Abstract
The re-emerging porcine epidemic diarrhea virus (PEDV) variant related diarrhea has been documented in China since late 2010 and now with global distribution. Currently, a virulent PEDV CH/YNKM-8/2013 and a CV777 vaccine strain-like AH-M have been successfully isolated from the clinical samples. To dissect out the underlying pathogenic mechanism of virulent PEDV and clarify the differences between virulent and CV777 vaccine strain-like PEDV infections, we performed an iTRAQ-based comparative quantitative proteomic study of Vero cells infected with both PEDV strains. A total of 661 and 474 differentially expressed proteins were identified upon virulent and CV777 vaccine strain-like isolates infection, respectively. Ingenuity Pathway Analysis was employed to investigate the canonical pathways and functional networks involved in both PEDV infections. Comprehensive studies have revealed that the PEDV virulent strain suppressed protein synthesis of Vero cells through down-regulating mTOR as well as its downstream targets 4EBP1 and p70S6K activities, which were validated by immunoblotting. In addition, the virulent strain could activate NF-κB pathway more intensively than the CV777 vaccine strain-like isolate, and elicit stronger inflammatory cascades as well. These data might provide new insights for elucidating the specific pathogenesis of PEDV infection, and pave the way for the development of effective therapeutic strategies.
Biological significance
Porcine epidemic diarrhea is now worldwide distributed and causing huge economic losses to swine industry. The immunomodulation and pathogenesis between PEDV and host, as well as the difference between virulent and attenuated strains of PEDV infections are still largely unknown. In this study, we presented for the first application of proteomic analysis to compare whole cellular protein alterations induced by virulent and CV777 vaccine strain-like PEDV infections, which might contribute to understand the pathogenesis of PEDV and anti-viral strategy development.
Keywords: Porcine epidemic diarrhea virus (PEDV), Isobaric tags for relative and absolute quantitation (iTRAQ), Proteomics, Pathway analysis
Graphical abstract
Highlights
-
•
Vero cells proteome was individually analyzed upon virulent and attenuated PEDV infections.
-
•
Many pathways and interactive networks were constructed based on differentially expressed proteins.
-
•
Virulent PEDV strain suppressed mTOR as well as its downstream targets 4EBP1 and p70S6K activities.
-
•
Virulent PEDV strain activated NF-κB pathway more intensively than the attenuated isolate.
1. Introduction
Porcine epidemic diarrhea (PED) is an acute and contagious enteric disease with high mortality in sucking piglets, causing huge economic losses to swine industry [1], [2]. The etiological agent, porcine epidemic diarrhea virus (PEDV), is an enveloped, single-stranded positive-sense RNA virus which belongs to Coronaviridae family [3]. The viral genome is approximately 28 kb, encodes at least seven open reading frames (ORFs). The two largest ORFs (ORF1a and ORF1b) located in the two-thirds of the genome downstream of the 5′UTR encode the viral replicase. The other one-third genome encodes the structural and accessory proteins including spike (S) glycoprotein, small envelope (E) protein, membrane (M) protein, nucleocapsid (N) protein, and accessory protein (ORF3) [4], [5], [6]. In late 2010, severe outbreak of PED has occurred in China even if the herds were vaccinated, which was caused by a variant of highly virulent PEDV [2], [4], [7]. The new PED outbreaks were also reported in the United States, Canada, Vietnam, and Korea [8], [9], [10], [11]. Although many relevant studies have been carried out, they are mainly focused on the viral isolation and molecular epidemiology survey [5], [12]. The pathogenic mechanism and immune regulation between PEDV and host, as well as the difference between virulent and attenuated strains of PEDV infection remain still largely unknown.
Proteomic approaches are important tools in the comprehensive analysis of host responses to viral infection, such as foot-and-mouth disease virus (FMDV) [13], Marek's disease virus (MDV) [14], severe acute respiratory syndrome-associated coronavirus (SARS-CoV) [15], infectious bronchitis virus (IBV) [16], and highly pathogenic avian influenza (HPAI) virus [17]. Isobaric tags for relative and absolute quantitation (iTRAQ) combined with LC–MS/MS analysis are an emerging quantitative proteomic method with great accuracy, sensitivity, separating capacity, and high throughput for protein identification and quantification [18]. It has been applied to many virus–host interaction studies, which include porcine circovirus type 2 (PCV2) [19], transmissible gastroenteritis virus (TGEV) [20], hepatitis B virus (HBV) [21], and porcine respiratory and reproductive syndrome virus (PRRSV) [22]. These studies have outlined the dynamic interaction between host and pathogen, and promote a better understanding of the pathogenesis involved in viral infection.
Until now, reports about the interaction between PEDV and host interactions are rarely covered. It is well known that Vero cells are most commonly used not only for PEDV isolation, propagation and basic research, but also for many different human pathogens, including SARS-CoV [23], influenza virus [24], and respiratory syncytial virus [25]. Vero cells are interferon-deficient when infected by viruses while they still have the interferon α/β receptor, permitting productive growth of pathogens and reducing bystander effect in uninfected cells [16]. Although this may limit the comprehensive understanding of innate immune responses of virus-infected Vero cells, a map of Vero cell proteome is tremendously valuable. Recently, the protein changes of Vero cells contributing to a virulent PEDV variant and a classic PEDV strain CV777 were investigated, respectively [26], [27]. However, comparative proteomic analysis focused on PEDV strains with different virulence under the same circumstances has not been reported. In this study, we employed a quantitative proteomic analysis based on iTRAQ coupled with LC–MS/MS, to capture the differential protein expression profiles of Vero cells upon infection with virulent and CV777 vaccine strain-like PEDV strains. These differentially expressed proteins were classified into many signaling pathways, and interacting networks involved in PEDV infection were also constructed. Importantly, we noticed that the virulent PEDV strain could suppress protein synthesis of infected Vero cells through mTOR and its downstream targets inactivation. Moreover, the virulent strain could induce stronger inflammatory responses than the CV777 vaccine strain-like strain. Overall, this is the first report on comparison of whole cellular protein alterations induced by virulent and CV777 vaccine strain-like strains of PEDV by application of iTRAQ based proteomic analysis, which might accelerate our understanding of the pathogenic mechanisms involved in different PEDV infections.
2. Materials and methods
2.1. Cells and viruses
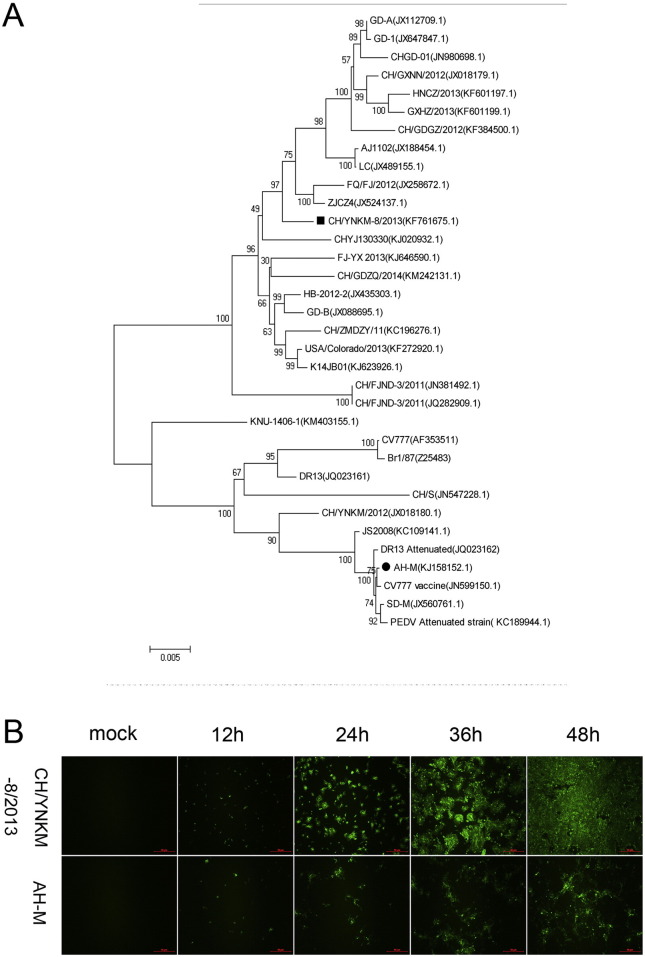
African green monkey kidney (Vero-E6) cells were purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C with 5% CO2. One PEDV strain CH/YNKM-8/2013 (Accession no. KF761675) was isolated from a sucking piglet with acute diarrhea. Another PEDV strain AH-M (Accession no. KJ158152) was isolated from piglets with mild diarrhea signs without mortality compared to the watery diarrhea and high mortality, and classified into the same cluster with CV777 vaccine and other attenuated strains. Phylogenetic analysis was constructed based on nucleotide sequences of full-length spike gene and depicted in Fig. 1A. The virus titers were determined by Reed–Muench method.
Fig. 1.
Phylogenetic analysis and virus infection. (A) Phylogenetic trees of PEDV based on nucleotide sequences of full-length spike genes. (B) Confirmation the proliferation of PEDV strain CH/YNKM-8/2013 or AH-M by IFA staining in infected Vero cells at 12, 24, 36, and 48 h pi, respectively. Mock-infected cells at 36 h pi were set as control. Scale bar = 50 μm.
2.2. Virus inoculation
Vero cells were cultured for nearly 24 h for 80% confluence and washed twice with serum-free medium. Then, the cells were infected with PEDV strains CH/YNKM-8/2013 and AH-M, respectively, at a multiplicity of infection (MOI) of 0.1 and incubated with serum-free DMEM containing 8 μg/mL trypsin (Invitrogen). The PEDV- or mock-infected cells were collected at 36 h postinfection (h pi). Each group was processed with three independent biological replicates. Viral propagation was confirmed by daily observation of the cytopathic effect (CPE), virus titer determination and indirect immunofluorescence assay (IFA) using a monoclonal antibody against PEDV S protein.
2.3. Protein isolation, digestion, and labeling with iTRAQ reagents
The PEDV- and mock-infected cell samples were collected with a cell scraper, centrifuged at 300 g for 10 min, and washed twice with ice-cold PBS containing 1 mM pervanadate and 1 mM sodium fluoride. The collected cells were lysed in 150 μL of RIPA lysis buffer containing 1 mM PMSF, and the soluble protein fraction was harvested by centrifugation at 12,000 g for 20 min at 4 °C after ultrasonication treatment for eight times. The protein concentration was determined by BCA Protein Quantitation Kit (Sangon Biotech, Shanghai, China). After reduction and cysteine-blocking as described in the iTRAQ protocol (AB Sciex, Concord, ON), solutions containing 100 μg protein were digested overnight at 37 °C with sequence grade modified trypsin (Promega, Madison, WI) and then labeled with different iTRAQ tags as follows: iTRAQ 113 (IT113) and iTRAQ 114 (IT114) for mock-infected samples; iTRAQ 115 (IT115) and iTRAQ 116 (IT116) for CH/YNKM-8/2013 strain infected samples; iTRAQ 117 (IT117) and iTRAQ 118 (IT118) for AH-M strain infected samples. The labeled samples were then mixed and dried with a rotary vacuum concentrator.
2.4. LC–MS/MS analysis
Strong cation exchange (SCX) chromatography was performed with a 20AB high-performance liquid chromatography (HPLC) system (Dionex Ultimate 3000, USA) (Phenomenex columns; Gemini-NX 3u C18 110A; 150 mm × 2.00 mm). The iTRAQ labeled peptides were redissolved in buffer A (20 mM HCOONH4, 2 M NaOH, pH 10), followed by a gradient elution with buffer B (20 mM HCOONH4, 2 M NaOH, 80% acetonitrile [ACN], pH 10) (5% B for 10 min, 5–15% B for 5 min, 15–50% B for 45 min, 50–90% B for 10 min, 90% B for 10 min, 90–5% B for 10 min) at a flow rate of 200 μL/min. The whole elution process was monitored by the absorbance of 214 nm and a total of 24 fractions were collected. Each fraction was acidulated with 50% TFA and lyophilized.
The fractions above were dissolved in aqueous solution containing 0.1% FA and 2% ACN, and then centrifuged at 12,000 g for 10 min at 4 °C. The peptides were introduced into a reverse-phase C18 trapping column (100 μm inner diameter × 100 mm, 3 μm resin from Michrom Bioresources, Auburn, CA) through the 20AB HPLC system (Dionex Ultimate 3000, USA). The mobile phases used were composed of solvent A (0.1% formic acid, 5% acetonitrile, and 95% H2O) and solvent B (0.1% formic acid, 95% acetonitrile, 5% H2O). The gradient run was from 5–40% B for 70 min with a flow rate of 300 nL/min. MS/MS was performed with a Triple TOF 5600 mass spectrometer (AB Sciex). During data acquisition, machines were set to the information dependent mode and MS spectra were acquired by the mass range of 400–1250 m/z (high resolution ≥ 30,000) with 250 ms accumulation time. A maximum of 20 MS/MS (resolution ≥ 15000) were chosen from each MS spectrum with 100 ms minimum accumulation time for each precursor and dynamic exclusion was fixed to 20 s.
2.5. Data analysis
Protein identification and quantification were performed with the ProteinPilot™ Software (Version: 4.5; Revision number: 1656; Applied Biosystems) using Paragon™ Algorithm (4.5.0.0, 1654) as searching engine. MS/MS data were searched against Macaca mulatta Uniprot database (August 15, 2014, containing 35,536 proteins, http://www.uniprot.org/proteomes/UP000006718). The cysteine alkylation by methyl methanethiosulfonate and biological modifications in the algorithm were programed for the search parameters. The protein confidence threshold cutoff was set to 1.3 (unused) to assume at least two peptides with 95% confidence interval, and all proteins identified must be above 95% confidence. In addition, false discovery rate (FDR) analysis was applied for the protein identification by searching against a concatenated reversed database. The peptide for quantification was automatically selected by Paragon™ algorithm to calculate the reporter peak area, error factor (EF) and p value. Proteins with fold change > 1.5 or < 0.67 and a pvalue < 0.05 were considered as significantly different expressions. Auto bias-corrected were executed to decrease the artificial error.
2.6. Immunofluorescence assay
CH/YNKM-8/2013 and AH-M infected Vero cells were fixed with cold ethanol at 12, 24, 36, and 48 h pi, respectively, followed by incubation with mouse monoclonal antibody against PEDV S protein for 1 h at 37 °C, and cells were washed three times with PBS before further incubation with FITC-conjugated anti-mouse IgG (Southern Biotech) in the dark. After washing five times with PBS, the images were captured under a fluorescence microscopy.
2.7. RNA extraction and real-time PCR analysis
Total cellular or viral RNA was extracted from the PEDV-infected and mock-infected Vero cells or supernatants using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. After the RNA was reverse-transcribed to cDNA using olige (dT) as the primer (Invitrogen), quantitative real-time PCR assay was performed using an Applied Biosystems ViiA 7 real-time PCR system. Relative quantitative real-time PCR was reacted in a 10 μL volume containing 50 ng of the cDNA template, FastStart DNA Master SYBR Green I Mix reagent kit (Roche) and 100 nM of each primer under conditions of 95 °C 10 min, then 40 cycles of 95 °C 30 s, 58 °C 30 s, and 72 °C for 30 s, and melting curves were obtained. Absolute quantitative real-time PCR was performed in a 10 μL volume containing cDNA template, FastStart Universal Probe Master (ROX) (Roche), 100 μM probe and primer. The cycle conditions were 50 °C 2 min, 95 °C 10 min, and 40 cycles of 95 °C 15 s, 56 °C 30 s, and 72 °C 31 s. For the standard curve, serial dilutions of a plasmid constructed by inserting the PEDV M gene into the pMD™18-T vector (TAKARA) were used to quantify the virus genomic copy number. Each cDNA sample was amplified in triplicate. The data analysis was performed using the Applied Biosystems ViiA 7 real-time PCR system software (Applied Biosystems).
2.8. Western blot analysis
The infected and mock-infected cells were collected at 36 h pi. Equivalent amounts of cell lysates from each sample were mixed with 5 × sample loading buffer and boiled for 10 min, then separated by 12% SDS-PAGE. The proteins were electrotransferred to 0.45 μm PVDF membranes (Millipore). Membranes were blocked with 5% (w/v) skim milk-TBST at room temperature for 2 h and then incubated overnight at 4 °C with primary antibodies to calnexin (Cell Signaling Technology, Inc.), phospho-p70S6K (Cell Signaling Technology, Inc.), annexin A1 (Protein-Tech, Inc.), and other antibodies purchased from ABclonal Technology. The membranes were washed with TBST and then incubated with horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG (ABclonal, Inc.) or goat anti-mouse IgG (ABclonal, Inc.) at 37 °C for 1 h. The signals were obtained using the clarity-enhanced chemiluminescence (ECL) reagent (Bio-Rad, Hercules, CA).
3. Results
3.1. Kinetics of PEDV multiplication in Vero cells
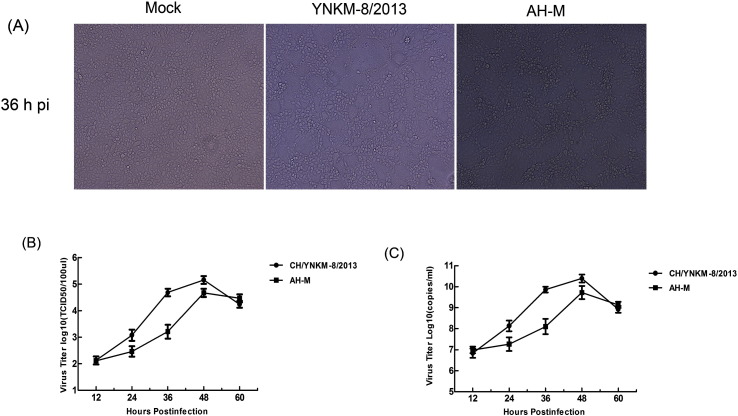
To determine the kinetics of PEDV propagation in Vero cells, CPEs and viral protein expressions, as well as viral titers were all monitored at different time-points after infection. As shown in SI Fig. 1(A), the CPEs caused by CH/YNKM-8/2013 were featured by vacuolation, formation of syncytia, and fusion of cells; while those by AH-M strain mainly involved cell aggregation, and net-like distribution. According to the PEDV growth curve in Vero cells, we obtained that both of these two strains peaked at 48 h pi, and then gradually declined (SI Fig. 1(B) and (C)). In addition, we also noticed that the cellular cytoskeleton of these two strains was severely collapsed at 48 h pi as a result of obvious CPE. The result of IFA indicated that PEDV began to replicate at 12 h pi, and most of the cells were infected at 36 h pi (Fig. 1B). In general, no significant host cell cytoskeleton collapse or membrane rearrangement observed but with high virus yield is regarded as the optimal time for proteomic analysis.[20], [28] Taken these substantial evidences into consideration, cell samples at 36 h pi were chosen for further proteomic analysis.
Supplementary Fig. S1.
The kinetics of PEDV CH/YNKM-8/2013 and AH-M infections in Vero cells, respectively.
3.2. Protein profile by iTRAQ combined with LC–MS/MS analysis
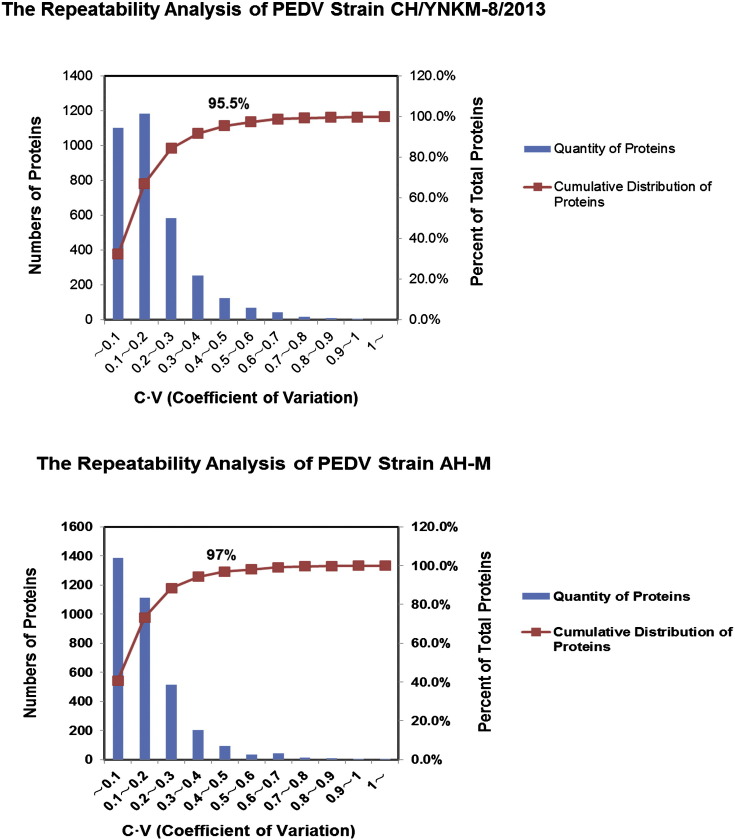
The protein extracts were prepared from PEDV- and mock-infected Vero cells as described in the experimental section. In total, 3406 and 3409 proteins were identified and quantified by iTRAQ coupled with LC–MS/MS analysis in Vero cells infected with PEDV strains CH/YNKM-8/2013 and AH-M, respectively. Significantly up/down regulated proteins were determined by a fold change > 1.5 or < 0.67 and a pvalue < 0.05. As for this, 354 proteins were significantly up-regulated and 307 proteins were markedly down-regulated during PEDV strain CH/YNKM-8/2013 infection (SI Table 1). Meanwhile, 295 up-regulated and 179 down-regulated proteins were generated in cells infected with PEDV strain AH-M (SI Table 1). Notably, three biological replicates among PEDV- or mock-infected groups were well-mixed when collecting samples and two technical replicates were carried to improve the reliability of our data. The repetitive analysis was shown in SI Fig. 2 (Supporting Information).
Supplementary Fig. S2.
Repeatability analysis of data obtained from iTRAQ coupled with LC–MS/MS proteomic approach.
3.3. Bioinformatics analysis of the Vero cell proteome
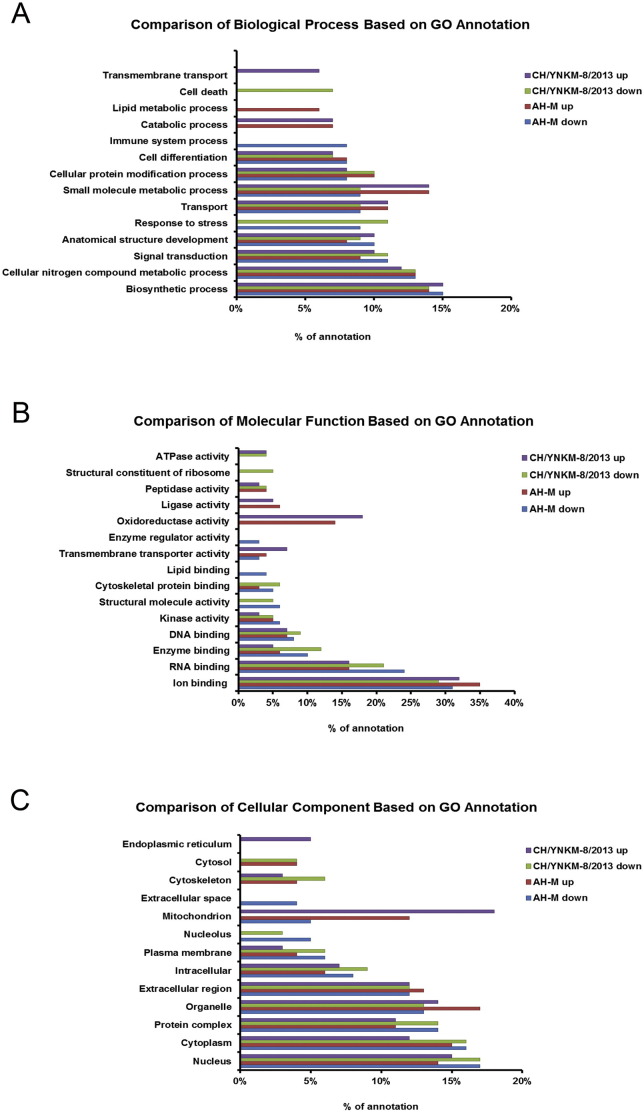
These differentially expressed proteins were assigned into different biological processes, molecular functions, and cellular components by searching the Gene Ontology and Uniprot databases. GO annotation comparison was performed to elucidate the characteristics of total altered proteins in Vero cells induced by CH/YNKM-8/2013 or AH-M infection, which might associate with virulence and pathogenicity. For biological process annotation, proteins in both groups were mainly involved in biosynthesis, metabolic and signal transduction processes, but “transmembrane transport” and “cell death” were only detected in CH/YNKM-8/2013 infection (Fig. 2A). The molecular function comparison indicated that ion and RNA binding accounted for large proportions in both PEDVs infection. Proteins associated with “ATPase activity” and “structural constituent of ribosome” were not identified in AH-M infected cells. Moreover, “structural molecule activity” relevant proteins were all down-regulated, while “oxidoreductase activity” relevant proteins were all up-regulated in both groups tested (Fig. 2B). In terms of cellular components annotation, proteins located in mitochondrion were both up-regulated, but endoplasmic reticulum associated proteins were merely detected with increased expression upon CH/YNKM-8/2013 infection. Nucleolus proteins identified were all decreased in both viruses infection cells (Fig. 2C). Together, the GO annotation comparison could provide a comprehensive overview of the differences regarding molecular characterization upon virulent and CV777 vaccine strain-like strains of PEDV infection, which might potentially involve in PEDV pathogenesis.
Fig. 2.
Comparison of GO term annotation upon the differentially expressed proteins in Vero cells infected with PEDV strain CH/YNKM-8/2013 or AH-M. (A) Biological processes, (B) Molecular functions, and (C) Cellular components.
As compared to the human genome, the monkey genome database was poorly annotated and many proteins were unassigned or uncharacterized. Here, gene identifications of the differentially expressed proteins were converted to human protein gi numbers, followed by uploading into the Ingenuity Pathways Analysis (IPA) tool. Function classifications, signaling pathways, and interacting networks were constructed based on the underlying biological evidence from the literature database.
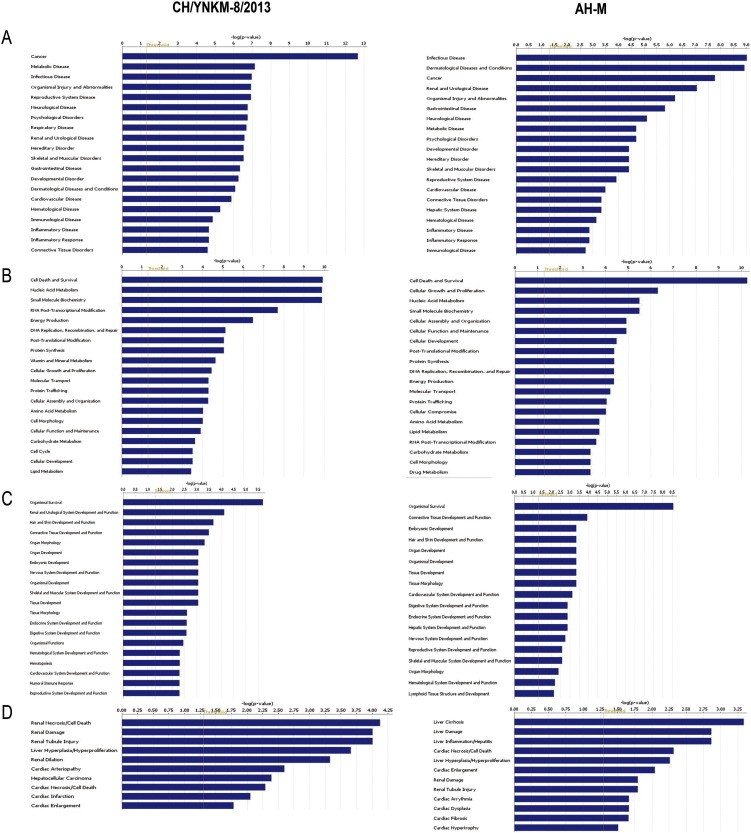
According to the function classification by IPA, these significantly differentially expressed proteins separately induced by CH/YNKM-8/2013 and AH-M, could be divided into four distinctive functional sets: (A) diseases and disorders; (B) molecular and cellular functions; (C) physiological system development and functions; (D) toxicity functions (shown in Fig. 3 and SI Table 2, p < 0.05). These proteins are involved in many biological processes and functions, including infectious disease, cancer, cell death and survival, cell growth and proliferation, post-translational modification, and organismal survival. It was noteworthy that several biological functions associated with gastrointestinal disease, digestive system development and function, and inflammatory disease were also mapped during PEDV infection, which were closely related with PED progression. What is more, the immunological disease, as well as inflammatory response and disease were more highlighted by CH/YNKM-8/2013 than by AH-M.
Fig. 3.
Functional characterization of significantly altered proteins after PEDV strains CH/YNKM-8/2013 and AH-M infection, respectively. (A) Diseases and disorders, (B) molecular and cellular functions, (C) physiological system development and functions, and (D) toxicity functions. More information is available in SI Table 2.
Functional characterization of significantly altered proteins after PEDV strains CH/YNKM-8/2013 and AH-M infection, respectively. (A) Diseases and disorders, (B) molecular and cellular functions, (C) physiological system development and functions, and (D) toxicity functions. More information is available in SI Table 2.
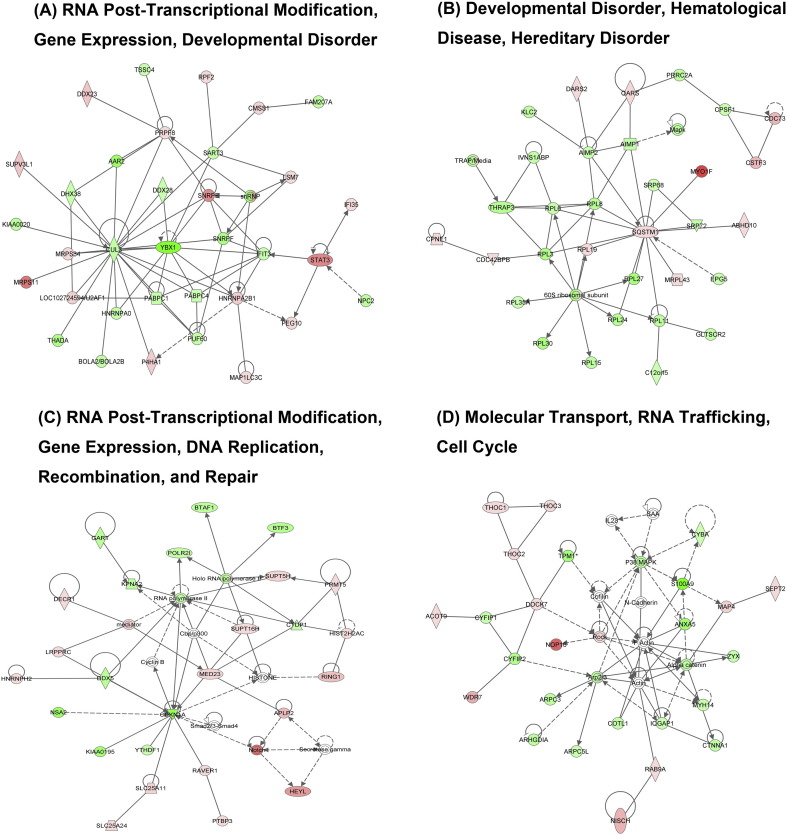
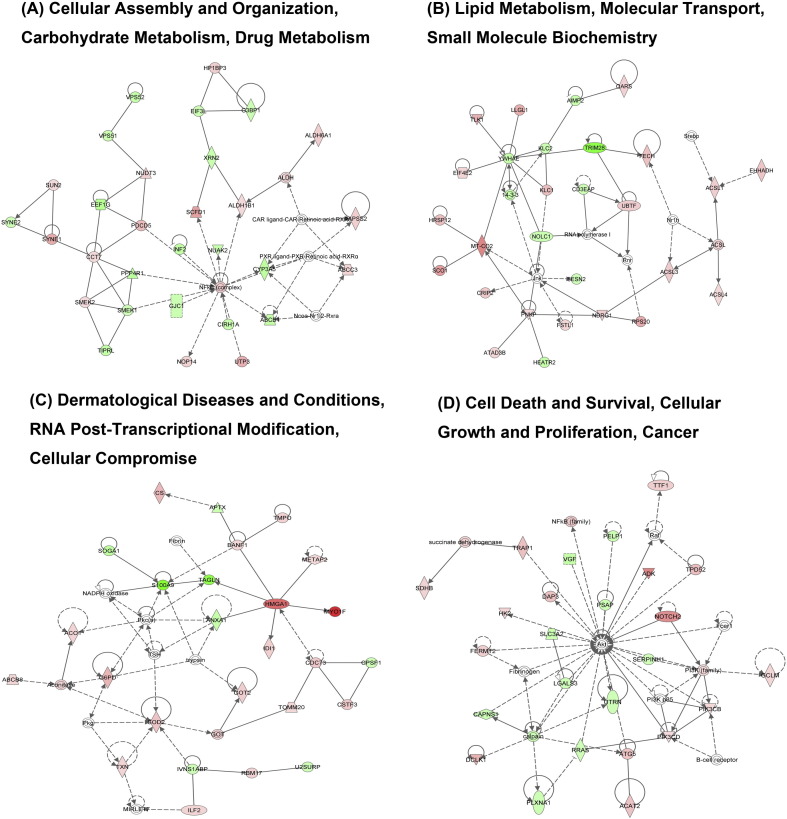
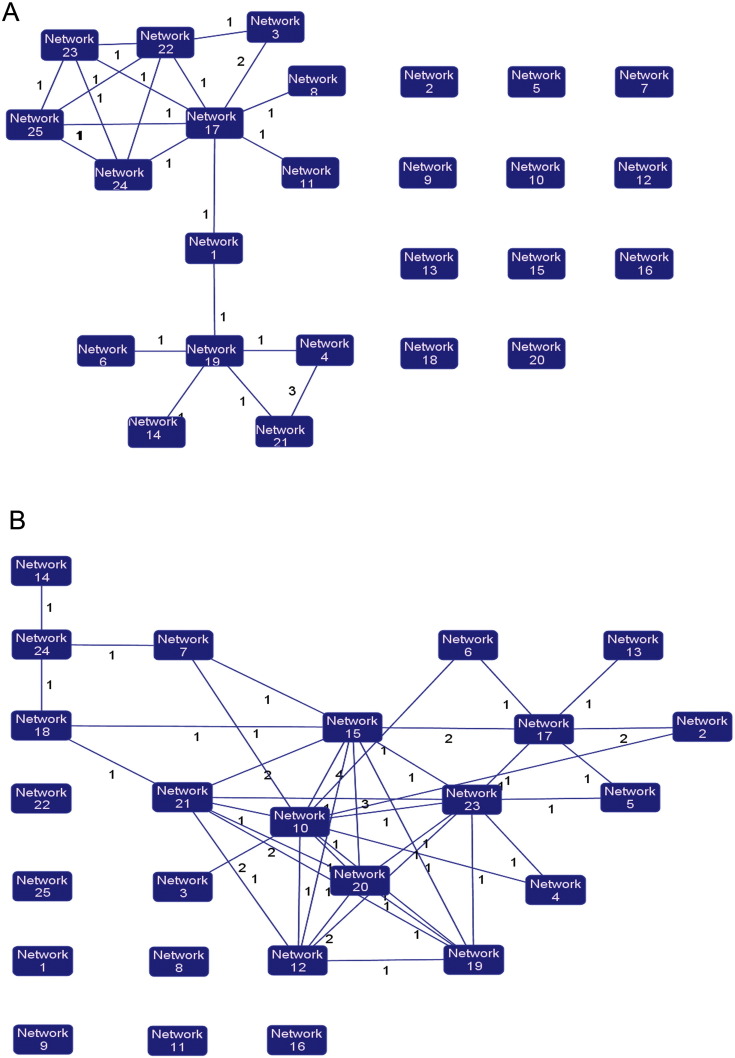
IPA tool was used to explore the potential specific functional networks for proteins that changed in abundance. In total, 25 protein networks were mapped based on these differentially regulated proteins upon CH/YNKM-8/2013 and AH-M infection, respectively (SI Table 4). To investigate the underlying biologically functional differences that may be related to viral infection, four strongly represented networks of interest were depicted here. For CH/YNKM-8/2013 infection, the four networks were (1) RNA Post-Transcriptional Modification, Gene Expression, Developmental Disorder (34 molecules; score 55; Fig. 4A); (2) Developmental Disorder, Hematological Disease, Hereditary Disorder (32 molecules; score 51; Fig. 4B); (3) RNA Post-Transcriptional Modification, Gene Expression, DNA Replication, Recombination, and Repair (26 molecules; score 35; Fig. 4C); (4) Molecular Transport, RNA Trafficking, Cell Cycle (25 molecules; score 34; Fig. 4D). Another four representative networks existed in Vero cells infected with PEDV strain AH-M were (1) Cellular Assembly and Organization, Carbohydrate Metabolism, Drug Metabolism (30 molecules; score 50; Fig. 5A); (2) Lipid Metabolism, Molecular Transport, Small Molecule Biochemistry (28 molecules; score 45; Fig. 5B); (3) Dermatological Diseases and Conditions, RNA Post-Transcriptional Modification, Cellular Compromise (26 molecules; score 38; Fig. 5C); (4) Cell Death and Survival, Cellular Growth and Proliferation, Cancer (25 molecules; score 38; Fig. 5D). Among the proteins present in these networks analysis, the up-regulated proteins are shown in shades of red and the down-regulated ones are in green. The proteins existed in the network but not identified in our analysis are depicted in white. It was worth mentioning that the biological processes involved in RNA post-transcriptional modification and developmental disorder were highlighted in PEDV strain CH/YNKM-8/2013 infection, while cellular assembly and organization, and metabolism were underscored in AH-M infection.
Fig. 4.
Ingenuity Pathway Analysis of proteins significantly altered in Vero cells upon PEDV strain CH/YNKM-8/2013 infection. Red, up-regulated proteins; green, down-regulated proteins; and white, proteins known to be in the network but not identified in this study. The color depth indicates the magnitude of the change in protein expression. The shapes are indicative of the molecular class. Lines with arrows connecting between the molecules indicate the molecular relationships. Solid lines indicate direct interactions and dashed lines indicate indirect interactions.
Fig. 5.
Ingenuity Pathway Analysis of proteins significantly altered in PEDV strain AH-M infected Vero cells. Red, up-regulated proteins; green, down-regulated proteins; and white, proteins known to be in the network but not identified in this study. The color depth indicates the magnitude of the change in protein expression. The shapes are indicative of the molecular class. Lines with arrows connecting between the molecules indicate the molecular relationships. Solid lines indicate direct interactions and dashed lines indicate indirect interactions.
3.4. Validation of protein identification and quantification by western blot
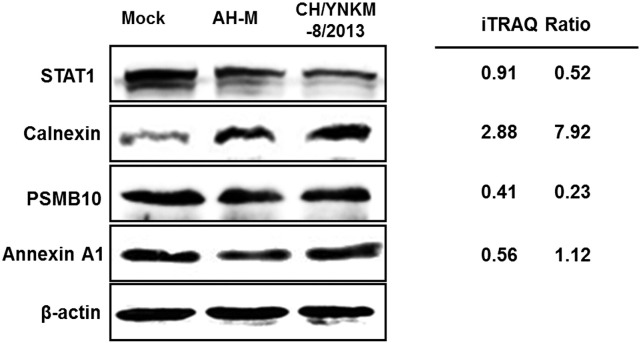
To confirm the differential expression of cellular proteome during PEDV infection, four proteins (STAT1, Calnexin, PSMB10, and Annexin A1) based on interest and different ratios were selected to be analyzed by western blot (shown in Fig. 6 ). Although, the verified proteins were relatively limited, the ratios from immunoblotting analysis were in accordance with those obtained from iTRAQ approach.
Fig. 6.
Validation of differentially expressed proteins with immunoblotting analysis. Immunoblotting analysis of proteins (STAT1, Calnexin, PSMB10, and Annexin A1) in PEDV-infected (CH/YNKM-8/2013 or AH-M) or mock-infected Vero cells. iTRAQ ratios (infection/mock) were shown on the right side. The β-actin protein was used as a control. Each experiment was repeated three times, and here are the representative results.
3.5. PEDV infection induces the disruption of translational regulation and inflammatory responses
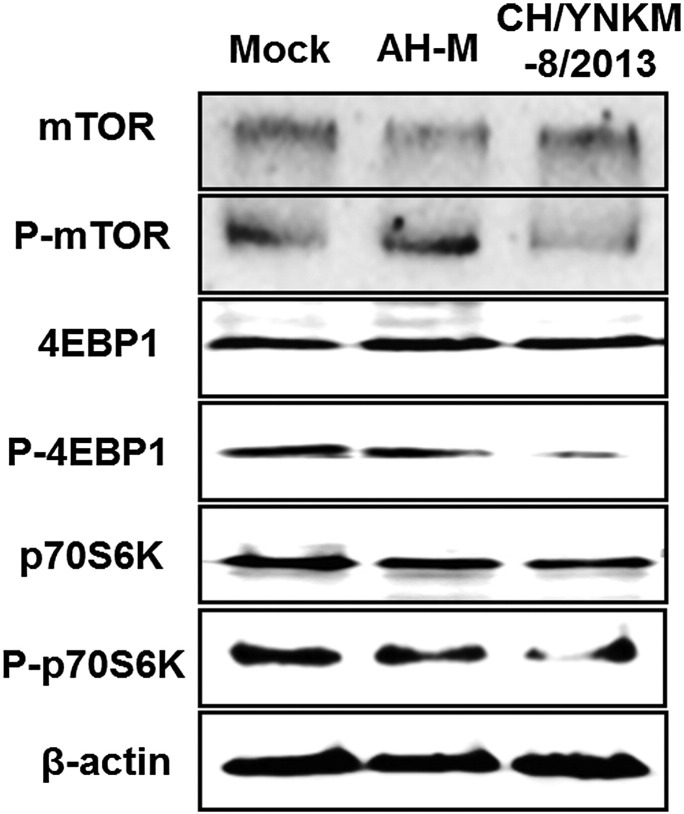
Several translational regulation related pathways were more highlighted for PEDV strain CH/YNKM-8/2013 infection by IPA, such as Mitochondrial Dysfunction (p value = 6.31 × 10− 15), EIF2 signaling (p value = 3.16 × 10− 10, z-score = − 2.67), regulation of eIF4 and p70S6K Signaling (p value = 9.33 × 10− 6, z-score = − 2.00), and mTOR Signaling (p value = 1.26 × 10− 2, z-score = − 2.00). EIF2 (eukaryotic initiation factor-2) initiation complex integrates a diverse of stimuli to regulate both global and specific mRNA translation. Eukaryotic translation initiation factor 4 (eIF4) and ribosomal protein S6 kinase (p70S6K), downstream effectors of mTOR (mammalian target of rapamycin), play critical roles in translational regulation. To assess the alteration of mTOR signaling during PEDV multiplication, the total expression and phosphorylation profiles of mTOR, 4EBP1 (eIF4E binding protein 1), and p70S6K were analyzed by western blot. As shown in Fig. 7 , the phosphorylation level of mTOR was decreased, followed by reduced expression of phosphorylated 4EBP1 and p70S6K mediated by CH/YNKM-8/2013 infection but not by AH-M. And this result was in agreement with the proteomic data acquired using iTRAQ labeled LC–MS/MS approach.
Fig. 7.
Virulent PEDV infection induced the inactivation of mTOR, 4EBP1 and p70S6K. Vero cells were infected with PEDV strain CH/YNKM-8/2013 or AH-M at 0.1 MOI or mock-infected. Cells were harvested and processed at 36 h pi for immunoblotting. The protein levels of total mTOR, 4EBP1, p70S6K, and phospho-mTOR (Ser2448), phospho-4EBP1 (Thr36), and phospho-p70S6K (Thr389) were detected in PEDV-infected or mock-infected Vero cells with available antibodies.
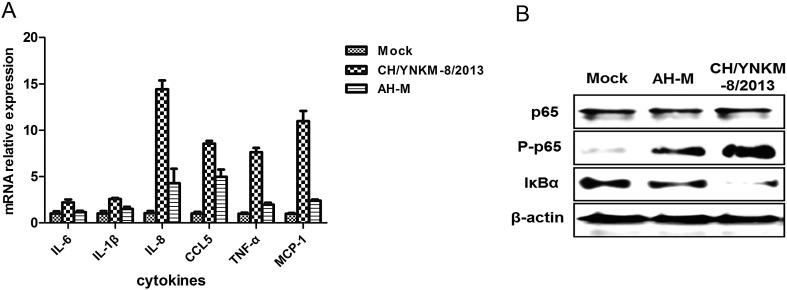
Another thrilling finding was that many immune regulation and inflammation relevant pathways were also constructed by IPA analysis, including NF-κB signaling pathway, JAK/STAT signaling pathway, PI3K/AKT pathway, IL-6 and IL-8 pathways. While considering the cytokines as indicators of immune dysfunction and inflammation, we examined the cytokine expressions of PEDV- or mock-infected Vero cells using quantitative real-time RT-PCR. It was demonstrated that the cytokines detected were more sharply increased after CH/YNKM-8/2013 infection (Fig. 8A). NF-κB signaling pathway is prevalently recognized to be closely associated with inflammatory responses. Its activation is accompanied by phosphorylation and nuclear translocation of p65, and degradation of IκBα. Immunoblotting analysis indicated that the phosphorylation of p65 was significantly enhanced. Meanwhile, IκBα was almost undetectable during CH/YNKM-8/2013 infection (Fig. 8B). No such obvious findings were presented upon AH-M strain infection. These substantial evidences were also coincided with the outcomes generated from proteomic analysis.
Fig. 8.
Confirmation of immune dysregulation in Vero cells upon PEDV infection with real-time RT-PCR or immunoblotting analysis. (A) Real-time RT-PCR analysis of cytokine expressions in Vero cells infected or mock-infected with PEDV at 0.1 MOI. The cells were collected at 36 h pi for real-time RT-PCR to analyze the relative expression of the specified cytokines mRNA. (B) PEDV infection induced the phosphorylation of p65 and degradation of IκBα. Vero cells were infected or mock-infected with PEDV strain CH/YNKM-8/2013 or AH-M at 0.1 MOI and harvested at 36 h pi for immunoblotting.
4. Discussion
To date, although the proteomic technology has been widely applied for virus–host interaction studies, little information has been available about the protein profile of host cells infected with PEDV. Here, iTRAQ combined with LC–MS/MS was applied for the comparative proteomic analysis of Vero cells infected with virulent and CV777 vaccine strain-like strains of PEDV, due to its superior performance in simultaneous comparison of multi-samples with wide dynamic protein abundance [29]. In this study, 661 and 474 differentially regulated proteins were identified in Vero cells infected by PEDV CH/YNKM-8/2013 and AH-M infection, respectively, on the basis of a fold change > 1.5 or < 0.67 and p value < 0.05. The immunoblotting results were also in accordance with the proteomic analysis. The differences regarding cellular responses were mainly involved in protein synthesis, immune regulation, cellular assembly and organization, signal transduction, and apoptosis. These data could provide comprehensive information to reveal the PEDV pathogenic mechanisms associated with virulent and CV777 vaccine strain-like strains of PEDV infection.
4.1. Cellular protein synthesis
The host protein synthesis machinery can be subverted by viruses to synthesize viral proteins and stifle host innate defense to facilitate its propagation [30]. One important finding in this study was that virulent PEDV infection suppressed ribosomal protein S6 kinase (p70S6K) and eIF4E-binding protein 1 (4EBP1) phosphorylation through mTOR complex inactivation. Similarly, eIF2 (eukaryotic initiation factor 2) signaling was also dramatically inhibited during virulent PEDV infection, which was documented to be advantageous for some virus propagation [31], [32], [33]. Mammalian target of rapamycin (mTOR), a key downstream effector of PI3K/AKT pathway, is commonly targeted by viral proteins for its central role in protein synthesis and cell growth with two different complexes (mTORC1 and mTORC2) [34], [35]. mTORC1 phosphorylates the p70S6K and 4EBP1, which enhance mRNA and cap-dependent translation, respectively [30], [36]. mTOR signaling was manipulated by many viruses to achieve high protein levels for both host and virus itself. For example, to facilitate viral protein synthesis, HCV non-structural protein 5A (NS5A) simultaneously activated mTORC1 and eIF4E through inactivation of 4EBP1 [37]. By up-regulating AKT level, human papillomavirus (HPV) activated mTORC1 which might be required for the initiation of viral replication [38]. Herpes simplex virus type 1 (HSV-1) [39], human cytomegalovirus (HCMV) [40], and vaccinia virus (VacV) [41] all augmented the content of mTOR following by 4EBP1 phosphorylation, thus promoting viral protein synthesis and viral replication.
In contrast, some viruses suppressed host translation machinery by down-regulated mTOR activity to contribute to cell cytopathology and apoptosis. Encephalomyocarditis virus [42], vesicular stomatitis virus [43], or adenovirus [44] infection restrained the cap-dependent translation by promoting the 4EBP1 hypophosphorylation. Influenza virus infection was also reported to decrease the abundance of phosphorylated eIF4E, which might assist to inhibit host translation [45]. Treated with rapamycin, a specific inhibitor of mTOR, the PRRSV titer was dramatically enhanced in host cells than the mock-treated [46]. In our study, the mTOR signaling and its downstream targets of 4EBP1 and p70 S6K were remarkably down-regulated in virulent PEDV CH/YNKM-8/2013 than that by the CV777 vaccine strain-like AH-M strain. It was noteworthy that the content of phosphatidylinositol-3-kinase (PI3K) was reduced in virulent PEDV infection, while increased in CV777 vaccine strain-like PEDV infection, which closely associate with the differences in mTOR signaling of Vero cells infected with these two strains. Particularly worth mentioning was that the virulent strain exhibited higher amount of viral protein and nucleic acids production compared to CV777 vaccine strain-like AH-M at the time-point tested. One possible explanation was that virulent PEDV subverted the translation apparatus of Vero cells to combat against innate antiviral responses for higher virus yield, resulting in earlier evident cytopathic effect and ultimately cell death. Besides, the difference in protein synthesis induced by the two viruses might be related with their different pathogenic mechanism in Vero cells. Furthermore, the mTOR pathway was intimately linked with cellular autophagy, which acted pivotal functions in viral replication and pathogenesis [47], [48]. The elaborate mechanism between autophagy and PEDV pathogenesis is currently under investigation in our laboratory.
4.2. Cellular immune response
During infection, the viruses initiate various strategies to suppress host immune system and many antiviral responses are conversely generated by host cells. The balance between host and virus confrontation determines the outcome of viral infection and disease progression [49]. The cell-mediated immune responses and inflammatory pathways remain poorly understood to PEDV infection, much less about the differences between virulent and CV777 vaccine strain-like AH-M strains. Previous studies have shown that PEDV infection could cause deficiency in the synthesis of type I interferon (IFN), and nucleocapsid (N) and papain-like protease 2 (PLP2) proteins functioned as IFN antagonist in vitro [50], [51]. Xu et al. reported that E and N proteins could activate NF-κB and aggrandize interleukin-8 expression using a porcine intestinal epithelial cell (IEC) line [52], [53]. Whereas it was also noticed that transient expression of N protein in HEK-293T cells could inhibit NF-κB signaling [50]. Besides, the recent report revealed that PEDV infection induced NF-κB activation through the TLR2, TLR3, and TLR9 pathways in porcine intestinal epithelial cells [54]. In our study, many immune regulatory pathways described above were constructed based on the differentially expressed proteins using IPA tool. To further investigate the differences regarding immunomodulation manipulated by virulent or CV777 vaccine strain-like PEDV infection, we examined the expression of specific inflammatory cytokines which were hallmarks of immune responses by real-time RT-PCR. It was noted that the expression of inflammatory cytokines detected were sharply up-regulated during virulent PEDV infection. As is well accepted that NF-κB acts at the crossroads of many signaling pathways, exerts central roles in regulation of the immune responses and cell survival/proliferation [55]. Immunoblotting analysis indicated NF-κB pathway was more significantly activated after virulent PEDV infection through p65 phosphorylation and IκBα degradation, while moderately activated after CV777 vaccine strain-like AH-M strain infection. Activation of NF-κB can facilitate replication of some viruses, such as TGEV and PRRSV [56], [57]. Yamamoto et al. noticed that inappropriate or over-activation of NF-κB could lead to inflammatory and autoimmune diseases [58]. Here, the more dramatically activation of NF-κB signaling in virulent PEDV strain infected cells might be due to the characteristics of virulence and pathogenicity, as well as stronger replication capability and infectivity of virulent strain than that by the CV777 vaccine strain-like strain, followed by more severe inflammatory cascades to aggravate Vero cells destruction. PEDV infection is marked by acute enteritis with mortality approaching 100% in sucking piglets up to one week of age, especially for virulent strain. We speculated that the acute enteritis caused by virulent PEDV might be associated with the excessive inflammatory cascades responses, which lead to devastating damage of epithelial cells in small intestine of the piglets.
STAT1 is a key member with great diversity of biological functions in the STAT family which mainly involves in JAK/STAT pathway. The phosphorylation and translocation of STAT1 lead to the transcription of interferon stimulated genes that provide an antiviral state for the host cells [59], [60]. It was reported that animals with STAT1 disruption were extremely susceptible to viral disease [61]. Robert J. et al. reported the mouse model deficient in STAT1 possessed an increased viral titer, lung pathology and susceptibility to SARS-CoV infection over the wild type mouse [59]. Our proteomic data showed that the STAT1 displayed no obvious change during CV777 vaccine strain-like PEDV infection, but notably decreased with 2-fold ratio in virulent PEDV infection, which was confirmed by western blot. STAT3, another important STAT family member, regulates cell proliferation, survival and anti-apoptosis process [62]. Moreover, the activation of STAT3 was closely associated with virus replication [63], [64]. In this study, STAT3 protein was found with up-regulation in both virulent (6.51-fold change) and CV777 vaccine strain-like (4.27-fold change) PEDV infected cells, which might facilitate cell survival and virus propagation. Take all these observations into account, the virulent PEDV might try to manipulate infected cells to sustain cell survival for maximizing viral replication and titers, coupled with cytopathologic changes and finally lead to dramatically pathological changes in host cells.
4.3. GTPase family and cellular maintenance
The GTPase family which contains three subfamilies, Rho, Rac and Cdc42, regulates various biological processes including the reorganization of actin cytoskeleton, vesicle trafficking, cell adhension and apoptosis [65]. It is well recognized that re-configuration of cytoskeleton is closely related with transcellular membrane trafficking and therefore facilitates viral replication [66]. Cytoskeletal changes have also been detected in proteomic studies of virus infections, such as infectious bursal disease virus (IBDV) [67], TGEV [20], and PCV2 [68]. Our proteomic study showed that the Rac and Rho signaling were significantly changed after virulent PEDV infection. Detailed analysis demonstrated that the alteration was caused by the differential expression of the key upstream regulator PI3K upon virulent and CV777 vaccine strain-like PEDV infection. In addition, integrin (another key upstream regulator), ARP2/3 (an initiation complex of actin polymerization), IQGAP and CYFIP1 which also played important roles in GTPase family, were all dramatically down-regulated in virulent PEDV infection. We inferred that virulent PEDV might manipulate these differentially displayed proteins to generate actin cytoskeleton networks collapse and cell–cell detachment in host cells, leading to an unstable cytoskeletal structure. The cytoskeletal disruption may contribute to virus multiplication and dissemination by facilitating its release and entry, conferring higher virus titer and infectivity to virulent strain. While no such obvious alterations were confirmed in CV777 vaccine strain-like AH-M infected cells, this might be resulted from the less severe cell pathological changes due to the lower viral titer and pathogenicity.
Viral infection can also convert the normal composition of host cells to promote viral propagation. Adherens junctions (AJs) and tight junctions (TJs) are essential components to maintain the integrity of the intestinal epithelium, which is crucial for proper functions of the barrier that regulates the absorption of nutrients and restricts uptake of luminal bacteria [69], [70]. It is widely acknowledged that porcine intestines are the primary site for PEDV multiplication, and structural alterations of adherens and tight junctions have been mentioned in intestinal epithelial cells infected with PEDV in vivo and in vitro [71], [72]. In our study, adherens and tight junctions were both found under dysregulation during virulent PEDV infection, while slight changes were induced by CV777 vaccine strain-like PEDV. Previous report indicated that PEDV infection lead to watery diarrhea and severe dehydration in piglets, and meanwhile accompanying with severe villous atrophy, a reduction in the villous height and crypt depth. These might be results of the interruption of digestion and absorption of nutrients and electrolytes, caused by the alteration of adherens and tight junctions in the process of PEDV infection. Furthermore, many significantly altered proteins covered in these signaling pathways were identified and quantified. The precise mechanism concerning host cellular structure alteration and diarrhea might be elucidated based on these substantial evidences.
4.4. Other changed proteins
Apoptosis is considered as an important component in regulating pathogenesis of many infectious diseases. In the process of virus infection, apoptosis inhibition can prevent premature cell death and thus maximize viral replication, while apoptosis induction can facilitate virus release and dissemination from infected cells [73], [74]. The initiation of apoptosis is usually controlled by the death receptor-mediated extrinsic pathway, and the mitochondrial-involved intrinsic pathway. In the present study, two apoptosis related proteins were detected and quantified. AIFM1, a mitochondrial apoptosis-inducing factor (AIF), functions as a key pro-apoptotic factor in the intrinsic apoptotic pathway by releasing from the mitochondria and translocating to the nucleus. Significant up-regulation of AIFM1 was confirmed in virulent PEDV infected cells and therefore apoptosis might be potentially induced. This was consistent with a previous report that PEDV infection could induce apoptosis through activation of mitochondrial apoptosis-inducing factor [75]. Besides, caspase-8 is critically involved in extrinsic apoptosis signaling pathway in mammals, which is triggered by stimulation of death receptors. Caspase 3, the main executioner caspase, can be activated by the initiator caspase, caspase 8, to bring about the apoptotic phenotype. The down-regulation of caspase-8 expression during virulent PEDV infection implied that an apoptosis inhibition mechanism might exist at early PEDV infection to achieve a higher virus titer, which was also confirmed by Zeng et al [26]. Based on these evidences, a mechanism that weakens virus-induced cell apoptosis to facilitate virus proliferation to a higher virus titer at early virus infection, and mitochondrial AIF-mediated apoptosis to favor virus dissemination and pathogenicity might coexist during virulent PEDV infection, just like the case of SARS-CoV [76].
Endoplasmic reticulum, the major site for protein transport and secretion, exerts a crucial role in guaranteeing the proper function of viral proteins. Calnexin is a molecular chaperone in ER, regulating the folding and maturation of glycosylated proteins [77]. Previous studies have demonstrated that calnexin strictly monitored the maturation of viral proteins, such as HIV gp160 and S protein of SARS-CoV, which were important for their infectivity [78], [79]. In addition, calnexin was also observed with increased expression in HCV infection [80]. Similarly, calnexin was significantly up-regulated by 7.92- and 2.88-fold change, in virulent and CV777 vaccine strain-like PEDV infections, respectively. This was confirmed by immunoblotting analysis. These evidences supported our hypothesis that the higher expression of calnexin might promote the maturation of S protein, resulting in conferring more severe infectivity on the virulent PEDV than the CV777 vaccine strain-like strain.
Taken together, this is the first attempt to explore the whole protein profiles individually infected by virulent and CV777 vaccine strain-like strains of PEDV through iTRAQ coupled with LC–MS/MS approach, followed by comparative analysis of proteomic characterization. Although the proteomic analysis here based on these differentially expressed proteins is descriptive, these substantial observations might lay the foundation for in-depth studies to elucidate the different pathogenesis and host responses upon virulent and CV777 vaccine strain-like PEDV infections.
The following are the supplementary data related to this article.
Supplementary Fig. S3.
Overview of specific functional networks upon PEDV strains CH/YNKM-8/2013 and AH-M infection, respectively.
Information for significantly differentially expressed proteins identified in Vero cells infected with PEDV strains CH/YNKM-8/2013 and AH-M, respectively.
Information for functional characterization, Pathway analysis, and Network analysis of differentially regulated proteins.
Transparency Document
Transparency document.
Acknowledgment
This work was supported by grants from the Natural Sciences Foundation of China (31272572) and the China Agricultural Research System(CARS-36). We thank Dr. Rui Luo, Huazhong Agricultural University for hiskind suggestions.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Liu X., Shi D., Shi H., Zhang X., Feng L. Complete genome sequence of a porcine epidemic diarrhea virus variant. J. Virol. 2012;86:3408. doi: 10.1128/JVI.07150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J., Rottier P.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Li H., Liu Y., Pan Y., Deng F., Song Y. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojkic D., Hazlett M., Fairles J., Marom A., Slavic D., Maxie G. The first case of porcine epidemic diarrhea in Canada. Can. Vet. J. 2015;56:149–152. [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y.K., Lim S.I., Lim J.A., Cho I.S., Park E.H., Le V.P. A novel strain of porcine epidemic diarrhea virus in Vietnamese pigs. Arch. Virol. 2015;160:1573–1577. doi: 10.1007/s00705-015-2411-5. [DOI] [PubMed] [Google Scholar]
- 11.Park S., Kim S., Song D., Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg. Infect. Dis. 2014;20:2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi J., Zeng S., Xiao S., Chen H., Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012;86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y., Yan G., Luo Y., Tong T., Liu X., Xin C. Quantitative proteomics by amino acid labeling in foot-and-mouth disease virus (FMDV)-infected cells. J. Proteome Res. 2013;12:363–377. doi: 10.1021/pr300611e. [DOI] [PubMed] [Google Scholar]
- 14.Chien K.Y., Blackburn K., Liu H.C., Goshe M.B. Proteomic and phosphoproteomic analysis of chicken embryo fibroblasts infected with cell culture-attenuated and vaccine strains of Marek's disease virus. J. Proteome Res. 2012;11:5663–5677. doi: 10.1021/pr300471y. [DOI] [PubMed] [Google Scholar]
- 15.Jiang X.S., Tang L.Y., Dai J., Zhou H., Li S.J., Xia Q.C. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics. 2005;4:902–913. doi: 10.1074/mcp.M400112-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmott E., Rodgers M.A., Macdonald A., McCrory S., Ajuh P., Hiscox J.A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell. Proteomics. 2010;9:1920–1936. doi: 10.1074/mcp.M900345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C., Zhang A., Guo J., Yang J., Zhou H., Chen H. Identification of human host proteins contributing to H5N1 influenza virus propagation by membrane proteomics. J. Proteome Res. 2012;11:5396–5405. doi: 10.1021/pr3006342. [DOI] [PubMed] [Google Scholar]
- 18.Herbrich S.M., Cole R.N., West K.P., Jr., Schulze K., Yager J.D., Groopman J.D. Statistical inference from multiple iTRAQ experiments without using common reference standards. J. Proteome Res. 2013;12:594–604. doi: 10.1021/pr300624g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Bai J., Lu Q., Zhang L., Jiang Z., Michal J.J. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine circovirus type 2. J. Proteome. 2013;79:72–86. doi: 10.1016/j.jprot.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 20.An K., Fang L., Luo R., Wang D., Xie L., Yang J. Quantitative proteomic analysis reveals that transmissible gastroenteritis virus activates the JAK-STAT1 signaling pathway. J. Proteome Res. 2014;13:5376–5390. doi: 10.1021/pr500173p. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J., Niu D., Sui J., Ching C.B., Chen W.N. Protein profile in hepatitis B virus replicating rat primary hepatocytes and HepG2 cells by iTRAQ-coupled 2-D LC–MS/MS analysis: insights on liver angiogenesis. Proteomics. 2009;9:2836–2845. doi: 10.1002/pmic.200800911. [DOI] [PubMed] [Google Scholar]
- 22.Luo R., Fang L., Jin H., Wang D., An K., Xu N. Label-free quantitative phosphoproteomic analysis reveals differentially regulated proteins and pathway in PRRSV-infected pulmonary alveolar macrophages. J. Proteome Res. 2014;13:1270–1280. doi: 10.1021/pr400852d. [DOI] [PubMed] [Google Scholar]
- 23.Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dauber B., Heins G., Wolff T. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J. Virol. 2004;78:1865–1872. doi: 10.1128/JVI.78.4.1865-1872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batonick M., Oomens A.G., Wertz G.W. Human respiratory syncytial virus glycoproteins are not required for apical targeting and release from polarized epithelial cells. J. Virol. 2008;82:8664–8672. doi: 10.1128/JVI.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng S., Zhang H., Ding Z., Luo R., An K., Liu L. Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics. 2015;15:1819–1828. doi: 10.1002/pmic.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D., Shi H., Guo D., Chen J., Shi D., Zhu Q. Analysis of protein expression changes of the Vero E6 cells infected with classic PEDV strain CV777 by using quantitative proteomic technique. J. Virol. Methods. 2015;218:27–39. doi: 10.1016/j.jviromet.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L.K., Chai F., Li H.Y., Xiao G., Guo L. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J. Proteome Res. 2013;12:2666–2678. doi: 10.1021/pr400011k. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q., Bai J., Zhang L., Liu J., Jiang Z., Michal J.J. Two-dimensional liquid chromatography–tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J. Proteome Res. 2012;11:2890–2903. doi: 10.1021/pr201266z. [DOI] [PubMed] [Google Scholar]
- 30.Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garaigorta U., Chisari F.V. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan R., Wang L., Graczyk T.M., Block T.M., Romano P.R. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 2002;76:9588–9599. doi: 10.1128/JVI.76.19.9588-9599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith J.A., Schmechel S.C., Raghavan A., Abelson M., Reilly C., Katze M.G. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma X.M., Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 35.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 36.Jastrzebski K., Hannan K.M., Tchoubrieva E.B., Hannan R.D., Pearson R.B. Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors. 2007;25:209–226. doi: 10.1080/08977190701779101. [DOI] [PubMed] [Google Scholar]
- 37.George A., Panda S., Kudmulwar D., Chhatbar S.P., Nayak S.C., Krishnan H.H. Hepatitis C virus NS5A binds to the mRNA cap-binding eukaryotic translation initiation 4F (eIF4F) complex and up-regulates host translation initiation machinery through eIF4E-binding protein 1 inactivation. J. Biol. Chem. 2012;287:5042–5058. doi: 10.1074/jbc.M111.308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchkovich N.J., Yu Y., Zampieri C.A., Alwine J.C. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh D., Mohr I. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 2004;18:660–672. doi: 10.1101/gad.1185304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh D., Perez C., Notary J., Mohr I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 2005;79:8057–8064. doi: 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaborowska I., Walsh D. PI3K signaling regulates rapamycin-insensitive translation initiation complex formation in vaccinia virus-infected cells. J. Virol. 2009;83:3988–3992. doi: 10.1128/JVI.02284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingras A.C., Svitkin Y., Belsham G.J., Pause A., Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connor J.H., Lyles D.S. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 2002;76:10177–10187. doi: 10.1128/JVI.76.20.10177-10187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi Q., Cuesta R., Schneider R.J. Tethering of eIF4G to adenoviral mRNAs by viral 100 k protein drives ribosome shunting. Genes Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feigenblum D., Schneider R.J. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J. Virol. 1993;67:3027–3035. doi: 10.1128/jvi.67.6.3027-3035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pujhari S., Kryworuchko M., Zakhartchouk A.N. Role of phosphatidylinositol-3-kinase (PI3K) and the mammalian target of rapamycin (mTOR) signalling pathways in porcine reproductive and respiratory syndrome virus (PRRSV) replication. Virus Res. 2014;194:138–144. doi: 10.1016/j.virusres.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Zhu B., Zhou Y., Xu F., Shuai J., Li X., Fang W. Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J. Virol. 2012;86:12003–12012. doi: 10.1128/JVI.01434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z., Jiang X., Liu D., Fan Z., Hu X., Yan J. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- 49.Pastorino B., Nougairede A., Wurtz N., Gould E., de Lamballerie X. Role of host cell factors in flavivirus infection: implications for pathogenesis and development of antiviral drugs. Antiviral Res. 2010;87:281–294. doi: 10.1016/j.antiviral.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao L., Ge X., Gao Y., Ren Y., Ren X., Li G. Porcine epidemic diarrhea virus infection induces NF-kappaB activation through the TLR2, TLR3, and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 2015;96:1757–1767. doi: 10.1099/vir.0.000133. [DOI] [PubMed] [Google Scholar]
- 55.Moynagh P.N. The NF-kappaB pathway. J. Cell Sci. 2005;118:4589–4592. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 56.Eleouet J.F., Chilmonczyk S., Besnardeau L., Laude H. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J. Virol. 1998;72:4918–4924. doi: 10.1128/jvi.72.6.4918-4924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S.M., Kleiboeker S.B. Porcine arterivirus activates the NF-kappaB pathway through IkappaB degradation. Virology. 2005;342:47–59. doi: 10.1016/j.virol.2005.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto Y., Gaynor R.B. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J. Clin. Invest. 2001;107:135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hogan R.J., Gao G., Rowe T., Bell P., Flieder D., Paragas J. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 2004;78:11416–11421. doi: 10.1128/JVI.78.20.11416-11421.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Germain M.A., Chatel-Chaix L., Gagne B., Bonneil E., Thibault P., Pradezynski F. Elucidating novel hepatitis C virus–host interactions using combined mass spectrometry and functional genomics approaches. Mol. Cell. Proteomics. 2014;13:184–203. doi: 10.1074/mcp.M113.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durbin J.E., Hackenmiller R., Simon M.C., Levy D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 62.Bowman T., Garcia R., Turkson J., Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 63.Waris G., Turkson J., Hassanein T., Siddiqui A. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J. Virol. 2005;79:1569–1580. doi: 10.1128/JVI.79.3.1569-1580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Okemoto K., Wagner B., Meisen H., Haseley A., Kaur B., Chiocca E.A. STAT3 activation promotes oncolytic HSV1 replication in glioma cells. PLoS One. 2013;8:e71932. doi: 10.1371/journal.pone.0071932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tapon N., Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr. Opin. Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 66.Taylor M.P., Koyuncu O.O., Enquist L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng X., Hong L., Shi L., Guo J., Sun Z., Zhou J. Proteomics analysis of host cells infected with infectious bursal disease virus. Mol. Cell. Proteomics. 2008;7:612–625. doi: 10.1074/mcp.M700396-MCP200. [DOI] [PubMed] [Google Scholar]
- 68.Fan H., Ye Y., Luo Y., Tong T., Yan G., Liao M. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals protein and pathway regulation in porcine circovirus type 2 infected PK-15 cells. J. Proteome Res. 2012;11:995–1008. doi: 10.1021/pr200755d. [DOI] [PubMed] [Google Scholar]
- 69.Hartsock A., Nelson W.J. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta. 1778;2008:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su C.W., Cao Y., Kaplan J., Zhang M., Li W., Conroy M. Duodenal helminth infection alters barrier function of the colonic epithelium via adaptive immune activation. Infect. Immun. 2011;79:2285–2294. doi: 10.1128/IAI.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung K., Eyerly B., Annamalai T., Lu Z., Saif L.J. Structural alteration of tight and adherens junctions in villous and crypt epithelium of the small and large intestine of conventional nursing piglets infected with porcine epidemic diarrhea virus. Vet. Microbiol. 2015;177:373–378. doi: 10.1016/j.vetmic.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao S., Gao J., Zhu L., Yang Q. Transmissible gastroenteritis virus and porcine epidemic diarrhoea virus infection induces dramatic changes in the tight junctions and microfilaments of polarized IPEC-J2 cells. Virus Res. 2014;192:34–45. doi: 10.1016/j.virusres.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomson B.J. Viruses and apoptosis. Int. J. Exp. Pathol. 2001;82:65–76. doi: 10.1111/j.1365-2613.2001.iep0082-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enjuanes L., Almazan F., Sola I., Zuniga S. Biochemical aspects of coronavirus replication and virus–host interaction. Annu. Rev. Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 75.Kim Y., Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. 2014;460–461:180–193. doi: 10.1016/j.virol.2014.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang B.S., Chan K.H., Cheng V.C., Yuen K.Y. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by SARS coronavirus and human coronavirus 229E. Hong Kong Med. J. 2009;15(Suppl. 9):23–26. [PubMed] [Google Scholar]
- 77.Bedard K., Szabo E., Michalak M., Opas M. Cellular functions of endoplasmic reticulum chaperones calreticulin, calnexin, and ERp57. Int. Rev. Cytol. 2005;245:91–121. doi: 10.1016/S0074-7696(05)45004-4. [DOI] [PubMed] [Google Scholar]
- 78.Jennelle L., Hunegnaw R., Dubrovsky L., Pushkarsky T., Fitzgerald M.L., Sviridov D. HIV-1 protein Nef inhibits activity of ATP-binding cassette transporter A1 by targeting endoplasmic reticulum chaperone calnexin. J. Biol. Chem. 2014;289:28870–28884. doi: 10.1074/jbc.M114.583591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukushi M., Yoshinaka Y., Matsuoka Y., Hatakeyama S., Ishizaka Y., Kirikae T. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J. Virol. 2012;86:11745–11753. doi: 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colman H., Le Berre-Scoul C., Hernandez C., Pierredon S., Bihouee A., Houlgatte R. Genome-wide analysis of host mRNA translation during hepatitis C virus infection. J. Virol. 2013;87:6668–6677. doi: 10.1128/JVI.00538-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information for significantly differentially expressed proteins identified in Vero cells infected with PEDV strains CH/YNKM-8/2013 and AH-M, respectively.
Information for functional characterization, Pathway analysis, and Network analysis of differentially regulated proteins.
Transparency document.