond P.W., Chan-Hui P.-Y., Mitcham J.L., Cieplak W., Stewart S.M., Grantham M.L. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R., Song A., Levin J., Dennis D., Zhang N.J., Yoshida H., Koriazova L., Madura L., Shapiro L., Matsumoto A. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antivir. Res. 2008;80:168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 56.Throsby M., van den Brink E., Jongeneelen M., Poon L.L.M., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.-M., Santelli E., Stec B., Cadwell G., Ali M. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang T.T., Tan G.S., Hai R., Pica N., Petersen E., Moran T.M., Palese P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bournazos S., Dilillo D.J., Ravetch J.V. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J. Exp. Med. 2015;207:2395. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dilillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Leon P.E., He W., Mullarkey C.E., Bailey M.J., Miller M.S., Krammer F., Palese P., Tan G.S. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc. Natl. Acad. Sci. 2016;113:E5944–E5951. doi: 10.1073/pnas.1613225113. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper illustrates that the ADCC induced by broadly reactive antibodies requires the well-known interaction between the Fc of the HA-bound antibodies with the FcγR on the effector cells, but also the interaction between the HA and its sialic acid receptor on effector cells.
- 62.Cox F., Kwaks T., Brandenburg B., Koldijk M.H., Klaren V., Smal B., Korse H.J.W.M., Geelen E., Tettero L., Zuijdgeest D. HA antibody-mediated FcγRIIIa activity is both dependent on FcR engagement and interactions between HA and sialic acids. Front. Immunol. 2016;7:1333–1342. doi: 10.3389/fimmu.2016.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krammer F., Palese P. Universal influenza virus vaccines: need for clinical trials. Nat. Immunol. 2014;15:3–5. doi: 10.1038/ni.2761. [DOI] [PubMed] [Google Scholar]
- 64.Jegaskanda S., Weinfurter J.T., Friedrich T.C., Kent S.J. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J. Virol. 2013;87:5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bakkouri El K., Descamps F., De Filette M., Smet A., Festjens E., Birkett A., Van Rooijen N., Verbeek S., Fiers W., Saelens X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J. Immunol. 2011;186:1022–1031. doi: 10.4049/jimmunol.0902147. [DOI] [PubMed] [Google Scholar]
- 66.Dilillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016;126 doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subedi G.P., Barb A.W., Subedi G.P., Barb A.W. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. MAbs. 2016;8:1512–1524. doi: 10.1080/19420862.2016.1218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park H.I., Yoon H.W., Jung S.T. The highly evolvable antibody Fc domain. Trends Biotechnol. 2016;34:895–908. doi: 10.1016/j.tibtech.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 69••.He W., Tan G.S., Mullarkey C.E., Lee A.J., Lam M.M.W., Krammer F., Henry C., Wilson P.C., Ashkar A.A., Palese P. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 2016;113:11931–11936. doi: 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, He and co-authors have analyzed how epitope specificity may influence ADCC of influenza virus infected cells. Of note, they showed that anti-head and NA-inhibiting antibodies reduced and enhanced, respectively, the ADCC mediated by broadly neutralizing anti-stem antibodies.
- 70.Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., Davis D.M., Strominger J.L., Yewdell J.W., Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 71.Bar-On Y., Glasner A., Meningher T., Achdout H., Gur C., Lankry D., Vitenshtein A., Meyers A.F.A., Mandelboim M., Mandelboim O. Neuraminidase-mediated, NKp46-dependent immune-evasion mechanism of influenza viruses. Cell Rep. 2013;3:1044–1050. doi: 10.1016/j.celrep.2013.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mozdzanowska K., Feng J., Eid M., Zharikova D., Gerhard W. Enhancement of neutralizing activity of influenza virus-specific antibodies by serum components. Virology. 2006;352:418–426. doi: 10.1016/j.virol.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 73.Feng J.Q., Mozdzanowska K., Gerhard W. Complement component C1q enhances the biological activity of influenza virus hemagglutinin-specific antibodies depending on their fine antigen specificity and heavy-chain isotype. J. Virol. 2002;76:1369–1378. doi: 10.1128/JVI.76.3.1369-1378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Terajima M., Cruz J., Co M.D.T., Lee J.-H., Kaur K., Wrammert J., Wilson P.C., Ennis F.A. Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J. Virol. 2011;85:13463–13467. doi: 10.1128/JVI.05193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huber V., Lynch J., Brockman-Schneider C.J., Le J., Metzger D. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. J. Immunol. 2001;166:7381–7388. doi: 10.4049/jimmunol.166.12.7381. [DOI] [PubMed] [Google Scholar]
- 76.Jegerlehner A., Schmitz N., Storni T., Bachmann M.F. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004;172:5598–5605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 77.Khurana S., Loving C.L., Manischewitz J., King L.R., Gauger P.C., Henningson J., Vincent A.L., Golding H. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci. Transl. Med. 2013;5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- 78.Rajão D.S., Chen H., Perez D.R., Sandbulte M.R., Gauger P.C., Loving C.L., Shanks G.D., Vincent A. Vaccine-associated enhanced respiratory disease is influenced by haemagglutinin and neuraminidase in whole inactivated influenza virus vaccines. J. Gen. Virol. 2016;97:1489–1499. doi: 10.1099/jgv.0.000468. [DOI] [PubMed] [Google Scholar]
- 79.Monsalvo A.C., Batalle J.P., Lopez M.F., Krause J.C., Klemenc J., Hernandez J.Z., Maskin B., Bugna J., Rubinstein C., Aguilar L. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat. Med. 2011;17:195–199. doi: 10.1038/nm.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nguyen-Van-Tam J.S., Venkatesan S., Muthuri S.G., Myles P.R. Neuraminidase inhibitors: who, when, where? Clin. Microbiol. Infect. 2015;21:222–225. doi: 10.1016/j.cmi.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 81.Balasingam S., Wilder-Smith A. Randomized controlled trials for influenza drugs and vaccines: a review of controlled human infection studies. Int. J. Infect. Dis. 2016;49:18–29. doi: 10.1016/j.ijid.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 82.Wollacott A.M., Boni M.F., Szretter K.J., Sloan S.E., Yousofshahi M., Viswanathan K., Bedard S., Hay C.A., Smith P.F., Shriver Z. Safety and upper respiratory pharmacokinetics of the hemagglutinin stalk-binding antibody VIS410 support treatment and prophylaxis based on population modeling of seasonal influenza A outbreaks. EBIOM. 2016;5:147–155. doi: 10.1016/j.ebiom.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramos E.L., Mitcham J.L., Koller T.D., Bonavia A., Usner D.W., Balaratnam G., Fredlund P., Swiderek K.M. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J. Infect. Dis. 2015;211:1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 84.de Jong M.D., Ison M.G., Monto A.S., Metev H., Clark C., O'Neil B., Elder J., McCullough A., Collis P., Sheridan W.P. Evaluation of intravenous peramivir for treatment of influenza in hospitalized patients. Clin. Infect. Dis. 2014;59:e172–e185. doi: 10.1093/cid/ciu632. [DOI] [PubMed] [Google Scholar]
- 85.Brandenburg B., Koudstaal W., Goudsmit J., Klaren V., Tang C., Bujny M.V., Korse H.J.W.M., Kwaks T., Otterstrom J.J., Juraszek J. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One. 2013;8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan G.S., Krammer F., Eggink D., Kongchanagul A., Moran T.M., Palese P. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J. Virol. 2012;86:6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tharakaraman K., Subramanian V., Viswanathan K., Sloan S., Yen H.-L., Barnard D.L., Leung Y.H.C., Szretter K.J., Koch T.J., Delaney J.C. A broadly neutralizing human monoclonal antibody is effective against H7N9. Proc. Natl. Acad. Sci. 2015;112:10890–10895. doi: 10.1073/pnas.1502374112. [DOI] [PMC free article] [PubMed] [Google Scholar]